Abstract
In this paper, samples of Cabernet Sauvignon wines produced in California have been analyzed on the basis of their elemental content and classified according to its geographical origin by the use of machine learning. Overall, 13 metals (Al, Cd, Co, Cr, Cu, Li, Mn, Ni, P, Pb, Rb, Sr, and Zn) were determined by inductively coupled plasma mass spectrometry (ICP-MS). We used two algorithms of variable selection in order to estimate the relevance of each metal to classification. Predictive models based on chemometric tools and machine learning algorithms were developed to differentiate origin of wine samples. Li and Sr were identified as the main responsible for the differentiation of samples. The application of Random Forest permitted to correctly classify all samples. A second analysis was performed by removing the variables Li and Sr to investigate the relevance of the others metals. We found that a group of seven variables (Cd, Ni, Mn, Pb, Rb, Co, Cu) which were able to discriminate the wines in 89% of accuracy by using Support Vector Machines. Results suggested that the developed methodology by advanced machine learning techniques is robust and reliable for the geographical classification of wine samples, and the study of the elements that characterize the regions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The fingerprinting of the content of trace metals in wines is a valuable method to authenticate the geographical origin of the same. The presence and concentration of metals in soil on which vines were grown enables their use to characterize the wines, i.e., the elements move from rock to soil and from soil to grape [1]. In particular, the wine authenticity has been extensively investigated because this beverage is an easily adulterated product and there exists an interest of consumers in foods strongly identified with a place of origin [1, 2].
The world wine production reached in 2018 a volume of 292.3 million of hectoliters [3]. California, the geographical origin of the wines analyzed in this study, is a world-renowned state for the ability to produce world class quality wine. Napa is a premier wine producing region producing a higher quality wines over the rest of California [4]. In this context, the authenticity of wines from California winery regions is an important issue. The multivariate data analysis and machine learning techniques are powerful tools to conduct quality control and wine authentication that have been used to discriminate wines from all around the world [1].
The Cabernet Sauvignon is by far the most important varietal for achieving high wine prices in California [4]. In spite of that, there are few researches that classified California wines produced with this grape variety. Californian wines made of grapes in different maturation states were classified by Umali et al. based on tannin content [5], and Hopfer and coworkers [6] classified the intraregional origin of red wines (Cabernet Sauvignon, Merlot and Pinot Noir) from North of California.
There are several studies as the mentioned above that had classified the geographical origin of red wines from different countries based on chemometrics and machine learning techniques [6,7,8,9,10,11,12]. However, these studies used two or more varieties and disregard this aspect to construct the classification model. The discrimination of wine-making origin of one variety allows to characterize the wine variety, providing information about the relationship between the variety and the origin, which can be useful to improve quality and avoid fraud. There are few studies that performed the geographical classification of one wine variety, such as the Cabernet Sauvignon [13], Malbec [14], and Sauvignon Blanc [15].
The most used classification techniques on the food authentication are the linear discriminant analysis, k-nearest neighbors, partial least squares-discriminant analysis and soft independent modeling by class analogy, and some variations of these ones [16,17,18]. The use of linear methods is easy to understand and is enough to obtain satisfactory results. Although most of the real-world datasets have several physical–chemical parameters, resulting in complex data with some nonlinearity, classical linear methods such as discriminant analysis cannot model this nonlinearity. Thus, nonlinear methods as advanced machine learning techniques are required to model complex problems [17, 19].
The present study brings a machine learning study for classification of Californian Cabernet Sauvignon wines from Napa and Paso Robles regions based on their elemental concentrations. We used seven classification algorithms (k-nearest neighbors, LDA, neural networks, partial least squares discriminant analysis, soft independent modeling class, random forest and support vector machines). The used methodology combines filter and wrapper-based feature selection procedures to characterize the wine-making regions. Although only 20 wine samples have been used to classify the geographical origin of Cabernet Sauvignon wines, the samples were collected from two wine regions (Napa and Paso Robles) and a similar number of samples, in the range of 15–24 samples, have been used in other chemometric studies with satisfactory results [20,21,22,23,24]. Our prime contributions in this research are:
We provide a classification model capable of predicting the geographical origin of Californian Cabernet Sauvignon wines from two specific wine-making regions;
We perform a comparative study on the performance of classical and advanced machine learning classification algorithms, which can offer theoretical contributions toward the comparison of these techniques on a real-world application;
We apply feature selection methods in order to recognize the most elements that discriminate the wines, providing a detailed view of the behavior of Napa and Paso Robles wines.
