Abstract
Cannflavins, flavonoids abundantly present in Cannabis sativa, possess a distinct chemical structure comprising a vanillyl group. Notably, the capsaicin structure also contains a vanillyl group, which is considered essential for interacting with the vanilloid receptor. The vanilloid receptor plays a crucial role in the perception of pain, heat, and inflammation and mediates the analgesic effects of capsaicin. Therefore, we postulated that prolonged exposure to cannflavin A (Can A) and cannflavin B (Can B) would provoke vanilloid receptor desensitization and hinder nocifensive responses to noxious thermal stimuli. C. elegans wild-type (N2) and mutants were exposed to Can A and Can B solutions for 60 min and then aliquoted on Petri dishes divided into quadrants for thermal stimulation. We then determined the thermal avoidance index for each C. elegans experimental group. Proteomics was performed to identify proteins and pathways associated with Can A or B treatment. Prolonged exposure to Can A and Can B hindered heat avoidance (32–35 °C) in C. elegans. No antinociceptive effect was observed 6 h post Can A or B exposure. Proteomics and Reactome pathway enrichment analyses identified hierarchical differences between Can A- and B-treated nematodes. However, both treatments were related to eukaryotic translation initiation (R-CEL-72613) and metabolic processes strongly associated with pain development. Our study aids in characterizing the pharmacological activity of cannflavins isolated from Cannabis sativa and outlines a possible application as pain therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
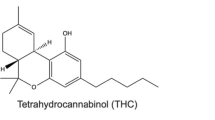
Chronic pain remains an evolving health concern worldwide (Mills et al. 2019; Cohen et al. 2021) and is considered a complex condition involving physical, mental, and emotional components (Finnerup 2019; Knotkova et al. 2021; Rosser et al. 2021). Thus, pain alleviation may require polymodal treatment strategies, including pharmacological, physical, and psychotherapies (Finnerup 2019). In recent years, partly owing to the ongoing opioid crisis, medicinal cannabis has gained momentum as a popular alternative for pain treatment, despite several controversies (Romero-Sandoval et al. 2018; Amin and Ali 2019; Ortiz et al. 2022; Procaccia et al. 2022). The findings of a study assessing brain magnetic resonance imaging (MRI) suggest that cannabis is not a painkiller but rather a pain distractor (Lee et al. 2013), which prompted several preclinical and clinical trials to explore the therapeutic benefits of cannabis for various applications (Pauli et al. 2020; Baratta et al. 2022; Procaccia et al. 2022). However, the therapeutic efficacy of cannabis in several health disorders remains unsubstantiated. Initially, various phytocannabinoids, more specifically Δ9-tetrahydrocannabinol and cannabidiol, extracted from Cannabis sativa, received considerable attention (Amin and Ali 2019). Using advanced analytical instruments, several components with high potency were extracted and characterized from C. sativa (Rea et al. 2019; Erridge et al. 2020; Bautista et al. 2021; Vita et al. 2022). Several non-cannabinoids, including prenylated flavones, have also been identified. Cannflavin A (Can A) and cannflavin B (Can B) are prenylated flavones isolated from C. sativa. Treatment with Can A and Can B was shown to inhibit prostaglandin E2 production in vitro (Barrett et al. 1986; Werz et al. 2014). Cannflavins exert antioxidant, anti-inflammatory, and neuroprotective effects (Radwan et al. 2008; Eggers et al. 2019; Bautista et al. 2021). However, we were particularly interested in Can A and Can B owing to the presence of a vanillyl functional group in their structure, as shown in Fig. 1. Vanilloids, including capsaicin (Cap), possess a vanillyl group essential for binding to the transient receptor potential vanilloid type 1 (TRPV1) receptor (Tominaga and Julius 2000; Jancsó et al. 2008; Yang et al. 2016; Privitera et al. 2017; Wortley et al. 2017; Martins et al. 2017; Moran 2017; Sultana et al. 2021).
Molecular structure of capsaicin, cannflavin A, and cannflavin B. Capsaicin is a transient receptor potential vanilloid type 1 (TRPV1) ligand. Ligand-receptor interactions are characteristically associated with the pharmacophore features, and the vanillyl group plays a fundamental role in interaction with the TRPV1
It is well-established that all animals display protective behaviors against noxious stimuli (e.g., mechanical, thermal, or chemical) to prevent damage (Mori 1999; Wittenburg and Baumeister 1999; Drew and Wood 2005). In mammals, primary afferent nociceptors sense potential tissue-damaging stimuli. The TRPV1 receptor is activated by proton (H+), endogenous and exogenous molecules, and noxious heat (Dickenson 1995; Caterina 2000; Tobin et al. 2002; Jancsó et al. 2008; Satheesh et al. 2016; Ohnishi et al. 2020). Moreover, TRPV1 is activated by vanilloids. Cap was the first vanilloid identified as a TRPV1 ligand. Importantly, prolonged Cap exposure can lead to analgesia (Tominaga and Julius 2000; Yoshimura and Yonehara 2001; Jordt and Julius 2002; Hui et al. 2003; Bölcskei et al. 2010; Anand and Bley 2011; Szolcsányi 2014; Abbas 2020). Accordingly, TRPV1 is an important target for the discovery and development of nonopioid pharmacotherapeutic agents aimed at pain management (Iftinca et al. 2021).
