Abstract
A novel milk-clotting enzyme (MCE) produced by Bacillus amyloliquefaciens GSBa-1 was purified and identified to belong to the peptidase M4 family, and the mature peptide with milk-clotting activity (MCA) was a neutral metalloproteinase with molecular mass of about 38 kDa. The optimal pH and temperature were determined to be at 5.5 and 57 °C for MCA, and at 7.0 and 57 °C for the proteolytic activity, respectively. The MCE exhibited slight autolysis that could be inhibited by Ca2+ and Na+. Hydrolysis of caseins revealed that κ-casein exhibited higher sensitivity to the MCE action than α- and β-casein. By in-gel tryptic digestion and LC–MS/MS analysis of the major peptide (about 13 kDa) generated from hydrolysis of κ-casein by the MCE, the cleavage site was identified to be at Lys 111–Lys 112, which was different from those of other MCEs reported earlier. The MCE from B. amyloliquefaciens GSBa-1 could serve as a novel milk coagulant for potential application in making cheese with desired proteolysis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Due to worldwide increase in cheese consumption and more demands for milk-clotting enzymes (MCEs), searching for different sources of MCEs including those of microbial origin has been a focus of many studies [1,2,3]. Bacteria taken as potential MCE producer candidates, including Bacillus species [4, 5] and fungi such as Aspergillus oryzae have been reported [6]. However, many microbial MCEs exhibit broad hydrolytic activity on caseins that often causes low cheese yields, loosen texture, and a bitter flavor of cheese. Therefore, efficient microbial MCEs must have the properties of high milk-clotting activity (MCA) but low unspecific proteolytic activity (PA).
Bacillus species has been widely investigated as it is known to produce various extracellular proteases for crucial industrial applications [7]. Bacillus MCEs with more production of MCA but lower PA compared with plant MCEs has attracted interest of researchers [3, 8]. The MCEs from some non-pathogenic strains of Bacillus species were applied in cheese making and resulted in yield reduction and bitter taste of the ripened cheese [4, 9]. The MCE from B. stearothermophilus had good thermal and pH stability, but the soft cheese made with this MCE possessed undesired organoleptic properties because of excessive proteolysis when compared with the cheese made with commercial coagulants [5]. Furthermore, the MCEs from B. amyloliquefaciens and Bacillus sp. P45 showed higher proteolysis in cheese leading to more peptide formation and softer cheese texture [10, 11]. Therefore, further understanding of the enzymatic and structural properties of the microbial MCEs, specially the proteolytic pattern and the mechanism involved is crucial to maintain cheese quality.
B. amyloliquefaciens GSBa-1 was isolated from the traditional starter called jiuqu, which contained a mixture of yeasts, molds, and bacteria. The rice wine made by fermentation of glutinous rice with jiuqu was used as a milk-clotting agent to make traditional Royal Cheese, a type of fresh cheese originally made for royal families since early Qing dynasty in China. B. amyloliquefaciens GSBa-1 isolated from jiuqu was confirmed to produce MCE [12]. The use of this MCE in making Mozzarella cheese was found to promote formation of free amino acids and volatiles due to its moderate proteolytic activity when compared with a commercial rennet [12, 13]. In this study, the MCE from B. amyloliquefaciens GSBa-1 was purified, identified, and enzymatically characterized. The proteolytic activity of the MCE, its hydrolytic pattern on milk proteins, and the specific cleavage site of κ-casein were studied. The present study would provide further understanding on the structure and properties of the microbial MCE, and its application in making cheese with desired proteolysis.
Materials and methods
Materials
B. amyloliquefaciens GSBa-1 was stored in freeze-dried form at − 80 °C, and it was activated consecutively for three times in LB medium (tryptone 10 g/L, yeast extract 5 g/L, sodium chloride 10 g/L; initial pH 7.2) at 37 °C for 24 h before use for preparation of the MCE. The skim milk powder, whey protein, and sodium caseinate were purchased from Fonterra Co-operative Group (Auckland, New Zealand). The individual α-casein, β-casein, and κ-casein were purchased from Sigma Aldrich (CA, USA).