Materials and methods
Instruments and apparatus
The determination of the elements was performed by ICP-MS (PerkinElmer NexIon 300D, PerkinElmer, Norwalk, CT, USA). ICP-MS operating conditions are shown in Table 1.
Reagents and standards
All reagents used were of analytical-reagent grade except HNO3, which was purified in a quartz sub-boiling still (Kürner) before use. A clean laboratory and laminar-flow hood capable of producing class 100 were used for preparing solutions. High-purity de-ionized water (resistivity 18.2 MΩ cm) obtained using a Milli-Q water purification system (Millipore, Bedford, MA, USA) was used throughout. All solutions were stored in high-density polyethylene bottles. Plastic materials were cleaned by soaking in 10% (v/v) HNO3 for 24 h, rinsed five times with Milli-Q water and dried in a class 100 laminar flow hood before use. All operations were performed on a clean bench. Multi-element stock solutions containing 1000 mg/L of each element were obtained from PerkinElmer (PerkinElmer, Norwalk, CT).
Wine samples
A total of 20 Cabernet Sauvignon wine samples, 10 from the region of Napa, California, USA, and 10 from the region of and Paso Robles, California, USA, were collected during the first quarter of 2016. The ICP-MS analysis determined the concentration of Al, Cd, Co, Cr, Cu, Li, Mn, Ni, P, Pb, Rb, Sr and Zn for each sample.
Instrumentation and analysis
A quadrupole inductively coupled plasma mass spectrometry instrument (q-ICP-MS, NexIon 300 Perkin Elmer, USA) equipped with Universal Cell Technology™ (UCT), for interference removal, was used for the determination of elements in wine samples. The method proposed by [25] was applied for sample analysis. Briefly, prior to ICP-MS analysis, samples were diluted 1:10 with 1% HNO3 and rhodium was added as internal standard (final concentration: 10 μg/L). Data quantitation was achieved with reference to matrix-matched multi-element standards that had been prepared in 1% ethanol. Isotopes determined by ICP-MS were 7Li, 27Al, 31P, 53Cr, 55Mn, 59Co, 60Ni, 65Cu, 66Zn, 85Rb, 88Sr, 111Cd, 208Pb.
Classification process
In this study, we organized the wine data in a matrix with dimension 20 × 14, 20 samples and 14 variables, 13 columns represented the chemical elementals, and one to represent the label (Napa and Paso Robles). We performed an analysis using algorithms considered as classical chemometric methods and machine learning algorithms originated from computer science field along with variable selection methods to characterize and to classify the origin of Cabernet Sauvignon wine samples. The seven classification algorithms used to classify the wine data are supervised machine learning (ML) techniques. The supervised ML uses pre-defined classes to learn through a training phase how data is organized into these classes [26], making possible to predict unlabeled samples based on the classification model. Figure 1 shows the flowchart of our study, including the data acquisition, feature selection and the training models process.
Linear discriminant analysis (LDA), k-nearest neighbors (K-NN), partial least squares discriminant analysis (PLS), and soft independent modeling by class analogy (SIMCA) are the most used chemometric tools [16,17,18]. LDA is the most studied and the oldest discrimination technique, proposed by Fisher [27]. This method searches for discriminant functions that achieve maximum discrimination among the classes by minimizing the within-class variance and maximizing the between-class variance.
KNN is a classifier which aims to group data by correlating inputs to similar outputs. The classification model uses as parameters the number of k neighbors and the distance between the data points (such as Euclidean distance, Manhattan distance, or Minkowski distance relation). PLS discriminant analysis is a classifier based on PLS regression technique, which uses a value between zero and one to predict the class for each sample. This technique uses an approach similar to principal component analysis and searches for the variables with a maximum covariance with the class labels [28]. SIMCA is a class-modeling classifier based on principal component analysis, which creates a separated model for each class [27]. These techniques were successfully used to classify from China [29], Argentina [7], and Washington State, USA [10].