Caenorhabditis elegans is a model organism relevant to human biology and disease (Brenner 1974; Markaki and Tavernarakis 2020). Given its well-characterized nocifensive behaviors, C. elegans is a suitable model for studying nociception. Nematodes are known to avoid noxious stimuli by changing swimming direction. The C. elegans genome encodes several genes associated with vanilloid receptors, including 5 TRPV orthologs (OSM-9, OCR-1, OCR-2, OCR-3, and OCR-4) (Montell 2003; Kahn-Kirby and Bargmann 2006; Schafer 2006; Xiao and Xu 2011). C. elegans TRPV orthologs OSM-9 and OCR-2 are closely linked to sensory transduction, with similarities to TRPV1 (Liedtke et al. 2007; Ezak and Ferkey 2011; Glauser et al. 2011; Ohnishi et al. 2020). Thus, C. elegans OSM-9 and OCR-2 channels demonstrate comparable activation and induce physiological responses similar to mammalian vanilloid receptors. Recently, we reported that prolonged exposure to Cap and other vanilloids can hamper the nocifensive response of C. elegans to noxious heat (32–35 °C). The antinociceptive effect was reversed 6 h after Cap exposure (Nkambeu et al. 2020, 2021; Salem et al. 2022). Further exploration suggested that Cap and eugenol can target the C. elegans vanilloid receptor OCR-2 (Nkambeu et al. 2023). Collectively, these studies demonstrate that known vanilloid receptor ligands generate antinociceptive effects in C. elegans corresponding to effects previously observed in experimental models of pain (Guenette et al. 2006, 2007a, b; Beaudry et al. 2010; Lionnet et al. 2010; Ferland et al. 2012; Szolcsányi 2014; Giaccari et al. 2021; Sultana et al. 2021).
Herein, we hypothesized that Can A and Can B may interact with vanilloid receptors in C. elegans (e.g., OSM-9, OCR-2) and produce an effect comparable to that of Cap. We believe that prolonged exposure to Can A or Can B solutions would hinder the nocifensive response of C. elegans to noxious heat. The objectives of the present study were to 1) characterize the effects of Can A and Can B on C. elegans thermal avoidance behavior and 2) perform proteomics to identify proteins and uncover biological pathways associated with Can A or Can B treatment.
Materials and methods
Chemicals and reagents
Chemicals and reagents used in the current study were purchased from Fisher Scientific (Fair Lawn, NJ, USA) or Millipore Sigma (St. Louis, MO, USA). Cap, Can A, and Can B were purchased from Toronto Research Chemicals (North York, ON, Canada).
C. elegans strains
C. elegans N2 (Bristol) was used as the reference strain. The null mutants tested included npr-19 (strain RB1668), npr-32 (strain RB1938), ocr-2 (strain JY243), and osm-9 (strain JY190). Selected mutants are associated to vanilloid and cannabinoid receptors. All nematode strains were acquired from the Caenorhabditis Genetics Center (CGC), University of Minnesota (Minneapolis, MN, USA). Nematodes were maintained and manipulated using previously described protocols (Brenner 1974; Margie et al. 2013). The nematodes were grown and maintained on nematode growth medium (NGM) agar and maintained in a refrigerated incubator at 22 °C. Unless other conditions were indicated, all manipulations were performed at room temperature (~ 22 °C).
C. elegans pharmacological manipulations
Cap was accurately weighed and dissolved in water (e.g., ultrapure type 1) at a concentration of 25 µM. The solution was briefly heated, vortexed, and sonicated for several minutes to dissolve Cap completely. Can A was accurately weighed and dissolved in acetone at a concentration of 1 mg/mL (2.3 mM), and Can B was accurately weighed and dissolved in methanol at a concentration of 1 mg/mL (2.7 mM). Can A and Can B were serially diluted to achieve concentrations of 10, 5, 1, and 0.25 µM in water. Nematodes were collected and washed according to Margie et al. (2013). Three days after growing and feeding on NGM, nematodes were collected and exposed to Cap, Can A, or Can B solutions without food. Seven milliliters of Cap, Can A, or Can B solution was dispensed to generate a small 2–3 mm solution film (the solution was partly absorbed by NGM), ensuring that the nematodes were swimming in the solution. Nematodes were treated with Cap, Can A, or Can B for 60 min, extracted, and carefully washed prior to behavioral assessments. Remnant (persistent) effects were tested following exposure to Can A or Can B. Nematodes were extracted, thoroughly washed, and dispensed on NGM devoid of Can A or Can B for 6 h prior to further behavioral testing (i.e., 6 h latency).
Thermal avoidance assays
The thermal avoidance method used was developed based on the previously described four-quadrant strategy (Margie et al. 2013). We have extensively used this method in previously published studies (Nkambeu et al. 2018, 2020, 2021; Salem et al. 2022). The relevant technical details are presented in Supplementary Figure S1. Briefly, Petri dishes (92 × 16 mm) were divided into four quadrants. The nematodes were off food during the experiments. A middle circle (1 cm in diameter) delimited an area where nematodes were dispensed but were also not considered. Quadrants included two heat stimulus areas (A and D) and two control areas (B and C). Sodium azide (0.5 M) was aliquoted to all quadrants to paralyze nematodes. Noxious heat was produced using an electronically heated metal tip (0.8 mm in diameter). The metal tips were positioned 2 mm above the NGM and generated a radial temperature gradient, resulting in a temperature of 32–35 °C, measured using an infrared thermometer. The stimulus temperature used was based on a previous report (Wittenburg and Baumeister 1999). Nematodes were isolated and washed, as described by Margie et al.(2013). Nematodes (typically 100 to 300 individuals) were aliquoted in the center of the middle circle of the marked Petri dish. After stimulation for 30 min, the Petri dish was maintained at 4 °C for at least 1 h. Using a digital stereomicroscope, nematodes were counted per quadrant. All nematodes in the inner circle were excluded. We used Eq. 1 to calculate the thermal avoidance index.
We also calculated the percent animal avoidance to determine nocifensive responses to noxious heat.