Purification of the MCE
After growth of B. amyloliquefaciens GSBa-1 in LB medium at 37 °C for 24 h, the culture was centrifuged (6000 rpm, 4 °C, 30 min). The supernatant was collected and cooled ethanol was added making its volume up to 70%. The precipitate was collected by centrifugal separation (10,400×g, 30 min), dissolved in distilled water, and freeze dried with a lyophilizer (OSTC, Beijing, China) to obtain the crude MCE. The dissolved and filtered (0.45 μm pore size) crude MCE in 50 mM Tris–HCl buffer (pH 6.8) was subjected to a DEAE Sepharose Fast Flow column (50 cm × 2 cm) previously equilibrated with the same Tris–HCl buffer. Elution was performed with a linear gradient of NaCl from 0 M, 0.2 M, 0.4 M, 0.6 M to 0.8 M at a flow rate of 2 mL/min in 50 mM Tris–HCl buffer (pH 6.8). The fractions containing high MCA were pooled, then desalted, and concentrated by ultrafiltration with molecular weight cutoff at 10 kDa. The purity of the purified MCE was confirmed by SDS-PAGE (12.5% separation gel and 4.5% spacer gel). The Kjeldahl method was used to determine the total protein content, and then the recovery rate and other related parameters were calculated.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis
The SDS-PAGE was performed according to the method of Laemmli [14]. It was performed at room temperature in Mini-Protean short plates (Bio-Rad Laboratories, Hercules, California) with a stacking gel of 4.5% and a separating gel of 12.5%. The step-voltage procedure was performed as following: voltage was kept constant at 70 V till the samples reached the junction between stacking gel and separating gel, and then decreased to 50 V till the samples reached the point of 1.5 cm away from the end. The protein on each gel was stained by 0.01% w/v Coomassie Brilliant Blue R250 (Sigma Chemical Co., St. Louis, MO) for 20 min and destained in a solvent of methanol:glacial acetic acid:deionized water at a ratio of 250:70:680 (v:v:v). The gel was digitally scanned and analyzed using Image Lab Software (Bio-Rad Laboratories).
Identification of the MCE
The purified MCE displayed as a protein band on SDS-PAGE was cut from the gel and diced to 1 mm3, transferred to a 1.5 mL micro-tube, and digested by trypsin according to the method from Zhao et al. [15]. Then the peptide segments of enzymolysis after decoloration and dehydration were extracted, and identified by LC–MS–MS (Thermo Fisher Scientific, Waltham, MA, USA). Samples (5 μL) were injected onto Ultimate 3000 system (Thermo Fisher Scientific, USA) for desalting prior to chromatographic separation on a 100 μm × 10 cm in-house made column packed with a reversed-phase ReproSil-Pur C18-AQ resin (3 μm, 120 Å, Dr. Maisch GmbH, Germany) using the following gradient at a flow rate of 300 nL/min: 0–5 min 5% solution B, 25 min 50% solution B, 30 min 90% solution B, 35 min 90% solution B, 45 min 10% solution B, 35 min 10% solution B, where solution A was 0.1% formic acid in water and solution B was 0.1% formic acid in 80% acetonitrile. The column eluate was ionized using Thermo Scientific Q Exactive Electrospray. MS parameters: resolution: 70,000, AGC target: 3e6, maximum IT: 40 ms, scan range: 350–1800 m/z. MS/MS parameters: resolution: 17,500, AGC target: 1e5, maximum IT: 60 ms, TopN: 20, NCE/stepped NCE: 27. Finally, the raw MS files were analyzed and searched against protein database (UniProt) based on the species of the samples using MaxQuant.
Assay of MCA and PA
Milk-clotting activity was determined according to the method of Arima et al. [16]. The method was based on the determination of time for milk clot formation. One unit was defined as the amount of enzyme that clotted 1 mL of a solution containing 0.1 g skim milk powder in 40 min at 35 °C. Briefly, 5 mL of a 10% (w/v) suspension of skim milk powder was used as the substrate and kept at 35 °C for 5 min. Then 0.5 mL of milk-clotting enzyme solution was added and mixed. The time between addition and appearance of clots in the milk solution was recorded and the clotting activity was calculated as follows: \({\text{MCA}}\;({\text{Su/mL}}) = {\raise0.7ex\hbox{${2400}$} \!\mathord{\left/ {\vphantom {{2400} {{\text{clotting}}\;{\text{time}}\;({\text{s}})}}}\right.\kern-0pt} \!\lower0.7ex\hbox{${{\text{clotting}}\;{\text{time}}\;({\text{s}})}$}} \times {\text{dilution}}\;{\text{factor}}.\)
The proteolytic activity was determined following the Folin–Lowry method as described by Zhao et al. [17]. Inactivation of the enzyme and precipitation of the non-hydrolyzed protein were carried out by using trichloroacetic acid (TCA). One unit of proteolytic activity was defined as the amount of enzyme to catalyze casein to produce 1 μg of tyrosine in 1 min at 40 °C.
Optimum temperature and pH for enzymatic activity
The influence of temperature and pH on the MCA and PA of the MCE was studied following the forementioned activity assay methods. The effect of different temperatures (from 22 to 82 °C) at pH 6.8 of the original milk and the effect of pH levels (from 5.5 to 11.0) at 37 °C were evaluated. For each test, the enzyme was added to the milk, which had already been heated to the set temperature or adjusted to the set pH.