The support vector machines (SVM), random forest (RF) and multilayer perceptron (MLP) are three popular techniques which have yielded good results in the recent machine learning and data mining literature. These algorithms are more computationally intense than classical chemometric techniques, and in some cases do not have a reproducible solution [16]. Besides that, these algorithms show a great potential and more advantages compared to classical ones.
SVM is a classifier that obtains an optimal hyperplane with maximum margin to separate the classes of samples being a most robust and accurate methods in all well-known data mining algorithms [30]. Moreover, it is a useful classification algorithm when few training data are available [31]. RF algorithm is a classifier that generates multiple decision trees. Classification occurred according to the most voted class among the trees [32]. MLP is a complex structure based on biological neurons that can model real-world complex relationships being able to predict unknown sample classes [33]. The training process of the MLP propagates feed-forward through the network, layer after layer, by computing the output of each neuron until the output layer. By means of backpropagation, whether the output is inconsistent the error is calculated and propagated backward to adjust the connection weights and result into a new output. These techniques were successfully used to classify wines from Spain [34], Merlot and others wines from South America [8, 35].
Feature selection
Feature selection (FS) is a data mining preprocessing step which selects a subset of variables from the input which can efficiently describe the input data while reducing effects from noise or irrelevant variables and still provide good predictions. FS methods are capable of improving learning performance, lowering computational complexity, building better generalizable models, and decreasing required variables to obtain the desired model [36].
We used a two-phase feature selection by combining filter and wrapper methods. The filter methods use as principle a score value to order the variable importance into a ranking. We used the F-score and Random Forest Importance to generate two importance rankings and to create feature subsets based on the importance score to use on the wrapper phase.
F-score [37] is a simple technique which measures the discrimination of two sets of real numbers. Given training vectors \({x}_{k}, k = \{1,..., m\}\) if the number of positive and negative instances are \({n}^{+}\) and \({n}^{-}\), respectively, then the F-score of the ith feature is defined as:
where \({\stackrel{-}{x}}_{i}, {{\stackrel{-}{x}}_{i}}^{(+)}, {{\stackrel{-}{x}}_{i}}^{(-)}\) are the average of the \(i\)th feature of the whole, positive, and negative data sets, respectively; \({{x}_{ki}}^{(+)}\)is the ith feature of the \(k\)th positive instance, and \({{x}_{ki}}^{(-)}\) is the ith feature of the \(k\)th negative instance. The numerator indicates the discrimination between the positive and negative sets, and the denominator indicates the one within each of the two sets. The larger the F-score is, the more likely this feature is more discriminative.
Random Forest Importance (RFI) provides a variable importance measures based on the RF classifier and the Gini index. The bootstrap sample of RF classifier retained about 2/3 of the original samples and 1/3 of replicate samples. The remaining 1/3 of original samples (the out-of-bag samples) is used to test the tree formed based on the bootstrap sample and to calculate the variable importance [32].
Wrapper methods select the best feature subset based on the performance of the features as input data to a classifier. We used an iterative forward-selection procedure according to the importance rankings. Thus, 13 feature subsets were generated for each filter feature selection method. Each feature subset was used as input data to the classifiers LDA, PLS discriminant analysis, KNN, SIMCA, SVM, MLP and RF.
Model evaluation
To evaluate the model’s predictive performance, we used the tenfold cross-validation repeated 10 times method. In \(k\)-fold cross-validation technique randomly split data set \(D\) into \(k\) subsets \({D}_{1}, {D}_{2}, \dots , {D}_{k}\) (the folds) of approximately equal size. The process of build the classification model occurs for \(k\) times, which the model was constructed with the training set (\(k-1\) folds, each fold at a time was left out) and prediction ability was tested on the samples of fold omitted. The model accuracy is obtained based on the correct classifications, divided by the number of instances in the dataset. The final estimate of accuracy (i.e., the model performance) is the mean of all estimates computed. This process was repeated 10 times.