Proteomic analysis
Nematodes (control, Can A- or Can B-treated) were extracted using a film of a liquid medium, centrifuged at 1,000 × g for 10 min, and carefully washed. We resuspended nematodes in a phosphate-buffered saline solution (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4) containing 1% (v/v) Triton X-100 and cOmplete™ protease inhibitor cocktail (Roche Diagnostic Canada, Laval, QC, Canada). Next, the suspensions were transferred to 1.5 mL homogenizer tubes containing 25 mg of 500-μm glass beads. Homogenization was achieved using a Bead Mill Homogenizer (Fisher Scientific) and included five bursts of 60 s each set at 5 m/s. Then, samples were centrifugated at 12,000 × g for 10 min. Protein concentrations were determined using the Bradford assay for each resulting sample. In total, 200 µg of protein was aliquoted, and ice-cold acetone precipitation (1:5, v/v) was used to isolate proteins. The protein pellets were dissolved in 100 µL of 50 mM TRIS–HCl buffer (pH 8.0). Protein pellets were dissolved by vigorous vortexing (2,800 rpm) and sonication. To improve reaction and digestion heat, we performed heat denaturing at 120 °C for 10 min using a heated reaction block. The protein solution was allowed to cool for 15 min. Protein reduction was performed with 20 mM dithiothreitol (DTT) at 90 °C for 15 min. Subsequently, cysteine alkylation was performed with 40 mM iodoacetamide protected from light for 30 min. Five micrograms of trypsin was aliquoted in each sample and incubated at 37 °C for 24 h. The enzymatic reaction was stopped by adding 10 µL (~ 10% v/v) of a 1% trifluoroacetic acid (TFA) solution. The samples were centrifuged at 12,000 × g for 10 min. Next, 100 µL of the supernatant was transferred to high-performance liquid chromatography (HPLC) vials for mass spectrometry (MS) analysis.
A label-free precursor ion-based MS1 quantification workflow was used. A nano-flow Thermo Scientific Vanquish Neo UHPLC system (San Jose, CA, USA) was set up for pre-concentration mode using a Thermo Scientific PepMap Neo 5 µm C18 300 µm × 5 mm trap cartridge in back-flush configuration. Rapid sample loading (20 μL/min flow rate) was performed on a trap cartridge. Tryptic peptides were separated using a mobile phase comprising 0.1% formic acid in water (A) and 0.1% formic acid in an 80% acetonitrile and 20% water mixture (B), using a 60-min gradient from 5 to 50% B (flow rate of 300 nL/min). A Thermo Scientific PepMap Neo C18 2 µm × 75 µm × 150 mm nano column was connected to a Thermo Nanospray Flex ion source hyphenated with a Thermo Scientific Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer. The Nanospray emitter was set to 2200 V (positive mode), and the ion transfer tube temperature was set at 50 °C. The mass spectrometer operated in the TOP-10 data-dependent acquisition mode. The acquisition method on the mass spectrometer comprised a high-resolution MS1 survey scan (m/z 375–1200) acquired at 70,000 resolution (FWHM), followed by up to 10 MS2 (acquisition of most intense precursor ions with an intensity threshold 1 × 104). Selected precursors were isolated with a 2.0 Da isolation width, activated by HCD (28 NCE), and fragments were detected using ORBITRAP at a resolution of 17,500 (FWHM). Data were processed using Thermo Proteome Discoverer (version 3.0) with false discovery rate (FDR) analysis performed using Percolator. Proteome Discoverer was used to survey MS1 and MS2 spectra against the C. elegans proteome extracted from UniProt (taxon identifier 6239) using SEQUEST-HT engine at 10 ppm of precursor mass tolerance and 0.02 Da of fragment mass tolerance. Search parameters were as follows: trypsin digestion with up to 2 missed cleavages, carbamidomethylation of cysteine as a static modification, and oxidation of methionine as a variable modification. The tryptic peptide length was set to a minimum of six residues. Post-processing of SEQUEST-HT results was performed using Percolator. The resulting protein hits were filtered at 1% FDR. The protein abundance was determined using the average precursor ion intensity of only unique peptides, with a p-value ≤ 0.05 deemed significant. Proteome Discoverer 3.0 was used to calculate p-values of protein abundance using ANOVA, followed by Tukey's HSD post-hoc test. All proteins with a p-value of > 0.05 and fold-change < 2 were excluded from enrichment analyses.
Bioinformatics
Volcano plots were created based on log2 ratios and p-values. Venn diagram analysis was performed to display the logical relationship between treatments. Enrichment analyses using Gene Ontology (GO) cellular components and Reactome databases were completed using Metascape (Zhou et al. 2019). Only differentially expressed proteins (DEPs) were used for enrichment analyses (i.e., absolute log2 ratio ≥ 1.0; p-value ≤ 0.05). Parent to root node analysis was performed using ClueGO (Bindea et al. 2009), an integrated Cytoscape application (Shannon et al. 2003), using the Reactome pathway databases.
Statistical analysis
Nematode phenotyping data were analyzed using the non-parametric Kruskal–Wallis test and Dunn’s post-hoc test. Statistical significance was set to p ≤ 0.05. GraphPad Prism version 9.5.1 (GraphPad Software, Inc., La Jolla, CA, USA) was used to perform statistical analyses and generate associated figures.
Results and discussion
Recent studies have demonstrated that C. elegans can be used to evaluate the antinociceptive effects of bioactive compounds, and proteomics can yield comprehensive datasets for studying biological processes and pathways (Nkambeu et al. 2020, 2021, 2023; Salem et al. 2022). Supplementary Figure S1 presents the thermal avoidance assay. Initially, we confirmed the presence of any bias in C. elegans behavior using our experimental system. As depicted in Fig. 2, we observed no bias in quadrant selection for all experimental groups tested. In the absence of a heat stimulus, nematodes were uniformly distributed after 30 min. Moreover, exposure to Can A or Can B for 60 min did not change nematode mobility or induce any behavioral preference.
