Abstract
Biodegradation is an eco-friendly measure to address plastic pollution. This study screened four bacterial isolates that were capable of degrading recalcitrant polymers, i.e., low-density polyethylene, polyethylene terephthalate, and polystyrene. The unique bacterial isolates were obtained from plastic polluted environment. Dermacoccus sp. MR5 (accession no. OP592184) and Corynebacterium sp. MR10 (accession no. OP536169) from Malaysian mangroves and Bacillus sp. BS5 (accession no. OP536168) and Priestia sp. TL1 (accession no. OP536170) from a sanitary landfill. The four isolates showed a gradual increase in the microbial count and the production of laccase and esterase enzymes after 4 weeks of incubation with the polymers (independent experiment set). Bacillus sp. BS5 produced the highest laccase 15.35 ± 0.19 U/mL and showed the highest weight loss i.e., 4.84 ± 0.6% for PS. Fourier transform infrared spectroscopy analysis confirmed the formation of carbonyl and hydroxyl groups as a result of oxidation reactions by enzymes. Liquid chromatography–mass spectrometry analysis showed the oxidation of the polymers to small molecules (alcohol, ethers, and acids) assimilated by the microbes during the degradation. Field emission scanning electron microscopy showed bacterial colonization, biofilm formation, and surface erosion on the polymer surface. The result provided significant insight into enzyme activities and the potential of isolates to target more than one type of polymer for degradation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The rapid increase in global plastic production from 1.5 million tons in the 1950s to 335 million tons in 2016 has generated enormous plastic waste (Lebreton et al. 2018). The continuous accumulation of plastic waste due to anthropogenic activities has deeply disturbed land and ocean and has contributed to increasing global warming and carbon footprint. Some of the most commonly and widely used plastic products are made of LDPE (low-density polyethylene), PVC (polyvinylchloride), PP (polypropylene), PET (polyethylene terephthalate), and PS (polystyrene) (Bond et al. 2018). Almost 79% of the plastic waste generated undergoes partial treatment before being released into the natural environment (Geyer et al. 2017). The synthetic plastic that degrades into macro- (> 5 mm), micro- (~ 1–5 μm), and nano-plastic (< 1 μm) particles move up in the food chain and create environmental and ecological risks (Teuten et al. 2009). Therefore, it is very important to take serious action to combat the challenge of plastic pollution and to overcome its impact. Complete recycling of plastic is still unachievable as only 9% to 12% of the total global plastic is recycled, while the rest of the plastic waste generated is either incinerated or dumped on land, ultimately settling down in nearby water bodies or oceans (Jadaun et al. 2022). Plastic waste accumulates in both terrestrial and aquatic ecosystems, posing a significant threat to animals. Ingestion of plastic materials by these animals leads to severe and often lethal effects on their bodies (Quero and Luna 2017; Wang et al. 2022). Plastic also alters the physical properties of the soil, plant root traits, and nutrient uptake (Rillig et al. 2019). Although the effect of plastic waste or microplastic on humans is still unexplored, exposure to microplastic via inhalation can lead to particle toxicity (Wright and Kelly 2017; Sun et al. 2021).