The predictions were organized in a confusion matrix to compute the accuracy, sensitivity and specificity based on the true positive (TP), true negative (TN), false positive (FP), and false negative (FN) prediction values. The accuracy is the percentage of the model that has been right in its predictions. Sensitivity refers to the percentage of correct answers regarding the positive class. Specificity is the percentage of correct answers regarding the negative class. These measures are computed as fallows:
Results and discussion
Trace elements of Napa and Paso Robles samples
The entire analysis was conducted using R software [38], which provides the packages that we use in this data analysis, including classification, variable selection and data visualization [39,40,41]. Table 2 shows the mean, minimum and maximum concentration of the determined elements (µg/L) in the wine samples from the two regions, and the p value obtained from the Kruskal–Wallis test to compare the means populations. In an initial observation, we can see that the levels of Al, Cd, Co, Cu, Li, Mn, and Sr were higher in Paso Robles than Napa. For the remaining variables (Cr, Ni, Rb, P, Pb, and Zn), the levels were not so different between Napa and Paso Robles regions. However, the Kruskal–Wallis test shows that there is only a statistically significant difference (p value < 0.05) between the Napa and Paso Robles groups for the variables Cd, Li, Mn and Sr.
After establishing reference ranges for 13 metals in Napa and Paso Robles wine samples, a variable importance was established for classification models construction.
Variable importance
Figure 2 shows the importance values assigned to each variable (i.e., the elements) according to the filter algorithms F-score (a), and RFI (b). These values represent their relative importance to determinate the sample labels. The higher the value is, more significant the variables are to discriminate the classes according to the metrics.
The Li and Sr elements were the first two most important variables in both methods in alternated orders. Mn and Cd elements are in the third and fourth orders in both rankings in alternated orders. These top four variables are the same variables which demonstrated a statistically significant difference based on the Kruskal–Wallis test. The remaining variables have different ranking positions.
After computing the relative importance of the variables, we generate the variable subsets which were used to build the classification models based on the wrapper methodology. Each subset is generated with those variables that achieved the top \(i\) score values, with \(i=\{1, 2, \dots , 13\}\). Subset #X1 has the variable with the higher importance according to the variable selection method X; subset #X2 has the two variables with the higher importance, and so forth. The last subset, #X13, contains all the original variables. Each subset was applied to the classification algorithms KNN, LDA, MLP, PLS, RF, SIMCA and SVM, along with tenfold cross-validation repeated 10 times.
Classification models
Figure 3 shows the results obtained from the application of generated subsets according to the F-score ranking on the classification models. The variable Sr by itself was capable of classify the origin of the wine samples in 95% of accuracy with the classifiers SVM and SIMCA. The mean concentration of this element is 682.49 ± 122.91 µg/L for the Napa samples and 1113.07 ± 182.82 µg/L, showing a significant difference between this elemental concentration (p value < 0.001), which explain the high prediction ability. The performance of the models generated by SIMCA decreased when adding other variables in the input subset. The results of RF models were the highest, followed by the SVM models. The highest sensitivity and specificity rates were also from the RF models.
By the use of Sr and Li, the RF achieved 97% of accuracy, and this value increases until achieved perfect classification with a group of a range of 6–11 variables (Sr, Li, Mn, Cd, Co, Cu, Zn, Rb, Cr, P, Al). The SVM model obtained 96.5% of accuracy with Sr and Li as input variables, and 99% of accuracy with 6 variables (Sr, Li, Mn, Cd, Co, Cu). The remaining classifiers achieved classification rates above 80% with up to 5 variables. The MLP, SIMCA, KNN and PLS resulted in prediction ability below 80% when using six or more variables.
Figure 4 shows the results obtained from the application of generated subsets according to the RFI ranking on the classification models. For this ranking, the Li was considered as the most important variable. A perfect classification rate was obtained by the use of just this variable as input to the classifiers SVM, RF, MLP, and KNN. The concentration values of this elemental for the Napa class is 9.56 ± 4.69 µg/L, and for the Paso Robles class is 61.48 ± 40.03 µg/L, showing a significant difference between the classes (p value < 0.001). This explains the classification rate as Li concentration value is higher on the Paso Robles than Napa samples.