Comparison of the mobility and bias of WT (N2) and selected mutant nematodes in plates divided into quadrants maintained at a constant temperature (22 °C) without the application of a stimulus (negative control). The selected Caenorhabditis elegans genotypes show no quadrant selection in the absence or presence of Can A or Can B. Can A, cannflavin A; Can B, cannflavin B; WT, wild-type
As discussed previously, sustained exposure to TRPV1 agonists can induce receptor desensitization, resulting in analgesic effects (or antinociceptive effects in C. elegans) (Pinho-Ribeiro et al. 2017); we confirmed this observation in C. elegans in our recent reports (Nkambeu et al. 2020, 2021; Salem et al. 2022). We postulate that Can A and Can B interact with C. elegans vanilloid receptors owing to the presence of a vanillyl moiety, as shown in Fig. 1. TRPV1 is a popular drug target for managing chronic and neuropathic pain (Iftinca et al. 2021). Can A and Can B are flavonoids, a family of molecules used for their antioxidant, analgesic, and anti-inflammatory properties (Barrett et al. 1986; Eggers et al. 2019; Erridge et al. 2020; Bautista et al. 2021). It has been suggested that flavonoid mediate their effects by suppressing oxidative stress, glial cell activation, and mitochondrial dysfunction (Matias et al. 2016). As shown in Figs. 3 and 4, 1-h exposure to Can A and Can B induced remarkable antinociceptive effects. Notably, the Can A- and Can B-induced antinociceptive effect was comparable with that of Cap, the best-known vanilloid receptor ligand. Although inconclusive, our data suggest that the observed effect was concentration-dependent. Following Can A or Can B exposure, the nematodes were carefully washed and transferred onto NGM devoid of Can A or Can B, maintained in an incubator at 22 °C for 6 h (i.e., residual effect/latency test), and reexamined for the thermal avoidance response. Six hours after exposure, the thermal avoidance response of previously treated and washed C. elegans was comparable with that of untreated nematodes at all concentrations tested for both compounds (Figs. 3 and 4). Thus, Can A or Can B had no remnant effects. These results suggest that the duration of the effect could be relatively short after administration.
Assessment of the pharmacological effects of cannflavin A (Can A) on thermal avoidance in Caenorhabditis elegans. Nematodes were exposed to Can A or capsaicin (Cap) for 60 min prior to behavior experimentation. Individual values and medians are displayed and derived from at least 12 independent experiments for each experimental group. Can A exhibits dose-dependent effects and markedly impedes thermal avoidance in C. elegans. **** p < 0.0001 (non-parametric Kruskal–Wallis test—Dunn's multiple comparisons test). No residual antinociceptive effects can be observed 6 h after exposure
Assessment of the pharmacological effects of cannflavin B (Can B) on thermal avoidance in Caenorhabditis elegans. Nematodes were exposed to Can A or capsaicin (Cap) for 60 min prior to behavior experimentation. Individual values and medians are displayed and derived from at least 12 independent experiments for each experimental group. Can B exhibits dose-dependent effects and markedly impedes thermal avoidance in C. elegans. **** p < 0.0001 (non-parametric Kruskal–Wallis test—Dunn's multiple comparisons test). No residual antinociceptive effects can be observed 6 h after exposure
Furthermore, we used specific mutants to identify potential vanilloid and cannabinoid receptor targets and comprehensively clarify the exposure–response relationship. To identify target receptors of Can A and Can B, experiments were performed using specific C. elegans mutants (ocr-2, osm-9, npr-19, and npr-32). As shown in Fig. 5, all tested mutants exhibited lower sensitivity to noxious heat than wild-type (WT; N2) nematodes; however, the difference in sensitivity was not statistically significant. To identify targets, we exposed mutants to 10 µM of Can A or Can B for 60 min prior to the heat avoidance experiments. As shown in Fig. 5, except for the ocr-2 mutant exposed to Can A (Fig. 5A), antinociceptive effects mediated by Can A and Can B were quantifiable in all examined mutants. The efficacy of Can A was hampered in the ocr-2 mutant, although it remains unclear whether Can A targets OCR-2 to induce the observed effects. However, the results obtained following mutant exposure to Can A or Can B indicate redundancy in receptor involvement and possible compensation. Single mutants remained sensitive to heat but were less sensitive than the WT (i.e., not statistically significant for all testes mutants), suggesting redundant receptor function. Based on the collected data, we concluded that the double mutant (e.g., ocr-2/osm-9) would be unresponsive to noxious heat. Notably, we previously reported on the application of certain vanilloids, including eugenol, and the results were inconclusive (Nkambeu et al. 2021). Vanilloids are known to target C. elegans TRPV channels OCR-2 and OSM-9, with considerable data supporting OCR-2 as the target (Nkambeu et al. 2020, 2023). Can A and Can B contain a vanillyl functional group, known to be responsible for interacting with vanilloid receptors, that may bind to vanilloid receptors in C. elegans to mediate their analgesic effects.
Identification of vanilloid receptor orthologs responsible for the Can A and Can B-induced antinociceptive effects. Individual values and medians are displayed and derived from at least 12 independent experiments for each experimental group. Mutants tested included ocr-2 (A), osm-9 (B), npr-19 (C) andnpr-32 (D). Can A antinociceptive effects may be mediated via vanilloid receptor OCR-2. ** p < 0.01, * p < 0.05 (nonparametric Kruskal-Wallis test - Dunn's multiple comparisons test)
As shown in Figs. 3 and 4, we noted significant antinociceptive effects after Can A or Can B exposure for 1 h. Both molecules impeded the nocifensive response of C. elegans to noxious heat. However, the pathways and biological processes underlying Can A or Can B activity remain to be elucidated. Consequently, we used MS-based proteomics and network biology to decipher the relationship between these pathways and drug responses. C. elegans exposed to Can A (10 µM) and Can B (10 µM) for 1 h were subjected to label-free proteomic analyses. Figure 6 shows volcano plots to illustrate the differential abundance of proteins, with the x-axis representing the log2 ratio and the y-axis plotting log10 (p-value). Boxes represent a twofold change with a p-value of ≤ 0.05. Numerous DEPs were identified; 113 upregulated and 154 downregulated proteins were identified following Can A exposure (Fig. 6A), and 134 upregulated and 147 downregulated proteins identified following Can B exposure (Fig. 6B). Tables S1 and S2 (supplementary file) summarize the fold changes and p-values for all identified DEPs. Proteins with a single high-scoring peptide hit were not excluded, as previously discussed (Gupta and Pevzner 2009). Examining the lists of DEPs using Venn diagrams (Fig. 6C), we found that DEPs were specific to each experimental group, with a significant degree of overlap between C. elegans exposed to Can A or Can B.