Out of many synthetic plastics released as waste, LDPE, PET, and PS are the most common types of plastic waste stream and constitute an approximate proportion of 20, 11, and 6%, respectively (Fotopoulou and Karapanagioti 2017). The natural degradation of plastic waste by hydrolysis, biodegradation, and photodegradation has attracted much attention in recent years (Cai et al. 2018). However, properties in the plastic like shape, size, hydrophobic nature, high molecular weight, crystallinity, and additives prolong the degradation process, if not negligible (Li et al. 2021). Inherently, microorganisms are identified as the decomposers of natural polymers by converting them into value-added products (Jadaun et al. 2022). Subsequently, microbes are also explored for the biodegradation of synthetic polymers (Danso et al. 2019; Jadaun et al. 2022; Zhang et al. 2022) such as LDPE by Acinetobacter pitti IRN19 (26.8% degradation efficiency at 30 °C for 4 weeks, Montazer et al. 2018), Enterobacter cloacae AKS7 (9% degradation efficiency when the polymer is processed at 30 °C for 45 days, Sarker et al. 2020), Halomonas sp. (1.72% degradation efficiency at 30 °C for 90 days, Khandare et al. 2021), Brevundimonas sp. MGS1 (4.64% degradation efficiency at 37 °C for 30 days), Arthrobacter sp. SW4 (2.62% degradation efficiency at 37 °C for 30 days), and Arthrobacter sp. PD2 (2.21% degradation efficiency at 37 °C for 30 days) (Singh et al. 2023). Similarly, degradation of PET film was observed using engineered Clostridium thermocellum (62% degradation efficiency at 60 °C for 14 days, Yan et al. 2021), and degradation of PS film was observed by Rhodococcus ruber C208 (0.8% degradation efficiency in 56 days, Mor and Sivan 2008), Exiguobacterium sp. YT2 (7.5% degradation efficiency in 60 days, Yang et al. 2015). The biodegradation of synthetic plastic can be explained as changes in the polymer structure and properties (such as reduction in weight and mechanical strength) under the transformative actions of microbial enzymes (Danso et al. 2019). During polymer degradation, microbes first adhere to the surface, thereby colonizing the surface. Microbes further secrete extracellular enzymes, that bind to the polymer surface and cause hydrolytic cleavage. The polymer is cleaved to dimers and monomers and mineralized to carbon dioxide and water, which are used by microbes as energy sources (Tokiwa et al. 2009). Compared to the traditional chemical recycling processes, biocatalytic recycling is eco-friendlier as it offers a definitive solution for clearing plastic waste, leading to a circular economy. Although the biodegradation of plastic waste is well-defined, low degradation efficiency, long degradation time, and lack of hydrolytic and functional groups in LDPE and PS make it more challenging for biodegradation. Considering the ubiquity of plastic waste in different ecosystems and the tremendous genetic diversity of microorganisms, it is important to identify microbes that can target a wide variety of plastics in the waste in a given biome. This will help to screen the target-specific approach for the degradation and will also help to identify the most efficient enzyme involved during the degradation of a wide variety of plastic waste. To achieve this, the present study was designed to explore microorganisms from one of the most plastic-contaminated sites i.e., sanitary landfill (sludge) and mangrove forests (sediment). Sanitary landfill sites are preferred to discover new microorganisms with plastic degrading ability due to their pre-exposure and natural ability to degrade structures similar to polymers (Song et al. 2015). Landfill ecosystems are rich in aromatic and halogenated hydrocarbons like benzene and tetrachloroethylene; microbes degrade and utilize these hydrocarbons for their growth (Meyer-Dombard et al. 2020). Similarly, mangrove forests offer one of the extreme environmental conditions, such as large quantities of carbon, high salinity, pH, and anoxic soil. The roots of mangrove forests often capture the plastic debris (a mixture of various types of plastics) that are washed off the shore and from waste generated at the terrestrial sites (Deng et al. 2021). The plastic waste disintegrates into microplastic and is colonized by the microorganisms that consequently degrade the plastic waste. Hence, mangrove sites also serve as the ideal niche for discovering unique microorganisms for the degradation of a broad variety of plastic. Although biodegradation of plastic is one of the extensively studied topics, most of the research papers are limited to structural changes, surface topography, and viability of microorganisms during degradation. Hence, questions such as the identification of microbial isolates which can have a broad host range, detection of some common enzymes that play a crucial role during degradation, and by-product analysis released during degradation can help to understand biodegradation in-depth and ensure the process of complete degradation.