The RF models achieved a classification rate with a range of 97.5–100% to all feature subsets. The remaining classifiers keep the classification rate on a specific range or decrease when adding new input variables. The highest sensitivity and specificity rates were also from the RF models for this importance ranking.
Based on these classification results it is possible to see that the Li and Sr are the two main elements responsible for discriminating between the wines from Paso Robles and Napa based on our dataset. Figure 5 shows the biplot of variables Li and Sr. The samples are grouped according to its respective class. These elementals were also found as important to discriminate other wines. Sr was one of the main elements to discriminate Tempranillo blanco wines from different zones of the AOC Rioja [42]. Strontium was also one of the indicators to discriminate soils and wines of the three major wine-producing regions in Romania (Mn, Cr, Sr, Ag and Co) [43].
Li was found as the main descriptor to classify wines from Argentina, Brazil, France, and Spain by using linear discriminant analysis [44]. Lithium was also one of the five elements which showed a significant vineyard effect (Be, Eu, Ga, Li, Si) to wines from regions of Northern California (closest to Napa region) [6]. The authors conclude that these elements were not changed at all during the wine-making process or changed to the same extent in all regions analyzed. Despite the limited set of our samples, these results showed that the concentrations of Li and Sr were significantly different among the Cabernet Sauvignon wines and could reliably discriminate the wines from Napa and Paso Robles regions.
Importance of others metals
A second analysis was performed by removing the variables Li and Sr to investigate the relevance of the others metals (Al, Cd, Co, Cr, Cu, Mn, Ni, P, Pb, Rb, and Zn). The relevance of other chemical elementals subset can be useful to characterize Napa and Paso Robles wines in situations where Li and Sr variables cannot be measured, and for demonstrating that hidden patterns can be found from advanced machine learning techniques.
Figure 6 shows the importance values assigned to each variable without Li and Sr. The F-score importance order is the same importance order of Fig. 2 without Li and Sr, as the F-score computation is performed by considering each variable at time. However, the RFI order is not the same of Fig. 2 as the importance order is computed based on the whole dataset. Both importance rankings show different importance values and ordering to each variable. New subsets of variables were generated based on these new importance rankings.
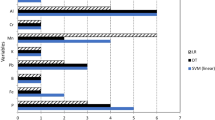
Figures 7 and 8 show the performance of the classification models to the feature subsets generated based on the F-score and RFI ranking. By removing the features Li and Sr it was not possible to classify the California wine-making regions with a good classification rate by the use of the classical chemometrics algorithms LDA, PLS-DA, SIMCA and KNN. All these classification models obtained a performance below 78% of accuracy.
However, SVM was able to classify the samples with 89% of accuracy using seven variables selected by RFI (Cd, Ni, Mn, Pb, Rb, Co, Cu). The best result based on the F-score ranking was composed of six variables, which achieved 83% of accuracy using six variables (Mn, Cd, Co, Cu, Zn, Rb). The combination of the chemical elements in these two subsets allowed SVM to classify the samples in a good performance. This fact indicates that these subsets are also capable of discriminating the wine-production regions, Napa and Paso Robles, without using the Li and Sr concentrations as input data to the classifiers.
These results suggest that advanced machine learning techniques are needed when dealing with complex information. The Li and Sr played an important role to discriminate the origin of Cabernet Sauvignon wines. However, classical techniques can model the data based on these elementals. The removal of these variables from the classification model and the use of advance algorithms allowed us to find information about the composition of wines and how the variables characterize the wine-producing regions.
According to a recent review, the combination of chemical information and mathematical models is the future of wine authentication [45]. The results of this study showed that beyond the mathematical model an ensemble of algorithms and critical analysis of the results is needed to improve the wine analysis to provide improved classification models that can result in useful information to wine authentication, improve quality and to avoid fraud.