Visualization of proteomic results using volcano plots. Volcano plots illustrate the differential abundances of proteins, with the x-axis showing the log2 ratio with respect to the control group (N2 maintained at 22 °C) and the y-axis representing − 1 × log10 (p-value). The boxes represent log2-fold-change and above with an adjusted p-value ≤ 0.05. The p-values were determined by a one-way ANOVA with post-hoc Tukey HSD. A. Caenorhabditis elegans exposed to 10 µM Can A for 1 h were compared with nematodes maintained at 22 °C. B. C. elegans exposed to 10 µM Can B for 1 h were compared to nematodes maintained at 22 °C. C. Venn diagram representing the degree of DEP overlap between the two experimental conditions under investigation. DEP, differentially expressed protein
Enrichment analysis of the Reactome pathways and GO Cellular Component databases revealed a high degree of enrichment for eukaryotic translation initiation (Reactome identifier: R-CEL-72613) for both treatments (Fig. 7A). Eukaryotic translation initiation factor 4E (eIF4E) is crucial for nociceptive plasticity and plays a major role in the development of chronic pain in animals, including humans (Uttam et al. 2018). Notably, eIF4E is an important effector of the ERK and mTORC1 signaling pathways involved in the development of pain. Accordingly, eIF4E-dependent mechanisms should be explored as new targets for developing novel drugs to alleviate pain. Notably, we have recently shown that treatment with Cap substantially enriched child (or sublevel) pathways connected to the pathway hierarchy of eukaryotic translation initiation (Reactome identifier R-CEL-72613) (Nkambeu et al. 2023). Hierarchical clustering revealed several common pathways following Can A and Can B treatment, with a large degree of enrichment in energy metabolism-associated Reactome pathways (e.g., citric acid [TCA] cycle and respiratory electron transport, citric acid cycle [TCA cycle], formation of ATP by chemiosmotic coupling). These Reactome pathways contain complex networks of chemical reactions essential for energy metabolism and lead to simple precursors that function as building blocks for anabolic reactions. The maintenance and regulation of anabolism are vital for enhancing the healing process. Notably, metabolic disruption can substantially impact neuronal plasticity (Formolo et al. 2022). Developing drugs that induce healthy neuroplastic changes represents a potentially attractive chronic pain treatment strategy (Sibille et al. 2016). The enrichment of GO cellular components provides information complementary to the Reactome pathways by identifying cellular compartments and structures. As shown in Fig. 7B, mitochondrial and child terms were significantly enriched following treatment with both Can A and Can B. These findings were highly compatible with the enrichment of energy metabolism associated with previously identified Reactome pathways. As shown in Fig. 7B, there was a high degree of cytosolic ribosome enrichment. Ribosomes are the core molecules essential for catalyzing RNA translation into proteins. Translational control is critical in chronic pain (Khoutorsky and Price 2018).
Functional characterization using Metascape combined with the Reactome (A) and Gene Ontology cellular component (B) databases. Analysis was performed with all DEPs (∣FC∣ ≥ 2 and p-value ≤ 0.05). Within the heatmap, the cell color densities were derived from -log10 (p-value). The rows are clustered based on the degree of overlap of the proteins within each set that led to the enrichment of each pathway. DEPs, differentially expressed proteins
Further analyses were performed using Can A- and B-specific DEPs. As shown in Fig. 6C, 114 and 128 specific DEPs were identified following treatment with Can A and Can B, respectively. For each treatment group, hierarchical cluster analysis was performed for specific DEPs and revealed certain distinct features. Treatment with Can A demonstrated enrichment of immune and adaptive immune systems (Fig. 8A and B). Cytokines and chemokines are key effectors in the development of chronic pain (Zhou et al. 2023). Immune cells are important modulators of neuronal activity. Furthermore, interferon signaling includes important effectors associated with nociception and acute and chronic pain (Tan et al. 2021). Therefore, we suggest that interferon signaling could be a potential new drug target for pain management. Other enriched pathways following Can A treatment were associated with catabolism and anabolism. Following Can B treatment, Reactome pathway enrichment using specific DEPs revealed the metabolism link process (Fig. 8A and C). Maintenance of anabolism and regulation of catabolism are the two hallmarks of chronic pain healing.
Functional characterization using Metascape combined with the Reactome. Analysis was performed with DEPs (∣FC∣ ≥ 2 and p-value ≤ 0.05) with no overlapping between the two experimental conditions under investigation. A) heatmap comparing experiment conditions. Parent to root node analysis of enriched Reactome terms derived from only specific DEPs observed following Caenorhabditis elegans exposition to Can A (B) and Can B (C). Node analyses were performed using ClueGO and CluePedia. Can A, cannflavin A; Can B, cannflavin B; DEPs, differentially expressed proteins
Conclusion
To the best of our knowledge, the present study is the first to demonstrate that controlled and prolonged exposure to Can A and Can B can induce antinociceptive effects. We established that these molecules successfully impede the nocifensive responses of C. elegans to noxious heat. Although we did not identify specific C. elegans vanilloid targets, functional redundancy between OCR-2 and OSM-9 was anticipated. Notably, we documented some hierarchical differences between Can A- and B-exposed nematodes. However, both groups were associated with eukaryotic translation initiation (R-CEL-72613) and metabolic processes, which are strongly associated with pain development and may be potential drug targets. Our findings improve the understanding of the pharmacological activity of cannflavins isolated from C. sativa. We postulate that cannflavins from C. sativa have remarkable antinociceptive effects and should be evaluated in animal models of pain.
Data availability
Data supporting the findings of this study are available from the corresponding author upon request.