Assay of various factors influencing the MCE activity
The activity of the MCE in the presence of metallic salts (KCl, NaCl, LiCl, ZnCl2, MgCl2, CaCl2, CuCl2, CdCl2, and PdCl2), organic solvents (methanol, ethanol, isopropanol, acetonitrile, and DMSO), denaturants (urea, GuHCl), detergents (Triton-X100 and SDS), protease inhibitor (EDTA and Pepstatin A) was studied. Prior to the assay, the MCE was incubated for 2 h in the presence of these different factors at the concentrations shown in Table 3. The remaining activity was calculated as a percentage of the activity of the untreated enzyme taken as 100%.
Different concentration of EDTA inhibitor from 0 to 1 mM/L was employed to test the tolerance of the MCE. The MCA and PA were determined after 30 min pre-incubation of the MCE with EDTA. The activity of the MCE in the absence of inhibitor was regarded as 100%.
Autolysis of the MCE in pure water with and without salt of sodium chloride and calcium chloride at the same concentration (0.05 mol/L) was determined. The MCE (0.01 mg/mL) was incubated at room temperature (25 °C), and the PA was measured after 0, 24, 48, 72, and 94 h of incubation using casein 1% (w/v) as substrate. The remaining activity was calculated as a percentage of activity of the MCE at 0 h.
Hydrolysis of caseins
For the hydrolysis of individual casein components by the MCE, α-, β-, and κ-casein were dissolved in 10 mM sodium phosphate buffer (pH 6.5) to obtain a protein concentration of 1 mg/mL. To evaluate the effect of the MCE concentration, the MCE (10 μL) was added to 0.1 mL individual casein solutions, and the reaction proceeded for 30 min at each MCE concentration of 0.78125 × 10−2, 1.5625 × 10−2, 3.125 × 10−2, 6.25 × 10−2, 1.25 × 10−1, 2.5 × 10−1, 5 × 10−1, and 1 mg/mL, respectively. To evaluate the effect of reaction time on hydrolysis of individual casein components, the individual aliquots of hydrolysates (the MCE at 3.125 × 10−2 mg/mL) were removed from the incubator at 10, 30, 60, 90, 180, 270, and 360 min, respectively. The hydrolysis was stopped and analyzed by SDS-PAGE as described above.
Analysis of cleavage site of κ-casein
The major peptide band obtained in the SDS-PAGE of the hydrolyzed κ-casein by the MCE was analyzed by in-gel tryptic digestion and LC–MS/MS using the same method referred in identification of the MCE.
Statistical analysis
All measurements in this study were performed with three parallels. The results were presented as the mean ± standard deviation. Significant differences between treatments were tested by ANOVA. Data analysis was performed using SPSS version 17.0 (SPSS, Inc., Chicago, IL).
Results and discussion
Purification and identification of the MCE
The culture filtrate (6 L) from B. amyloliquefaciens GSBa-1 with total MCA of 7660.74 SU and specific activity of 1.45 SU mg−1 protein was subjected to purification by addition of cold ethanol till 70% to precipitate the MCE, and 66.21-fold of purification with 87.72% of recovery of the MCE was obtained (Table 1). Further purification by DEAE Sepharose FF by stepwise increasing gradient elution with NaCl from 0, 0.2, 0.4, 0.6 to 0.8 M revealed four protein peaks (Fig. 1a). The first peak was tested with high MCA and low PA, and this purification step resulted in 450.70-fold purification with 51.35% recovery of the total activity, indicating a rather high degree of purification. SDS-PAGE of the purified fraction confirmed high purity of the MCE as shown by a single protein band with a molecular mass about 38 kDa (Fig. 1b).