Conclusion
To our knowledge, this is the first paper to analyze the origin of Cabernet Sauvignon wine samples from California by the use of machine learning techniques and ICP-MS. A first analysis identified that among the 13 elements found in the composition of wine samples, the Li and Sr are the variables with major discriminating power for origin samples according to the F-score and RFI. The concentration of these elementals is higher in wines produced on Paso Robles than Napa, explaining the high performance of the classifier.
A second data analysis allowed us to identify others chemical compounds that characterize the regions. We found that the variables Cd, Ni, Mn, Pb, Rb, Co, and Cu, can classify the geographical origin in 89% of accuracy by using SVM based on the collected samples from Napa and Paso Robles. The results demonstrate that feature selection and its critical analysis to remove some variables from the classification model is useful to identify the chemical elements that characterize the wine-producing regions of California, Napa and Paso Robles. Moreover, it also showed that in face of complex food data the use of advanced machine learning techniques is needed. The used methodology is useful to identify the characteristics of others wines and food products, and from others regions. For future studies, we expect that some limitations found in our present research can be addressed, such as the expansion of wine data. Future research could be expanded to include wines from other regions and varieties, and to model chemical information obtained from other analytical methods.
References
Versari A, Laurie VF, Ricci A, Laghi L, Parpinello GP (2014) Progress in authentication, typification and traceability of grapes and wines by chemometric approaches. Food Res Int 60:2–18. https://doi.org/10.1016/j.foodres.2014.02.007
Luykx DMAM, van Ruth SM (2008) An overview of analytical methods for determining the geographical origin of food products. Food Chem 107:897–911. https://doi.org/10.1016/j.foodchem.2007.09.038
I.O. of V. and Wine, State of the Vitiviniculture World Market (2019) https://www.oiv.int/public/medias/6679/en-oiv-state-of-the-vitiviniculture-world-market-2019.pdf. Accessed 1 Aug 2019
Hira A, Swartz T (2014) What makes Napa Napa? The roots of success in the wine industry. Wine Econ Policy 3:37–53. https://doi.org/10.1016/j.wep.2014.02.001
Umali AP, Ghanem E, Hopfer H, Hussain A, Kao YT, Zabanal LG, Wilkins BJ, Hobza C, Quach DK, Fredell M, Heymann H, Anslyn EV (2015) Grape and wine sensory attributes correlate with pattern-based discrimination of Cabernet Sauvignon wines by a peptidic sensor array. Tetrahedron 71:3095–3099. https://doi.org/10.1016/j.tet.2014.09.062
Hopfer H, Nelson J, Collins TS, Heymann H, Ebeler SE (2015) The combined impact of vineyard origin and processing winery on the elemental profile of red wines. Food Chem 172:486–496. https://doi.org/10.1016/j.foodchem.2014.09.113
Fabani MP, Arrúa RC, Vázquez F, Diaz MP, Baroni MV, Wunderlin DA (2010) Evaluation of elemental profile coupled to chemometrics to assess the geographical origin of Argentinean wines. Food Chem 119:372–379. https://doi.org/10.1016/j.foodchem.2009.05.085
Soares F, Anzanello MJ, Fogliatto FS, Marcelo MCA, Ferrão MF, Manfroi V, Pozebon D (2018) Element selection and concentration analysis for classifying South America wine samples according to the country of origin. Comput Electron Agric 150:33–40. https://doi.org/10.1016/j.compag.2018.03.027
Šelih VS, Šala M, Drgan V (2014) Multi-element analysis of wines by ICP-MS and ICP-OES and their classification according to geographical origin in Slovenia. Food Chem 153:414–423. https://doi.org/10.1016/j.foodchem.2013.12.081