References
Abbas MA (2020) Modulation of TRPV1 channel function by natural products in the treatment of pain. Chem-biol Interact 330:109178. https://doi.org/10.1016/j.cbi.2020.109178
Amin MR, Ali DW (2019) Pharmacology of medical cannabis. Adv Exp Med Biol 1162:151–165. https://doi.org/10.1007/978-3-030-21737-2_8
Anand P, Bley K (2011) Topical capsaicin for pain management: therapeutic potential and mechanisms of action of the new high-concentration capsaicin 8% patch. Br J Anaesth 107:490–502. https://doi.org/10.1093/bja/aer260
Baratta F, Pignata I, Enri LR, Brusa P (2022) Cannabis for medical use: analysis of recent clinical trials in view of current legislation. Front Pharmacol 13:888903. https://doi.org/10.3389/fphar.2022.888903
Barrett ML, Scutt AM, Evans FJ (1986) Cannflavin A and B, prenylated flavones from Cannabis sativa L. Experientia 42:452–453. https://doi.org/10.1007/bf02118655
Bautista JL, Yu S, Tian L (2021) Flavonoids in Cannabis sativa: biosynthesis, bioactivities, and biotechnology. ACS Omega 6:5119–5123. https://doi.org/10.1021/acsomega.1c00318
Beaudry F, Ross A, Lema PP, Vachon P (2010) Pharmacokinetics of vanillin and its effects on mechanical hypersensitivity in a rat model of neuropathic pain. Phytother Res 24:525–530. https://doi.org/10.1002/ptr.2975
Bindea G, Mlecnik B, Hackl H et al (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093. https://doi.org/10.1093/bioinformatics/btp101
Bölcskei K, Tékus V, Dézsi L et al (2010) Antinociceptive desensitizing actions of TRPV1 receptor agonists capsaicin, resiniferatoxin and N-oleoyldopamine as measured by determination of the noxious heat and cold thresholds in the rat. Eur J Pain (London, England) 14:480–486. https://doi.org/10.1016/j.ejpain.2009.08.005
Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77:71–94. https://doi.org/10.1093/genetics/77.1.71
Caterina MJ (2000) Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science 288:306–313. https://doi.org/10.1126/science.288.5464.306
Cohen SP, Vase L, Hooten WM (2021) Chronic pain: an update on burden, best practices, and new advances. Lancet 397:2082–2097. https://doi.org/10.1016/s0140-6736(21)00393-7
Dickenson A (1995) Spinal cord pharmacology of pain. Br J Anaesth 75(2):193–200. https://doi.org/10.1093/bja/75.2.193
Drew LJ, Wood JN (2005) Worm sensation! Mol Pain 1:8. https://doi.org/10.1186/1744-8069-1-8
Eggers C, Fujitani M, Kato R, Smid S (2019) Novel cannabis flavonoid, cannflavin A displays both a hormetic and neuroprotective profile against amyloid β-mediated neurotoxicity in PC12 cells: Comparison with geranylated flavonoids, mimulone and diplacone. Biochem Pharmacol 169:113609. https://doi.org/10.1016/j.bcp.2019.08.011
Erridge S, Mangal N, Salazar O et al (2020) Cannflavins - From plant to patient: A scoping review. Fitoterapia 146:104712. https://doi.org/10.1016/j.fitote.2020.104712
Ezak MJ, Ferkey DM (2011) A functional nuclear localization sequence in the C. elegans TRPV channel OCR-2. Plos One 6:e25047. https://doi.org/10.1371/journal.pone.0025047
Ferland CE, Beaudry F, Vachon P (2012) Antinociceptive effects of eugenol evaluated in a monoiodoacetate-induced osteoarthritis rat model. Phytother Res 26:1278–1285. https://doi.org/10.1002/ptr.3725
Finnerup NB (2019) Nonnarcotic methods of pain management. New Engl J Med 380:2440–2448. https://doi.org/10.1056/nejmra1807061
Formolo DA, Cheng T, Yu J et al (2022) Central adiponectin signaling – A metabolic regulator in support of brain plasticity. Adv Neurol 8:79–96. https://doi.org/10.3233/bpl-220138
Giaccari LG, Aurilio C, Coppolino F et al (2021) Capsaicin 8% patch and chronic postsurgical neuropathic pain. J Pers Medicine 11:960. https://doi.org/10.3390/jpm11100960
Glauser DA, Chen WC, Agin R et al (2011) Heat avoidance is regulated by transient receptor potential (TRP) channels and a neuropeptide signaling pathway in Caenorhabditis elegans. Genetics 188:91–103. https://doi.org/10.1534/genetics.111.127100
Guenette SA, Beaudry F, Marier JF, Vachon P (2006) Pharmacokinetics and anesthetic activity of eugenol in male Sprague-Dawley rats. J Vet Pharmacol Ther 29:265–270. https://doi.org/10.1111/j.1365-2885.2006.00740.x
Guénette SA, Hélie P, Beaudry F, Vachon P (2007a) Eugenol for anesthesia of African clawed frogs (Xenopus laevis). Vet Anaesth Analg 34:164–170. https://doi.org/10.1111/j.1467-2995.2006.00316.x
Guénette SA, Ross A, Marier J-F et al (2007b) Pharmacokinetics of eugenol and its effects on thermal hypersensitivity in rats. Eur J Pharmacol 562:60–67. https://doi.org/10.1016/j.ejphar.2007.01.044
Gupta N, Pevzner PA (2009) False discovery rates of protein identifications: A strike against the two-peptide rule. J Proteome Res 8:4173–4181. https://doi.org/10.1021/pr9004794
Hui K, Liu B, Qin F (2003) Capsaicin activation of the pain receptor, VR1: multiple open states from both partial and full finding. Biophys J 84:2957–2968. https://doi.org/10.1016/s0006-3495(03)70022-8
Iftinca M, Defaye M, Altier C (2021) TRPV1-targeted drugs in development for human pain conditions. Drugs 81:7–27. https://doi.org/10.1007/s40265-020-01429-2