Therefore, the present study was designed to identify plastic-degrading microorganisms that can degrade common polymers that constitute the maximum share in waste generation i.e., LDPE, PET, and PS. Sludge and sediment samples collected from Berjaya Sanitary Landfill, Bukit Tagar, Selangor, Malaysia, and Malaysian mangroves were screened to isolate bacteria that can degrade the three different polymers in the absence of carbon for 4 weeks. The degradation of LDPE, PET, and PS was analyzed by determining the viability of cells, changes in the functional group of the polymers, surface erosion, weight loss of the polymers, enzyme production (laccase and esterase activity), and end-product analysis to confirm the biodegradation. The outcome of the study will highlight enzymatic activities and oxidation and hydrolysis reactions involved in the degradation of the recalcitrant polymers LDPE, PET, and PS. The end product analysis will confirm the formation of small molecules released during the degradation of polymers which can help to understand the biodegradation mechanism in a better way.
Materials and methods
Polymer materials
Polyethylene (low density, pellet, CAS number 428043), polystyrene (pellet, CAS number 331651) and polyethylene terephthalate (granules, CAS number 429252) and culture media (Maximum Recovery Diluent, Reasoner’s 2A agar, and Bushnell Haas Broth) were purchased from Sigma-Aldrich, USA. Solvents and chemicals utilized throughout the experiments were of analytical grade (excluding methanol, and HPLC grade). For the degradation study, the polymers were surface sterilized in 70% ethanol in a glass petri dish and dried at 55 °C in an oven dryer (Memmert, Malaysia). The pre-treatment of the three polymers was conducted with ultraviolet (UV) radiation (30 W, with a UV intensity of 125 µW/cm2) for 1 h in the Biosafety Cabinet, Microbiology Lab, School of Science, Monash University, Malaysia.
Soil and leachate sample collection
Bacterial isolates were isolated from the leachate samples collected from a sanitary landfill in Selangor (3o30′10′′ N 101o28′24′′E) and soil samples were collected from the mangrove forest of Carey Island, Selangor (2°49′28′′N, 101°20′25′′E), a state on the west coast of Peninsular Malaysia. Both mangrove and landfill projects have the following myABS permit application; Ref: 131933 and Ref: 604107, respectively. Both the samples were placed in sterile plastic bags and transported to the laboratory for the isolation of pure microbial cells.
Isolation and screening of bacteria for biodegradation assay
Leachate and soil samples (1 mL) were diluted with 9 mL of maximum recovery diluent (isotonic diluent containing peptone 1.0 g/L and sodium chloride 8.5 g/L) at room temperature (Montazer et al. 2018). The serially diluted diluents were plated on R2A (Reasoner’s 2A agar) and incubated at 37 °C overnight to obtain pure individual colonies. Pure colonies were assessed for morphological characterization and Gram stain analysis (Binnerup et al. 1998). Pure cultures were suspended in 50% (v/v) glycerol/nutrient broth (Himedia, India, Catalogue number MM244) and stored at − 80 °C for long-term experimental studies. The microbial cells were further screened for lipolytic enzyme activity qualitatively (following the standard protocol of Jaiganesh and Jaganathan 2018). Isolates that exhibited an orange halo zone indicated the hydrolysis reaction and were consequently selected for the biodegradation assay. All the experiments were carried out in triplicate (n = 3).