a Purification of milk-clotting enzyme (GSBa-1 protease) from B. amyloliquefaciens GSBa-1 by DEAE Sepharose Fast Flow chromatography; b purity and molecular weight determination of GSBa-1 protease by SDS-PAGE, M: marker, lane 1: crude enzyme precipitated by 70% ethnol, lane 2: purified enzyme by DEAE Sepharose Fast Flow chromatography; c mass spectrum of the peptide segment corresponding to the enzyme from B. amyloliquefaciens GSBa-1; d amino acid sequence of GSBa-1 protease
Identification of the MCE by LC–MS–MS of the digested MCE sample with trypsin showed that it belonged to peptidase M4 (WP_032874795.1) since there were 5 peptide fragments (Table 2) detected to be homologous sequences from peptidase M4, a neutral metalloproteinase with 521 amino acids and a molecular mass of 56.831 kDa (Fig. 1c). This peptidase M4 (WP_032874795.1) was demonstrated to contain five domains: the signal peptide (1–27 amino acid residues), fungalysin/thermolysin propeptides motif (FTP) (78–126 amino acid residues), peptidase propeptides-YPEB domain (PepSY) (139–215 amino acid residues), M4 domain (225–372 amino acid residues), and M4-C domain (375–520 amino acid residues). Then the mature structure of the MCE from B. amyloliquefaciens GSBa-1 containing M4 and M4-C domains from peptidase M4, with a molecular mass around 38 kDa and milk-clotting activity, was formed from shear action by proteases or auto-digestion. Similarly, the MCE from B. subtilis was identified to belong to another peptidase M4 (WP-069149540.1), and the mature peptide with a molecular mass of 42 kDa and milk-clotting activity was also formed probably due to autodigestion [18]. Other MCEs from Bacillus sp. P45 and B. amyloliquefaciens D4 had molecular mass of 27 kDa and 58.2 kDa, respectively [19, 20]. A serine protease with milk-clotting activity from B. licheniformis USC13 had a molecular mass of 62 kDa and the processed mature form of 34 kDa [21].
Temperature and pH optima of the MCE
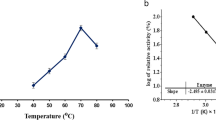
The pH and temperature dependence of both the MCA and PA of the MCE from B. amyloliquefaciens GSBa-1 were studied as shown in Fig. 2. The MCA of the MCE decreased with increasing pH from 5.5, and it completely lost the activity at pH 8.5, while the maximal PA of the MCE was at pH 7.0 with decreased activity under both acidic and alkaline conditions (Fig. 2a). Therefore, slightly acidic condition would be favorable for the MCE to be applied in cheese making to obtain more efficient milk coagulation but less proteolysis of cheese. Acidification of milk, e.g., to pH 5.5 or pH 6.0, was known to cause a certain degree of demineralization of casein micelles and decrease of electrostatic repulsion between micelles that created more favorable condition for milk coagulation [22]. The decreased PA of the MCE at acidic pHs could be due to change of the ionic microenvironment surrounding the enzyme that led to binding of the enzyme with proteins or partial irreversible inactivation of the enzyme caused by destruction of the spatial structure or changes in the active sites of the enzyme. Figure 2b indicated that the MCE was relatively heat resistant with both its maximal MCA and PA at 57 °C, and decreased MCA and PA were observed at 62 °C. Further increase from 62 °C up to 82 °C caused milk coagulation mainly because the heat induced denaturation of milk protein, but not the activity of the MCE. In addition, the MCE could also be denatured or partially denatured at the temperatures higher than 62 °C, and thus further evaluation of the MCE activities at the higher temperatures was not performed.
The optimum pH (5.5, 7.0) and temperature (57 °C) for the MCA and PA of the MCE from B. amyloliquefaciens GSBa-1 were different from those of other microbial MCEs reported earlier. The MCE from B. licheniformis 5A1 had optimum pH at 5.5, but with higher optimum temperature (70 °C) for MCA [23]. The MCE from B. subtilis natto had optimum pH at 6.0 and temperature of 60 °C for MCA [7]. The MCE from Enterococcus faecalis TUA 2495 L showed an increase of MCA with a decrease of pH from 7.8 to 5.8, but the PA was optimal at pH 8.0–9.0, and the optimum temperature was 70 °C for MCA and 50 °C for PA [24]. Other MCEs from Aspergillus oryzae MTCC 5341 and Nocardiopsis sp. exhibited maximal MCA at pH 6.3, 7.5, and at 55 °C, respectively [6, 25]. Whereas, the optimum temperature for Mucor miehei protease was higher than 63 °C and the optimum pH was 4.7 [26], and the MCA of the enzyme from Penicillium oxalicum was maximal at pH 4.0–5.0 and 65 °C [27]. Therefore, MCEs from various microbes achieved their maximal MCA generally under acidic pH and different temperature conditions.