Jancsó G, Dux M, Oszlács O, Sántha P (2008) Activation of the transient receptor potential vanilloid-1 (TRPV1) channel opens the gate for pain relief. Brit J Pharmacol 155:1139–1141. https://doi.org/10.1038/bjp.2008.375
Jordt S, Julius D (2002) Molecular basis for species-specific sensitivity to “hot” chili peppers. Cell 108:421–430
Kahn-Kirby AH, Bargmann CI (2006) TRP channels in C. elegans. Annu Rev Physiol 68:719–736. https://doi.org/10.1146/annurev.physiol.68.040204.100715
Khoutorsky A, Price TJ (2018) Translational control mechanisms in persistent pain. Trends Neurosci 41:100–114. https://doi.org/10.1016/j.tins.2017.11.006
Knotkova H, Hamani C, Sivanesan E et al (2021) Neuromodulation for chronic pain. Lancet 397:2111–2124. https://doi.org/10.1016/s0140-6736(21)00794-7
Lee MC, Ploner M, Wiech K et al (2013) Amygdala activity contributes to the dissociative effect of cannabis on pain perception. Pain 154:124–134. https://doi.org/10.1016/j.pain.2012.09.017
Liedtke WB, Heller S, Sze JY (2007) The TRPV Channel in C. elegans Serotonergic Neurons. CRC Press
Lionnet L, Beaudry F, Vachon P (2010) Intrathecal eugenol administration alleviates neuropathic pain in male Sprague-Dawley rats. Phytother Res 24:1645–1653. https://doi.org/10.1002/ptr.3174
Margie O, Palmer C, Chin-Sang I (2013) C. elegans</em> Chemotaxis Assay. Journal of visualized experiments : JoVE 1–6. https://doi.org/10.3791/50069
Markaki M, Tavernarakis N (2020) Caenorhabditis elegans as a model system for human diseases. Current Opin Biotechnol 63:118–125. https://doi.org/10.1016/j.copbio.2019.12.011
Martins D, Silva M, Tavares I (2017) TRPV1 in pain control from the brain. Oncotarget 8:16101–16102. https://doi.org/10.18632/oncotarget.13316
Matias I, Buosi AS, Gomes FCA (2016) Functions of flavonoids in the central nervous system: astrocytes as targets for natural compounds. Neurochem Int 95:85–91. https://doi.org/10.1016/j.neuint.2016.01.009
Mills SEE, Nicolson KP, Smith BH (2019) Chronic pain: a review of its epidemiology and associated factors in population-based studies. Brit J Anaesth 123:e273–e283. https://doi.org/10.1016/j.bja.2019.03.023
Montell C (2003) The venerable inveterate invertebrate TRP channels. Cell Calcium 33:409–417
Moran MM (2017) TRP channels as potential drug targets. Annu Rev Pharmacol Toxicol 58:309–330. https://doi.org/10.1146/annurev-pharmtox-010617-052832
Mori I (1999) Genetics of chemotaxis and thermotaxis in the nematode Caenorhabditis elegans. Ann Rev Genet 33:399–422. https://doi.org/10.1146/annurev.genet.33.1.399
Nkambeu B, Salem JB, Beaudry F (2021) Eugenol and other vanilloids hamper Caenorhabditis elegans response to noxious heat. Neurochem Res 46:252–264. https://doi.org/10.1007/s11064-020-03159-z
Nkambeu B, Salem JB, Beaudry F (2020) Capsaicin and its analogues impede nocifensive response of Caenorhabditis elegans to noxious heat. Neurochem Res 45:1851–1859. https://doi.org/10.1007/s11064-020-03049-4
Nkambeu B, Salem JB, Beaudry F (2023) Antinociceptive activity of vanilloids in Caenorhabditis elegans is mediated by the desensitization of the TRPV channel OCR-2 and specific signal transduction pathways. Neurochem Res 48:1900–1911. https://doi.org/10.1007/s11064-023-03876-1
Nkambeu B, Salem JB, Leonelli S et al (2018) EGL-3 and EGL-21 are required to trigger nocifensive response of Caenorhabditis elegans to noxious heat. Neuropeptides 73:41–48. https://doi.org/10.1016/j.npep.2018.11.002
Ohnishi K, Saito S, Miura T et al (2020) OSM-9 and OCR-2 TRPV channels are accessorial warm receptors in Caenorhabditis elegans temperature acclimatisation. Sci Rep 10:18566. https://doi.org/10.1038/s41598-020-75302-3
Ortiz YT, McMahon LR, Wilkerson JL (2022) Medicinal cannabis and central nervous system disorders. Front Pharmacol 13:881810. https://doi.org/10.3389/fphar.2022.881810
Pauli CS, Conroy M, Heuvel BDV, Park S-H (2020) Cannabidiol drugs clinical trial outcomes and adverse effects. Front Pharmacol 11:63. https://doi.org/10.3389/fphar.2020.00063
Pinho-Ribeiro FA, Verri WA, Chiu IM (2017) Nociceptor sensory neuron–immune interactions in pain and inflammation. Trends Immunol 38:5–19. https://doi.org/10.1016/j.it.2016.10.001
Privitera R, Birch R, Sinisi M et al (2017) Capsaicin 8% patch treatment for amputation stump and phantom limb pain: a clinical and functional MRI study. J Pain Res 10:1623–1634. https://doi.org/10.2147/jpr.s140925
Procaccia S, Lewitus GM, Feder CL et al (2022) Cannabis for medical use: versatile plant rather than a single drug. Front Pharmacol 13:894960. https://doi.org/10.3389/fphar.2022.894960
Radwan MM, ElSohly MA, Slade D et al (2008) Non-cannabinoid constituents from a high potency Cannabis sativa variety. Phytochemistry 69:2627–2633. https://doi.org/10.1016/j.phytochem.2008.07.010
Rea KA, Casaretto JA, Al-Abdul-Wahid MS et al (2019) Biosynthesis of cannflavins A and B from Cannabis sativa L. Phytochemistry 164:162–171. https://doi.org/10.1016/j.phytochem.2019.05.009
Romero-Sandoval EA, Fincham JE, Kolano AL et al (2018) Cannabis for chronic pain: Challenges and considerations. Pharmacother J Hum Pharmacol Drug Ther 38:651–662. https://doi.org/10.1002/phar.2115