Orellana S, Johansen AM, Gazis C (2019) Geographic classification of US Washington State wines using elemental and water isotope composition. Food Chem 1:100007. https://doi.org/10.1016/j.fochx.2019.100007
Geana EI, Popescu R, Costinel D, Dinca OR, Ionete RE, Stefanescu I, Artem V, Bala C (2016) Classification of red wines using suitable markers coupled with multivariate statistic analysis. Food Chem 192:1015–1024. https://doi.org/10.1016/j.foodchem.2015.07.112
Šperková J, Suchánek M (2005) Multivariate classification of wines from different Bohemian regions (Czech Republic). Food Chem 93:659–663. https://doi.org/10.1016/j.foodchem.2004.10.044
da Costa NL, Castro IA, Barbosa R (2016) Classification of Cabernet Sauvignon from two different countries in South America by chemical compounds and support vector machines. Appl Artif Intell 30:679–689. https://doi.org/10.1080/08839514.2016.1214416
Urvieta R, Buscema F, Bottini R, Coste B, Fontana A (2018) Phenolic and sensory profiles discriminate geographical indications for Malbec wines from different regions of Mendoza, Argentina. Food Chem 265:120–127. https://doi.org/10.1016/j.foodchem.2018.05.083
Cozzolino D, Cynkar WU, Shah N, Smith PA (2011) Can spectroscopy geographically classify Sauvignon Blanc wines from Australia and New Zealand? Food Chem 126:673–678. https://doi.org/10.1016/j.foodchem.2010.11.005
Brereton RG (2015) Pattern recognition in chemometrics. Chemom Intell Lab Syst 149:90–96. https://doi.org/10.1016/j.chemolab.2015.06.012
Jiménez-Carvelo AM, González-Casado A, Bagur-González MG, Cuadros-Rodríguez L (2019) Alternative data mining/machine learning methods for the analytical evaluation of food quality and authenticity—a review. Food Res Int. https://doi.org/10.1016/j.foodres.2019.03.063
Callao MP, Ruisánchez I (2018) An overview of multivariate qualitative methods for food fraud detection. Food Control 86:283–293. https://doi.org/10.1016/j.foodcont.2017.11.034
Li H, Liang Y, Xu Q (2009) Support vector machines and its applications in chemistry. Chemom Intell Lab Syst 95:188–198. https://doi.org/10.1016/j.chemolab.2008.10.007
Ríos-Reina R, Morales ML, García-González DL, Amigo JM, Callejón RM (2018) Sampling methods for the study of volatile profile of PDO wine vinegars. A comparison using multivariate data analysis. Food Res Int 105:880–896. https://doi.org/10.1016/j.foodres.2017.12.001
Moreno J, Moreno-García J, López-Muñoz B, Mauricio JC, García-Martínez T (2016) Use of a flor velum yeast for modulating colour, ethanol and major aroma compound contents in red wine. Food Chem 213:90–97. https://doi.org/10.1016/j.foodchem.2016.06.062
Agazzi FM, Nelson J, Tanabe CK, Doyle C, Boulton RB, Buscema F (2018) Aging of Malbec wines from Mendoza and California: evolution of phenolic and elemental composition. Food Chem 269:103–110. https://doi.org/10.1016/j.foodchem.2018.06.142
Andreu-Navarro A, Russo P, Aguilar-Caballos MP, Fernández-Romero JM, Gómez-Hens A (2011) Usefulness of terbium-sensitised luminescence detection for the chemometric classification of wines by their content in phenolic compounds. Food Chem. https://doi.org/10.1016/j.foodchem.2010.08.014
La Torre GL, La Pera L, Rando R, Lo Turco V, Di Bella G, Saitta M, Dugo G (2008) Classification of Marsala wines according to their polyphenol, carbohydrate and heavy metal levels using canonical discriminant analysis. Food Chem. https://doi.org/10.1016/j.foodchem.2008.02.071
Martin AE, Watling RJ, Lee GS (2012) The multi-element determination and regional discrimination of Australian wines. Food Chem 133:1081–1089. https://doi.org/10.1016/j.foodchem.2012.02.013
Witten I, Frank E, Hall M, Pal C (2016) Data mining: practical machine learning tools and techniques. Morgan Kaufmann, San Francisco (ISBN: 978-0-12-804291-5)
Bevilacqua M, Bucci R, Magrì AD, Magrì AL, Nescatelli R (2013) Classification and class-modelling. Data Handl Sci Technol 28:171–233. https://doi.org/10.1016/B978-0-444-59528-7.00005-3
