Abstract
The generation of growth curves is a common and established method for determining the properties of bacterial strains. Such properties can be growth rates in enrichment broths or resistance to antibiotics. Enrichment methods are the gold standard for enumerating bacteria and determining growth properties. Newer methods can use agents that are metabolized during bacterial growth, which leads to a shift in fluorescence. This principle is used, for example, to measure viable bacterial cells. In this work, a real-time thermal cycler was used to cultivate bacteria and measure the change in fluorescence during bacterial growth to determine the generation time of different bacterial strains. The ability of the real-time diagram enables a significant simplification and increase in cost efficiency in the generation of bacterial growth curves. It was also possible to monitor the susceptibility of bacteria to antibiotic resistance, which opened another important application of this technique.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cultivation of bacteria has been done with essentially unchanged methods for a long time. Using agar plates, qualitative and quantitative results can be obtained. Many bacteria strains can or must be cultivated in liquid media at different conditions (e.g. temperature, time, additives, anaerobe or aerobe). An advantage of liquid media cultivation is that the handling can be easier allowing more sample material and a higher sample throughput compared to the plating technics. In cases where a high sensitivity is not necessary, the cultivation can be miniaturized, and dilution steps can be integrated to enable simple quantification by e.g. Most Probable Number (MPN) calculations applied in more recent and automated microbiological analysis. Often at the end of the cultivating time either a visual counting of the colony-forming units (cfu) is applied to quantify the bacteria on plates or the change in colour is measured in limiting dilution assays.
In the past culture media were optimised e.g. [1]. Considering the growth rate and specificity. Many of these optimization steps are based on trial and error, requiring a lot of experimental data. Therefore, many attempts were undertaken to gain these data in a more efficient way. 96 well plate-based systems were propagated [2,3,4,5] or microreactors [8] were used [3] and growth was monitored using fluorescence dyes [6, 7].
The growth of bacteria follows duplication rules equal to the duplication of target DNA during each thermal cycle of the polymerase chain reaction (PCR). Real-time thermocyclers are specially constructed and programmed to adjust temperature exactly during cycling and for continuous measurement of the change in fluorescence using a fluorimeter. Based on such parallels, we tried to use a thermocycler to grow bacteria in liquid media and to chart their growth using the software from the real-time thermocycler. Each cycle takes a certain time and the growth is reflected by the colour change in the fluorescence of the metabolized agent [9,10,11]. In the present study the generation time of bacteria was determined comparing the growth curves from assays spiked with different bacteria loads. In addition, the approach described here was used to measure the influence of antibiotics on bacterial growth. To our knowledge this is the first time such a real-time cultivation of bacteria is described, showing its potential to select suitable media and detect antibiotic load in a very versatile way. Compared to the classical plating methods, the approach described here reduces time and effort to gather information on growth characteristics of bacteria strains.
Materials and methods
Bacteria strains
The following strains were used for the experiments: Bacillus cereus DSM 31 and DSM 4313, E. coli ATCC 25922, Staphylococcus aureus ATCC 25923 and ATCC BAA 1026, Clostridium. perfringens ATCC 13124, Pseudomonas. aeruginosa ATCC 27853 NCTC, Vibrio parahaemalyticus ATCC 17802, Enterococcus faecalis ATCC 19433
Lenticule® Discs from the Health Protection Agency (HPA) Culture Collections were used for the following strains: Salmonella enteritidis NCTC 6676, Listeria monocytogenes NCTC 11994 and Bacillus Subtilis (BGA) spore suspension, Merck No. 110649.
Cultivation media and dye to track cell growth
Enrichment Media ALB:
Per litre: 13.8 g Bolton Broth (Oxoid) + 28.7 g Fraser Broth Base (Oxoid) mixed with 1 L distilled water followed by sterilisation (15 min at 121 ℃).