Molecular identification of the bacterial isolates
The four pure culture isolates (two obtained from landfill and two obtained from mangrove soil) were obtained by screening of the esterase enzyme. All the isolates were grown in nutrient broth overnight at 37 °C. The bacterial cells were centrifuged at 6000 rpm for 2 min to pellet down the cells, and the supernatant was discarded. The bacterial pellet was used for DNA isolation. Genomic DNA isolation was performed using the VIVANTIS GF-1 nucleic acid isolation kit following the manufacturer’s protocol. DNA quality and concentration were determined using the BioDrop UV/VIS spectrophotometer by checking the absorbance ratio at 260/280 nm. High-quality genomic DNA was subjected to PCR amplification of the 16S rRNA gene. The gene was amplified using the 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′TACGGYTACCTTGTTACGACTT-3′) universal primers. The PCR conditions used were (i) 1 cycle of initial denaturation at 94 °C for 2 min; (ii) 34 cycles at 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 30 s for the denaturation, annealing and extension steps, respectively; and (iii) 1 cycle at 72 °C for 7 min for the final extension of the amplified DNA. The PCR products were then purified using the QIAquick PCR purification kit (Qiagen) and submitted for Sanger sequencing with the help of a commercial service provider (Apical Scientific Sdn. Bhd., Selangor, Malaysia). The DNA sequences obtained were analyzed using the software Molecular Evolutionary Genetic Analysis (MEGA) version 7.0 (Kim et al. 2012). The sequences were trimmed for ambiguous nucleotides and assembled at the overlapping regions. The 16S rRNA gene sequences were then analyzed using the NCBI BLAST tool and EzBioCloud server (http://www.ezbiocloud.net/eztaxon) to identify the closest neighbors (Yoon et al. 2017; Larkin et al. 2007). The 16S rRNA gene sequences were aligned with the closest taxa using ClustalW (Larkin et al. 2007). Using the aligned 16S rRNA gene sequences, phylogenetic trees were built by the neighbor-joining (NJ) algorithm (Saitou and Nei 1987). The robustness of phylogenetic inference from the tree topology was estimated by 1000 bootstrap replicates. The 16S rRNA gene sequences of the bacterial isolates were submitted to GenBank (http://www.ncbi.nlm.nih.gov), and the accession numbers are listed in Table 1.
Biodegradation of LDPE, PET, and PS
The four bacterial isolates were cultivated in nutrient broth (HiMedia Labs) (overnight at 37 °C with agitation at 120 rpm) until the cultures attained log phase (absorbance of 1.00 at 600 nm). The microbial counts of the inoculum were recorded as colony-forming units (CFU/mL) (Kim et al. 2023). The initial microbial count of the isolates was recorded as 3.8 × 106 CFU/mL for MR5 and 4.8 × 106 CFU/mL for MR10, 5.1 × 106 CFU/mL for BS5, 4.5 × 106 CFU/mL for TL1, at the beginning of the biodegradation experiment. The biodegradation experiment setup contained Bushnell Hass (BH) medium and UV-pretreated polymers (LDPE, PET, and PS) (55 ± 0.3 mg), and the experiment setup contained (i) polymers with bacterial isolates, (ii) only bacterial isolates, and (iii) only polymers. All the flasks were kept in the incubator shaker (Lab Companion, USA) at 37 °C at 90 rpm for 4 weeks. The degradation assay was carried out in triplicates (n = 3). The BHB media was screened for the growth of bacterial isolates (by determining the OD value at 600 nm, microbial viable count, and cell dry weight) in 7 day intervals for 4 weeks.
Determination of bacterial growth and dry cell weight
The growth of the four bacterial isolates (MR5, MR10, BS5, and TL1) was measured by determining the absorbance (Optical density) at 600 nm and microbial viable count (CFU/mL) after 7 days intervals for 4 weeks using a UV spectrophotometer (Tecan Spark 10 M, Austria) (Banerjee et al. 1993). Baseline correction of the instrument was done using the absorbance of the media without bacterial culture. In addition to bacterial growth, one mL of BHB broth was withdrawn from each experiment setup and centrifuged (12000 rpm, 5 min at 25 °C) to collect the cell pellets. The pellets were dried at 45 °C (oven dryer, Memmert, Malaysia) and were weighed to estimate the dry weight of the bacterial cells (Ren et al. 2019).