Activity of the MCE as influenced by various factors
The MCA and PA of the MCE from B. amyloliquefaciens GSBa-1 as influenced by various factors were studied, including EDTA, pepstatin A, various metal ions, organic solvents, and protease inhibitors. As shown in Table 3, both the MCA and PA were inactivated by EDTA but not affected by pepstatin A, indicating that the MCE was probably a metalloprotease. The PA of the MCE was also not obviously inhibited by the monovalent cations (K+, Na+, and Li+) and divalent cations (Zn2+, Mg2+, and Ca2+), but the MCA of the MCE was improved by these cations, especially Ca2+ that increased the MCA by 120%. However, both the MCA and PA were potentially decreased by heavy metal ions such as Cu2+ and Pb2+ and partially by Cd2+, suggesting that these heavy metal ions might damage the active conformation of the enzyme. Similarly, El-Tanboly et al. [28] also reported the inactivation of the microbial MCE by Cu2+. No obvious inhibitory effect on MCA and PA was found with 50% (v/v) methanol, ethanol, isopropanol, acetonitrile, and DMSO with the remaining activities over 90%, indicating that the active center of the enzyme was not located in the sensitive area to these organic solvents. The inhibitory effect of the organic solvents on the catalytic activity of protease was reported earlier mainly due to their impact on assimilating the essential water molecules, thus destroying the water structure around the protein [29]. The denaturants such as urea and GuHCl did not affect the PA of the MCE, but both urea and GuHCl decreased the MCA with the remaining activity of 80.06% and 96.24%, respectively. However, treatment of the MCE with Triton X-100 increased the MCA by 5% and decreased the PA by about 6%, while complete loss of the MCA and only 11.27% of PA remained after treatment with SDS, probably due to changes of the protein conformation or interfacial properties of the MCE upon treatment by the denaturants that affected the catalytic properties of the enzyme [30].
To confirm the MCE from B. amyloliquefaciens GSBa-1 to be a metalloprotease, the effect of EDTA at different concentrations on the MCA and PA of the MCE was further investigated (Fig. 3a). Increase in the concentration of EDTA caused apparent decrease in both the MCA and PA, which was totally lost at 0.25 mM or higher concentration of EDTA. Previously, the MCE from Termitomyces clypeatus MTCC 5091 was also inhibited by EDTA, confirming the enzyme to be a metalloprotease [31].
Most proteases undergo autodigestion which is dependent on the enzyme concentration and temperature, and the autolysis is mostly prominent at low concentration of the enzyme [32, 33]. Therefore, to further investigate the storage stability of the MCE, the autolytic property of the MCE was studied with addition of Ca2+ or Na+ at ambient temperature (about 25 °C) at a concentration of 0.01 mg/mL. As shown in Fig. 3b, the PA of the MCE showed a clear decrease, and over half of the activity lost after 96 h. However, in the presence of Na+, the autolysis process was much slower, and about 80% of the PA remained after 96 h. Addition of Ca2+ increased resistance of the MCE to autolysis, and the PA was maintained without decrease during 96 h of storage. Previously, the MCE from B. licheniformis 5A5 was found to decrease to about half of its activity after 3 days of storage at room temperature (25–30 °C) [4]. The MCE from B. sphaericus NRC 24 retained about 82% of its activity in the first 10 days, but totally lost activity after 30 days of storage at room temperature [34]. The protective effect of Ca2+ against autolysis of protease during storage as observed in this study was not reported earlier.
Hydrolysis of caseins by the MCE
To understand the proteolytic action of the MCE on casein, hydrolysis of the casein components, namely α-, β-, and κ-casein, with different concentrations of the MCE was investigated, revealing different hydrolysis patterns among the three casein components (Fig. 4a–c). The hydrolysis degree of the casein components increased with the increase of the MCE up to 1 mg/mL. Obvious hydrolysis of κ-casein and β-casein with clear bands of the degraded products, but only slight degradation of α-casein was observed at low concentration (0.78125 × 10−2 mg/mL) of the MCE (lane 2 in Fig. 4a–c). Complete hydrolysis of the casein components with disappearance of the corresponding bands was observed at 2.5 × 10−1 mg/mL of the MCE for α-casein (lane 7, Fig. 4a), and 1.25 × 10−1 mg/mL for κ-casein (lane 6, Fig. 4c). Two distinct bands (10–15 kDa) of the hydrolysis products from β-casein, and one distinct band (about 13 kDa) from κ-casein appeared, indicating more specific hydrolysis toward these two casein components by the MCE. However, higher concentration of the MCE (0.5 mg/mL and more) could totally degrade caseins including their hydrolytic products, but not β-casein since faint bands from this component were still visible (lane 8, 9, Fig. 4b).