Bajoub A, Medina-Rodríguez S, Gómez-Romero M, Ajal EA, Bagur-González MG, Fernández-Gutiérrez A, Carrasco-Pancorbo A (2017) Assessing the varietal origin of extra-virgin olive oil using liquid chromatography fingerprints of phenolic compound, data fusion and chemometrics. Food Chem 215:245–255. https://doi.org/10.1016/j.foodchem.2016.07.140
Ouyang Q, Zhao J, Chen Q (2013) Classification of rice wine according to different marked ages using a portable multi-electrode electronic tongue coupled with multivariate analysis. Food Res Int 51:633–640. https://doi.org/10.1016/j.foodres.2012.12.032
Xue H, Yang Q, Chen S (2009) SVM: support vector machines. In: Top ten algorithms data min, pp. 37–59
Jurado JM, Alcázar Á, Palacios-Morillo A, De Pablos F (2012) Classification of Spanish DO white wines according to their elemental profile by means of support vector machines. Food Chem 135:898–903. https://doi.org/10.1016/j.foodchem.2012.06.017
Breiman L (2001) Random forests. Mach Learn 45:5–32. https://doi.org/10.1023/A:1010933404324
Zhang GP (2000) Neural networks for classification: a survey. IEEE Trans Syst Man Cybern 30:451–462. https://doi.org/10.1109/5326.897072
Gómez-Meire S, Campos C, Falqué E, Díaz F, Fdez-Riverola F (2014) Assuring the authenticity of northwest Spain white wine varieties using machine learning techniques. Food Res Int 60:230–240. https://doi.org/10.1016/j.foodres.2013.09.032
Costa NL, Llobodanin LAG, Castro IA, Barbosa R (2019) Using Support Vector Machines and neural networks to classify Merlot wines from South America. Inf Process Agric 6:265–278. https://doi.org/10.1016/j.inpa.2018.10.003
Chandrashekar G, Sahin F (2014) A survey on feature selection methods. Comput Electr Eng 40:16–28. https://doi.org/10.1016/j.compeleceng.2013.11.024
Chen Y-W, Lin C-J (2006) Combining SVMs with various feature selection strategies. In: Featur. extr. Springer, pp. 315–324. https://doi.org/10.1007/978-3-540-35488-8_13
R Core Team (2016) R: a language and environment for statistical computing. R Found. Stat. Comput, Vienna
Kuhn M (2015) Caret: classification and regression training. Astrophys Source Code Libr
Romanski P, Kotthoff L, Kotthoff ML (2018) Package Fselector: selecting attributes, repos. CRAN 18
Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer, New York
Pérez-Álvarez EP, Garcia R, Barrulas P, Dias C, Cabrita MJ, Garde-Cerdán T (2019) Classification of wines according to several factors by ICP-MS multi-element analysis. Food Chem 270:273–280. https://doi.org/10.1016/j.foodchem.2018.07.087
Geana I, Iordache A, Ionete R, Marinescu A, Ranca A, Culea M (2013) Geographical origin identification of Romanian wines by ICP-MS elemental analysis. Food Chem 138:1125–1134. https://doi.org/10.1016/j.foodchem.2012.11.104
Yamashita GH, Anzanello MJ, Soares F, Rocha MK, Fogliatto FS, Rodrigues NP, Rodrigues E, Celso PG, Manfroi V, Hertz PF (2019) Hierarchical classification of sparkling wine samples according to the country of origin based on the most informative chemical elements. Food Control 106:106737. https://doi.org/10.1016/j.foodcont.2019.106737
Villano C, Lisanti MT, Gambuti A, Vecchio R, Moio L, Frusciante L, Aversano R, Carputo D (2017) Wine varietal authentication based on phenolics, volatiles and DNA markers: state of the art, perspectives and drawbacks. Food Control 80:1–10. https://doi.org/10.1016/j.foodcont.2017.04.020
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
da Costa, N.L., Ximenez, J.P.B., Rodrigues, J.L. et al. Characterization of Cabernet Sauvignon wines from California: determination of origin based on ICP-MS analysis and machine learning techniques. Eur Food Res Technol 246, 1193–1205 (2020). https://doi.org/10.1007/s00217-020-03480-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-020-03480-5