Enrichment Media PEPG:
Buffered Pepton Water, Merck, cat no. 13880
Enrichment Media Tryptic Soy Broth (TSB): Merck, cat no. 5459
Antibiotics used:
Penicillin, Sigma-Aldrich, cat no. 13750
Gentamicinsulfat Sigma-Aldrich, cat no. G1914
Streptomycinsulfat Sigma-Aldrich, cat no. C6501
Clenbuterol hydrochlorid Sigma-Aldrich, cat no. C5423
Chloramphenicol Sigma-Aldrich, cat no. C0378
Neomycinsulfat Sigma-Aldrich, cat no. N1876
Tetracycline hydrochlorid Sigma-Aldrich, cat no. 87130
Sulfadimidin Sigma-Aldrich, cat no. S6256
Sulfadimethoxine Sigma-Aldrich, cat no. S7007
Dye to track cell growth: PrestoBlue cell viability reagent, Invitrogen, cat no. A13261. Function: PrestoBlue is permeable through the membrane of living cells and reduced by them into a fluorescent dye by reduction of the resazurin based PrestoBlue solution. This shifts the colour from blue to red and can be monitored by a fluorimeter (emission/excitation nm: 535–560/590–615).
Real-time cultivation procedure
Assay composition (25 μl total volume):
18 μl cultivation medium + 2 μl cell viability reagent PrestoBlue + 5 μl Bacteria suspension
For all assay’s, tubes of 100 μl volume have been used.
Incubation was performed on a Rotorgene 6000 real-time system (Corbett, Australia) according to the following protocols:
Incubation conditions to chart unspecific bacterial growth for 48 h (aerobe mesophilic bacteria): main incubation at 37 ℃, 288 cycles each 600 s, data acquisition for channel orange (filter setting: excitation 585 nm, detection 610 nm), at the end of each cycle. Final deactivation of the bacteria at 95 ℃ for 5 min. One cycle is equal to 10 min.
Enrichment of Salmonella Enteritidis, Listeria monocytogenes and Escherichia coli O157 for 8.3 h: main incubation at 30 ℃, 500 cycles each 60 s, data acquisition for channel orange (excitation 585 nm, detection 610 nm) at the end of each cycle. Final inactivation of the bacteria at 95 ℃ for 5 min. One cycle is equal to 1 min.
Incubation conditions using Bacillus subtilis as antibiotic sensitive organism for 6 h:
incubation at 30 ℃, 360 cycles each 60 s, data acquisition for channel red (excitation 625 nm, detection 660 nm), at the end of each cycle. Final deactivation of the bacteria at 95 ℃ for 5 min. One cycle is equal to 1 min.
Calculation of generation time
For bacteria, the generation time is the time when bacteria duplicate during the exponential phase of growth [12]. In consequence, a 1:10 diluted bacterial culture takes 2 3.32 (equal to 1/log2) generation times to reach the same signal as the undiluted culture (bacterial density in solution and/or degradation of a dye like PrestoBlue indicating the growth).
Log: base 10
T0: time when sample with undiluted bacterial load reaches a signal threshold line
T1:10: time when the 1:10 diluted bacterial load reaches a signal threshold line
\(\Delta time\) = difference of T1:10 and T0
\(generation time \left(min\right)=\Delta time(min)\times log2\) [12]
This equation estimates the generation time and is valid for optimal growth in the exponential growth phase.
Results and discussion
Real-time cultivation of bacteria dilution series to determine growth rates
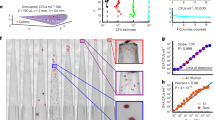
Different bacterial strains were cultivated with real-time procedure using two different cultivation media ALB and PEPG. The goal of the study was on the one hand to develop and evaluate an easy approach to determine growth rates of bacteria and on the other hand to assess which of the two media is suitable to cultivate the tested bacteria strains. The Results show that both media are comparable considering the growth rate for the tested bacteria strains (Fig. 1; Tables 1 and 2). The undiluted Bacillus cereus culture reached the threshold line after 97 min. The 1:10 diluted culture needed 41 min more time. Using the equation described above, a generation time of 12.5 min could be calculated. When the same calculation is done for the 1:100 (14.8 min) and 1:1′000 dilutions (19.6 min), a mean generation time of 15.6 min (Table 1) can be calculated. We attribute the systematically increase of the generation time as a result of the decreasing availability of nutrition factors over the time. The 1:10′000 culture did not reach the threshold line and thus could not be considered for calculation. The same measurements have been done for Enterococcus faecalis, Escherichia coli, Vibrio parahaemalyticus, Stapylococcus aureus, Listeria monocytogenes, Salmonella enteritidis and Pseudomonas aeruginosa. All the calculated generation times were comparable to published generation times for optimal growth conditions except for Listeria. For Listeria, the published generation time is twice as long. However, it needs to be considered, that generation times are dependent on all factors influencing bacterial growth. Therefore, bacterial growth rate can vary considerably between the different experimental settings.