SDS-PAGE of the hydrolysates of α-casein (a), β-casein (b), and κ-casein (c) by the MCE from B. amyloliquefaciens GSBa-1 with increasing concentration (M: marker, 1–9: concentration of the MCE of 0, 0.78125 × 10−2, 1.5625 × 10−2, 3.125 × 10−2, 6.25 × 10−2, 1.25 × 10−1, 2.5 × 10−1, 5 × 10−1, 1 mg/mL); SDS-PAGE of the hydrolysates of α-casein (d), β-casein (e), and κ-casein (f) by the MCE with increasing hydrolysis time (M: marker, 1–8: 0,10, 30, 60, 90, 180, 270, and 360 min)
At the moderate concentration (3.125 × 10−2 mg/mL) of the MCE, the hydrolysis pattern of the casein components dependent on the hydrolysis time up to 360 min was further studied (Fig. 4d–f). As expected, more proteins were hydrolyzed with increasing hydrolysis time, and similar bands (10–15 kDa) which resulted from hydrolysis of β-casein and κ-casein as described above were also observed. After hydrolysis for 180 min, complete hydrolysis of κ-casein was observed as indicated by disappearance of the corresponding band (lane 6–8 in Fig. 4f), and most α- and β-casein were also hydrolyzed as shown by the residue faint bands (lane 6–8 in Fig. 4d, e), suggesting that α- and β-casein were more resistant than κ-casein to hydrolysis by the MCE. In addition, compared with α- and β-casein, κ-casein was more easily hydrolyzed in the first 10 min since the hydrolyzed product with a specific band at about 13 kDa was rapidly generated, and this band was still discernible till 360 min (Fig. 4f).
The results described above indicated that compared with α- and β-casein, κ-casein exhibited higher sensitivity to the MCE action, and the enzyme also demonstrated specificity on κ-casein to obtain a peptide product about 13 kDa. This was of significance in terms of possible mechanism of the MCE action through initial specific hydrolysis on κ-casein to destabilize casein micelles to induce milk coagulation. In cheese industry, the specific degradation of κ-casein by rennet was considered the main factor that affected milk clotting, and slower hydrolysis of α- and β-casein, as shown in this study, facilitated the formation of firm curds. Previously, other microbial rennet-like enzymes from Termitomyces clypeatus MTCC 5091 and Enterococcus faecalis TUA2495L were also shown to be preferentially active toward κ-casein, and the degradation of α- and β-casein proceeded slowly [24, 31], whereas a metalloproteinase with milk-clotting activity from Bacillus subtilis exhibited high specificity to the substrate β-casein [2]. The MCE separated from glutinous rice wine demonstrated different degradation degrees for α-CN, β-CN, and κ-CN [35].
Main cleavage site of κ-casein by the MCE
To determine the main cleavage site of κ-casein, the main peptide band (about 13 kDa in Fig. 4c) derived from hydrolysis of κ-casein by the MCE from B. amyloliquefaciens GSBa-1 was digested by trypsin, and the peptide fragments were analyzed by peptide mass fingerprinting using LC–MS–MS. The peptide fragments generated from in-gel trypsin treatment are listed in Table 4 according to the sequence of κ-casein. It obviously displayed that most peptide fragments ended with a lysine or arginine because of the trypsin digestion. As shown in Fig. 5, the main cleaved peptide from κ-casein started from the N-terminal end of the κ-casein to the 111th amino acid residue, and no peptide fragments from 112 Lys to the C-terminal were observed, proving the cleavage site of κ-casein at Lys 111–Lys 112. The molecular weight of this main peptide from κ-casein containing 111 amino acids was 12.922 kDa, which was consistent with the size (13 kDa) of the band observed in the SDS-PAGE analysis (Fig. 4c).
Rennet-induced milk coagulation generally involves initial destabilization of casein micelles by the enzymatic cleavage of certain peptide bonds on κ-casein, and the cleavage sites vary with the source of the enzyme. Cleavage of Phe105–Met106 of bovine κ-casein was involved in milk coagulation by several commercial MCEs such as bovine chymosin, and the microbial MCEs from Rhizomucor miehei and Rhizomucor pusillus [36, 37]. The metalloproteinase from Paenibacillus spp. BD3526 cleaved κ-casein at the Met106–Ala107 bond [9]. The milk-clotting protease extracted from glutinous rice wine had preferred cleavage site on bovine κ-casein at Thr94–Met95 [35]. The main cleavage site on κ-casein by other microbial MCEs from Cryphonectria parasitica and Endothia parasitica was found to be at Ser104–Phe105 [37, 38]. In this study, the Lys111–Lys112 peptide bond on bovine κ-casein was susceptible to be attacked by the MCE from B. amyloliquefaciens GSBa-1. This preferable cleavage site of peptide bonds containing lysine on κ-casein was not reported earlier for microbial rennet-like proteases, confirming that the MCE from B. amyloliquefaciens GSBa-1 was a new milk-clotting enzyme.