Rosser BA, Fisher E, Eccleston C et al (2021) Psychological therapies delivered remotely for the management of chronic pain (excluding headache) in adults. Cochrane Database Syst Rev 8(8):CD007407. https://doi.org/10.1002/14651858.cd0138635
Salem JB, Nkambeu B, Arvanitis DN, Beaudry F (2022) Resiniferatoxin hampers the nocifensive response of Caenorhabditis elegans to noxious heat, and pathway analysis revealed that the Wnt signaling pathway is involved. Neurochem Res 47:622–633. https://doi.org/10.1007/s11064-021-03471-2
Satheesh NJ, Uehara Y, Fedotova J et al (2016) TRPV currents and their role in the nociception and neuroplasticity. Neuropeptides. https://doi.org/10.1016/j.npep.2016.01.003
Schafer WR (2006) Proprioception: a channel for body sense in the worm. Curr Biol 16:R509–R511. https://doi.org/10.1016/j.cub.2006.06.012
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Sibille KT, Bartsch F, Reddy D et al (2016) Increasing neuroplasticity to bolster chronic pain treatment: a role for intermittent fasting and glucose administration? J Pain 17:275–281. https://doi.org/10.1016/j.jpain.2015.11.002
Sultana A, Singla RK, He X et al (2021) Topical capsaicin for the treatment of neuropathic pain. Curr Drug Metab 22:198–207. https://doi.org/10.2174/1389200221999201116143701
Szolcsányi J (2014) Capsaicin as a therapeutic molecule. Prog Drug Res Fortschritte Der Arzneimittelforschung Progrès Des Recherches Pharm 68:1–37. https://doi.org/10.1007/978-3-0348-0828-6_1
Tan P-H, Ji J, Yeh C-C, Ji R-R (2021) Interferons in pain and infections: emerging roles in neuro-immune and neuro-glial interactions. Front Immunol 12:783725. https://doi.org/10.3389/fimmu.2021.783725
Tobin D, Madsen D, Kahn-Kirby A et al (2002) Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron 35:307–318
Tominaga M, Julius D (2000) Capsaicin receptor in the pain pathway. Jpn J Pharmacol 83:20–24. https://doi.org/10.1254/jjp.83.20
Uttam S, Wong C, Price TJ, Khoutorsky A (2018) eIF4E-dependent translational control: a central mechanism for regulation of pain plasticity. Front Genet 9:470. https://doi.org/10.3389/fgene.2018.00470
Vita SD, Finamore C, Chini MG et al (2022) Phytochemical analysis of the methanolic extract and essential oil from leaves of industrial hemp Futura 75 cultivar: isolation of a new cannabinoid derivative and biological profile using computational approaches. Plants 11:1671. https://doi.org/10.3390/plants11131671
Werz O, Seegers J, Schaible AM et al (2014) Cannflavins from hemp sprouts, a novel cannabinoid-free hemp food product, target microsomal prostaglandin E2 synthase-1 and 5-lipoxygenase. PharmaNutrition 2:53–60. https://doi.org/10.1016/j.phanu.2014.05.001
Wittenburg N, Baumeister R (1999) Thermal avoidance in Caenorhabditis elegans: an approach to the study of nociception. Proc Natl Acad Sci U S A 96:10477–10482
Wortley MA, Birrell MA, Belvisi MG (2017) Drugs affecting TRP channels. Handb Exp Pharmacol 237:213–241. https://doi.org/10.1007/164_2016_63
Xiao R, Xu XZS (2011) C. elegans TRP channels. Adv Exp Med Biol 704:323–339. https://doi.org/10.1007/978-94-007-0265-3_18
Yang BH, Piao ZG, Kim Y-B et al (2016) Activation of vanilloid receptor 1 (VR1) by eugenol. J Dent Res 82:781–785. https://doi.org/10.1177/154405910308201004
Yoshimura M, Yonehara N (2001) Influence of capsaicin cream in rats with peripheral neuropathy. Pharmacol Res 44:105–111. https://doi.org/10.1006/phrs.2001.0830S1043-6618(01)90830-8[pii]
Zhou WBS, Shi XQ, Liu Y et al (2023) Unbiased proteomic analysis detects painful systemic inflammatory profile in the serum of nerve-injured mice. Pain 164:e77–e90. https://doi.org/10.1097/j.pain.0000000000002695
Zhou Y, Zhou B, Pache L et al (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10:1523. https://doi.org/10.1038/s41467-019-09234-6
Funding
M. Lahaise received a National Sciences and Engineering Research Council of Canada Undergraduate Student Research Award. This project was funded by the National Sciences and Engineering Research Council of Canada (F. Beaudry Discovery Grant No. RGPIN-2020–05228). The laboratory equipment was funded by the Canadian Foundation for Innovation (CFI), the Fonds de Recherche du Québec (FRQ), and the Government of Quebec (F. Beaudry CFI John R. Evans Leaders grant nos. 36706 and 42043). F. Beaudry holds the Canada Research Chair in Metrology of Bioactive Molecules and Target Discovery (grant no. CRC-2021–00160). This research was partially supported by funding from the Canada Research Chairs Program.
Author information
Authors and Affiliations
Contributions
M. Lahaise, F. Boujenoui, and F. Beaudry conceived and designed this study. M. Lahaise and F. Boujenoui conducted the experiments. M. Lahaise, F. Boujenoui, and F. Beaudry conducted data analysis and wrote the manuscript. All the authors have read and approved the final manuscript. All authors declare that all data were generated in-house and that no paper mill was used.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lahaise, M., Boujenoui, F. & Beaudry, F. Cannflavins isolated from Cannabis sativa impede Caenorhabditis elegans response to noxious heat. Naunyn-Schmiedeberg's Arch Pharmacol 397, 535–548 (2024). https://doi.org/10.1007/s00210-023-02621-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00210-023-02621-3