Bacillus cereus strain dilution series 1:10, 1:100, 1:1′000, 1:10′000 and no bacterial load cultivated in ALB liquid media. Y-axis fluorescence signal, X-axis time (min). Data acquisition in the orange channel. Growth changes the spectral characteristic leading to an increase of fluorescence in the orange channel. For each Δ time the generation time was calculated
Real-time cultivation of Bacillus subtilis to trace the influence of different antibiotics on the bacterial growth
Bacillus subtilis was cultivated as described and exposed to different concentrations of 9 different antibiotics. As expected, the growth of Bacillus subtilis was inhibited in a concentration depended manner (Fig. 2) by all the antibiotics. The cultures treated with no or ineffective doses of antibiotics grew and degraded the dye showing a sigmoid decrease of the fluorescence. The slope of the curves decreased with increasing antibiotic concentrations. For Neomycinsulfat growth inhibition could be observed with the addition of 0.32 mg/ml. For Penicillin, Streptomycinsulfat, Clenbuterol, Chloramphenicol and Sulfadimidin 0.1 mg/ml resulted in growth inhibition. The most effective antibiotic was Tetracycline, where only 0.002 mg/ml was necessary to inhibit growth (Fig. 2G). A total inhibition of growth is assumed, in cases where there is no or only a slight linear decrease of the curve.
Bacillus subtilis strain with dilution series of different antibiotic cultivated in TSB liquid media containing PrestoBlue. Y-axis fluorescence signal, X-axis time (min). High antibiotic concentrations inhibit the growth of Bacillus subtilis (straight, slowly descendent lines). The fluorescence signal of samples without antibiotic decrease early and lose fluorescence signal after about 270 min. Data acquisition in the red channel. Growth changes the spectral characteristic leading to a decrease of fluorescence in the red channel. a Penicillin (mg/ml) 1, 0.32, 0.1, 0.032, 0.01, 0.0032, 0.001, none. Concentrations of 0.1 mg/ml or higher lead to inhibition of growth (no decrease of the graph). b Gentamicinsulfat (mg/ml) 0.1, 0.032, 0.01, 0.0032, 0.001, 0.00032, 0.0001, none. Concentrations of 0.01 mg/ml or higher lead to inhibition of growth (no decrease of the graph). c Streptomycinsulfat (mg/ml) 1, 0.32, 0.1, 0.032, 0.01, 0.0032, 0.001, none. Concentrations of 0.1 mg/ml or higher lead to inhibition of growth (no decrease of the graph). d Clenbuterol hydrochlorid (mg/ml) 1, 0.32, 0.1, 0.032, 0.01, 0.0032, 0.001, none. Concentrations of 0.1 mg/ml or higher lead to inhibition of growth (no decrease of the graph). e Chloramphenicol (mg/ml) 0.1, 0.032, 0.01, 0.0032, 0.001,0.00032, 0.0001, none. Concentrations of 0.1 mg/ml or higher lead to inhibition of growth (no decrease of the graph). f Neomycinsulfat (mg/ml) 1, 0.32, 0.1, 0.032, 0.01, 0.0032, 0.001, none. Concentrations of 0.32 mg/ml or higher lead to inhibition of growth (no decrease of the graph). g Tetracycline hydrochlorid (mg/ml) 0.2, 0.064, 0.02, 0.0064, 0.002,0.00064, 0.0002, none. Concentrations of 0.002 mg/ml or higher lead to inhibition of growth (no decrease of the graph). h Sulfadimidin (mg/ml) 1, 0.32, 0.1, 0.032, 0.01, 0.0032, 0.001, none. Concentrations of 0.1 mg/ml or higher lead to inhibition of growth (no decrease of the graph). i Sulfadimethoxin (mg/ml) 1, 0.32, 0.1, 0.032, 0.01, 0.0032, 0.001, none. Concentrations of 0.1 mg/ml or higher lead to inhibition of growth (no decrease of the graph)
Conclusion