Conclusions
In the present study, a novel MCE from B. amyloliquefaciens GSBa-1 was purified and identified to belong to peptidase M4 (WP_032874795.1), and the mature peptide with MCA formed from this protein had molecular mass of about 38 kDa. The optimal pH and temperature of the MCE were 5.5 and 57 °C for MCA, and 7.0 and 57 °C for PA, respectively. The activity of the MCE was not significantly affected by various metallic salts, organic solvents, denaturants, and detergents tested, but heavy metal ions and SDS destroyed the activity. The MCE was confirmed to be a metalloprotease as indicated by the concentration-dependent inhibition with EDTA. The MCE showed certain degree of autolysis that could be inhibited by Ca2+ and Na+. The MCE could hydrolyze casein with κ-casein to be more susceptive to the MCE action, which was advantageous for application of the MCE in cheese making to effectively induce milk coagulation. The main cleavage site on κ-casein by the MCE was identified to be at Lys 111-Lys 112, which was not reported earlier. The MCE could be an effective choice of milk coagulant in cheese making.
References
Ding Z, Liu S, Gu Z, Zhang L, Zhang K, Shi G (2011) Production of milk-clotting enzyme by Bacillus subtilis B1 from wheat bran. Afr J Biotechnol 10:9370–9378
Li Y, Liang S, Zhi D, Chen P, Su F, Li H (2012) Purification and characterization of Bacillus subtilis milk-clotting enzyme from Tibet Plateau and its potential use in yak dairy industry. Eur Food Res Technol 234:733–741
Kumari NR, Bhushan B, Pal A, Panwar A, Malhotra S (2016) Purification, physico-chemico-kinetic characterization and thermal inactivation thermodynamics of milk clotting enzyme from Bacillus subtilis MTCC 10422. LWT Food Sci Technol 65:652–660
Ahmed SA, Helmy WA (2012) Comparative evaluation of Bacillus licheniformis 5A5 and Aloe variegata milk-clotting enzymes. Braz J Chem Eng 29:69–76
Ahmed SA, Wehaidy HR, Ibrahim OA, Abd El Ghani S, El-Hofi MA (2016) Novel milk-clotting enzyme from Bacillus stearothermophilus as a coagulant in UF-white soft cheese. Biocatal Agric Biotechnol 7:241–249
Vishwanatha KS, Appu Rao AG, Singh SA (2010) Production and characterization of a milk-clotting enzyme from Aspergillus oryzae MTCC 5341. Appl Microbiol Biotechnol 85:1849–1859
Wu F, Chang C, Shih I (2013) Optimization of the production and characterization of milk clotting enzymes by Bacillus subtilis natto. SpringerPlus 2:1–10
Zhang W, He X, Liu H, Guo H, Ren F, Gao W, Wen P (2013) Statistical optimization of medium components for milk-clotting enzyme production by Bacillus amyloliquefaciens D4 using wheat bran-an agro-industry waste. J Microbiol Biotechnol 23:1084–1091
Hang F, Wang Q, Hong Q, Liu P, Wu Z, Liu Z, Zhang H, Chen W (2016) Purification and characterization of a novel milk-clotting metalloproteinase from Paenibacillus spp. BD3526. Int J Biol Macromol 85:547–554
An Z, He X, Gao W, Zhao W, Zhang W (2014) Characteristics of miniature cheddar-type cheese made by microbial rennet from Bacillus amyloliquefaciens: a comparison with commercial calf Rennet. J Food Sci 79:214–221
Lemes AC, Pavón Y, Lazzaroni S, Rozycki S, Brandelli A, Kalil SJ (2016) A new milk-clotting enzyme produced by Bacillus sp. P45 applied in cream cheese development. LWT Food Sci Technol 66:217–224
Teng J, Liu Z, Su M, Gao H, Zheng Y, Li L, Zheng Z, Zhang J, Yang Z (2018) Study on the application of Bacillus amyloliquefaciens GSBa-1 rennet in Mozzarella Cheese. Food Sci (Chin) 22:85–92
Teng J, Zhao X, Yang Y, Zhang J, Zhao A, Jiang Y, Li L, Zheng Z, Yang Z (2017) Isolation and identification of microbial strains producing Rennet from Jiuqu, a traditional Chinese fermentation starter. Food Sci (Chin) 38:23–28
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Li Z, Scott K, Hemar Y, Otter D (2018) Protease activity of enzyme extracts from tamarillo fruit and their specific hydrolysis of bovine caseins. Food Res Int 109:380–386
Arima KYJ, Iwasaki S (1970) The acidic ptoteases. Methods Enzymol 30:446–459
Zhao X, Wang J, Zheng Z, Zhao A, Yang Z (2015) Production of a milk-clotting enzyme by glutinous rice fermentation and partial characterization of the enzyme. J Food Biochem 39:70–79
Meng F, Chen R, Zhu X, Lu Y, Nie T, Lu F, Lu Z (2018) Newly effective milk-clotting enzyme from Bacillus subtilis and its application in cheese making. J Agric Food Chem 66:6162–6169
Daroit DJ, Corrêa APF, Segalin J, Brandelli A (2010) Characterization of a keratinolytic protease produced by the feather-degrading Amazonian bacterium Bacillus sp. P45. Biocatal Biotransfor 28:370–379
He X, Ren F, Guo H, Zhang W, Song X, Gan B (2011) Purification and properties of a milk-clotting enzyme produced by Bacillus amyloliquefaciens D4. Korean J Chem Eng 28:203–208
Ageitos JM, Vallejo JA, Sestelo AB, Poza M, Villa TG (2007) Purification and characterization of a milk-clotting protease from Bacillus licheniformis strain USC13. J Appl Microbiol 103:2205–2213
Gastaldi E, Lagaude A, De La Fuente BT (1996) Micellar transition state in casein between pH 5.5 and 5.0. J Food Sci 61:59–64
Esawy MA, Combet-Blanc Y (2006) Immobilization of Bacillus licheniformis 5A1 milk-clotting enzyme and characterization of its enzyme properties. World J Microbiol Biotechnol 22:197–200
Sato S, Tokuda H, Koizumi T, Nakanishi K (2004) Purification and characterization of an extracellular proteinase having milk-clotting activity from Enterococcus faecalis TUA2495L. Food Sci Technol Res 10:44–50
Cavalcanti MTH, Teixeira MFS, Lima Filho JL, Porto ALF (2004) Partial purification of new milk-clotting enzyme produced by Nocardiopsis sp. Bioresour Technol 93:29–35
Martin P, Raymond MN, Bricas E, Ribadeau Dumas B (1980) Kinetic studies on the action of Mucor pusillus, Mucor miehei acid protease and chymosins A and B on a synthetic chromophoric hexapeptide. BBA-Biomembranes 612:410–420
Hashem AM (2000) Purification and properties of a milk-clotting enzyme produced by Penicillium oxalicum. Bioresour Technol 75:219–222
El-Tanboly ES, El-Hofi M, Youssef BY, El-Desoki W, Ismail A (2013) Utilization of salt whey from Egyptian Ras (Cephalotyre) cheese in microbial milk clotting enzymes production. Acta Sci Pol Technol Aliment 12:9–19
Sugihara A, Tani T, Tominaga Y (1991) Purification and characterization of a novel thermostable lipase from Bacillus sp. J Biochem 109:211–216
Naeem A, Fatima S, Hasan Khan R (2006) Characterization of partially folded intermediates of papain in presence of cationic, anionic, and nonionic detergents at low pH. Biopolymers 83:1–10
Majumder R, Banik SP, Khowala S (2015) Purification and characterisation of κ-casein specific milk-clotting metalloprotease from Termitomyces clypeatus MTCC 5091. Food Chem 173:441–448
Yadav RP, Patel AK, Jagannadham MV, Neriifolin S (2012) A dimeric serine protease from Euphorbia neriifolia Linn.: purification and biochemical characterization. Food Chem 132:1296–1304
Afsharnezhad M, Shahangian SS, Sariri R (2019) A novel milk-clotting cysteine protease from Ficus johannis: purification and characterization. Int J Biol Macromol 121:173–182
El-Bendary MA, Moharam ME, Ali TH (2009) Efficient immobilization of milk clotting enzyme produced by Bacillus sphaericus. Pol J Food Nutr Sci 59:67–72
Jiang T, Chen L, Xue L, Chen L (2007) Study on milk-clotting mechanism of rennet-like enzyme from glutinous rice wine: proteolytic property and the cleavage site on κ-casein. J Dairy Sci 90:3126–3133
García V, Rovira S, Teruel R, Boutoial K, Rodríguez J, Roa I, López MB (2012) Effect of vegetable coagulant, microbial coagulant and calf rennet on physicochemical, proteolysis, sensory and texture profiles of fresh goats cheese. Dairy Sci Technol 92:691–707
Drohse HB, Foltmann B (1989) Specificity of milk-clotting enzymes towards bovine kappa-casein. Biochim Biophys Acta 995:221–224
Jacob M, Jaros D, Rohm H (2011) Recent advances in milk clotting enzymes. Int J Dairy Technol 64:14–33
Acknowledgements
The National Key Research and Development Program of China (2018YFC1604302), National Natural Science Foundation of China (Grant no. 31871823), and Beijing Talent Cultivation Quality Construction—First-class Professional Construction (Municipal Level)—Food Science and Engineering (PXM2019_014213_000010) are kindly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, X., Cai, M., Yang, ZJ. et al. Purification and characterization of a novel milk-clotting enzyme produced by Bacillus amyloliquefaciens GSBa-1. Eur Food Res Technol 245, 2447–2457 (2019). https://doi.org/10.1007/s00217-019-03361-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-019-03361-6