The off-label application of a real-time thermocycler to monitor bacterial growth is presented. In this setting the PCR-mix is replaced by cultivation media in which bacteria are growing. Obviously, for the used real-time System, the bacterial growth is similar enough to the amplification of PCR products so that the software could be applied without modifications. In consequence, it could be shown that using a real-time thermocycler, growth characteristics can be monitored with little hands-on time. Overnight experiments can be performed without extended presence of laboratory personal. Growth curves were dose dependent and could be used to determine generation times in different media. The determined growth times corresponded to values of the literature. The influence of antibiotics could be visualized by this real-time cultivation. Application of antibiotics had a clear impact on growth and was reflected in the growth charts. Although, the proof of principle was demonstrated and may enable to conduct the described assays in a significant more cost-efficient way, results are preliminary and require confirmation. Other types of thermocyclers like block cyclers and more dense plates (e.g. 384 well plates) in conjunction with pipetting robots may lead to further increase of efficiency.
References
Tsai HN, Hodgson DA (2003) Development of a synthetic minimal medium for listeria monocytogenes. Appl Environ Microbiol 69(11):6943–6945
Xu ZN, Shen WH, Chen XY, Lin JP, Cen PL (2005) A high-throughput method for screening of rapamycin-producing strains of Streptomyces hygroscopicus by cultivation in 96-well microtiter plates. Biotechnol Lett 27(15):1135–1140
Côte J, Kamen AA, André G (1993) Protein-free culture medium improvement: testing additives and their interactive effects in 96-well plates. Appl Microbiol Biotechnol 39(4–5):577–584
Yu H, Alexander CM, Beebe DJ (2007) A plate reader-compatible microchannel array for cell biology assays. Lab Chip 7:388–391
Kensy F, Zang E, Faulhammer C, Tan RK, Büchs J (2009) Validation of a high-throughput fermentation system based on online monitoring of biomass and fluorescence in continuously shaken microtiter plates. Microb Cell Fact 8:31
Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M (2004) Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother 48:1803–1806
Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR (1990) New colorimetric cytotoxicity assay for anticancer-drug screening. JNCI J Natl Cancer Institut 82(13):1107–1112
Maharbiz MM, Holtz WJ, Howe RT, Keasling JD (2004) Microbioreactor arrays with parametric control for high-throughput experimentation. Biotechnol Bioeng 85(4):376–381
Räz B, Iten M, Grether-Bühler Y, Kaminsky R, Brun R (1997) The Alamar Blue® assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Trop 68(2):139–147
Lall N, Henley-Smith CJ, De Canha MN, Oosthuizen CB, Berrington D (2013) Viability reagent, prestoblue, in comparison with other available reagents, utilized in cytotoxicity and antimicrobial assays. Int J Microbiol 2013:5 (Article ID 420601)
O'Brien J, Wilson I, Orton T, Pognan F (2000) Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem 267(17):5421–5426
Powell EO (1956) Growth rate and generation time of bacteria, with special reference to continuous culture. J General Microbiol 15:492–511
Van Merode AE, Van der Mei HC, Busscher HJ, Waar K, Krom BP (2006) Enterococcus faecalis strains show culture heterogeneity in cell surface charge. Microbiology 152:807–814
International Commission on Microbiological Specifications for Foods, Microorganisms in Foods; Characteristics of Microbial Pathogens (1996) Blackie Academic &Professional. New York London ISBN 041247350X
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Compliance with ethics requirements
This article does not contain any studies with human or mammalian subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Köppel, R., Tolido, I., Ledermann, R. et al. Use of a real-time thermocycler for charting bacterial growth. Eur Food Res Technol 246, 2093–2099 (2020). https://doi.org/10.1007/s00217-020-03558-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-020-03558-0