Determination of weight loss of LDPE, PET and, PS
Weight loss of the LDPE, PET and PS were determined after 4 weeks of incubation. The polymers were suspended in the sodium dodecyl sulfate (SDS) (2%) for 30 min followed by washing with sterile distilled water for 20 min to remove the excess bacterial cells from the surface. The granules were dried overnight (50 °C in oven, Memmert, Malaysia) to ensure complete drying before measuring the weight loss (Harshvardhan and Jha 2013). The weight loss of the polymers was calculated using the following formula:
Quantitative analysis of laccase and esterase during biodegradation
Quantitative estimation of laccase enzyme activity was performed in 7 day intervals for 4 weeks of incubation from each experiment set (Khandare et al. 2021). The BHB broth was centrifuged (6000 rpm for 5 min at room temperature) to obtain cell-free supernatant, which was further used as an enzyme against the respective substrate to quantify enzyme activities. The reaction mixture contained 2 mM guaiacol (1 mL), 10 mM sodium acetate buffer (pH 4.6, 3 mL) and cell-free supernatant (1 mL). The reaction mixture was incubated at 30 °C for 15 min (Papinutti and Martínez 2006). Cell-free supernatant from the control flask served as a blank to analyze the non-enzymatic degradation of the substrate. The absorbance of the reaction mixture was recorded at 450 nm. One unit of the laccase activity was defined as 1 µmol guaiacol transformed into product per minute at 30 °C.
Quantitative estimation of esterase activity was determined quantitatively following protocol by Kay et al. (1993). The reaction mixture contained 0.8 mL phosphate buffer (0.1 M, pH 7.0), 134 µL of substrate PNP (p-nitrophenyl butyrate), and 0.066 mL of cell-free supernatant and incubated at 30 °C for 30 min. The reaction mixture was observed for the color change, and absorbance was measured using a spectrophotometer at 410 nm. Cell-free supernatant from the control flask served as a blank to analyze the non-enzymatic degradation of the substrate. One unit of esterase activity was defined as the amount of enzyme required to release 1 µM of PNP per minute at 30 °C.
Characterization of the degraded polymer by FTIR spectroscopy
LDPE, PET, and PS recovered from each treatment after 4 weeks of treatment were washed with sterile water and disinfected with ethanol (70%) to remove excess bacterial cells from the surface. The granules were dried overnight (50 °C in oven, Memmert, Malaysia), and analyzed using attenuated total reflection Fourier-transform infrared spectroscopy (Perkin Elmer FT-IR C106361). The changes in the functional groups were identified based on the peaks detected in the spectra. The absorbance was determined in the mid-IR region of 400–4000 cm−1 wavenumber (modified from Albertsson and Karlson 1990).
Field emission scanning electron microscopy analysis of degraded polymer
Polymers (LDPE, PET, and PS) were recovered after 4 weeks of incubation from each treatment to observe the bacterial colonization and changes in the structural morphology using Field emission scanning electron microscopy (FESEM). Each polymer sample was washed with 2% SDS (Sodium dodecyl sulfate) solution followed by rinsing in distilled water for 30 min to remove excess bacterial cells from the surface and dried (50 °C overnight, Lab dryer, Model FDD-1000D, Malaysia). Dried polymers were gold-coated (Quorum, Q150R Rotary Pump Coater, UK) to increase the surface conductivity, and images were obtained from the FESEM microscope (Hitachi SU8010, USA).
Detection of compounds by liquid chromatography–mass spectrometry analysis
Liquid chromatography–mass spectrometry (LC-MS) analysis was performed using cell-free supernatant after 4 weeks of degradation assay. BHB broth (30 mL) was collected from each experiment set and centrifuged at 12,000 rpm for 10 min at room temperature. The supernatant was collected and freeze-dried. Freeze-dried samples were dissolved in methanol (1 mg/mL) and sonicated for 10 min. The resultant supernatant (1 µL) was used for LC–MS (Agilent 1290 Infinity LC system coupled to Agilent 6520 Accurate-Mass Q-TOF mass spectrometer with dual ESI source, School of Medicine, Monash University Malaysia). The column used was Agilent Eclipse XDB-C18 Narrow-bore, 150 mm × 2.1 mm, 3.5 micron (P/N 930990–902), at 25 °C, the flow rate was 0.5 mL/min and the run time was 30 min. Data analysis was carried out using Agilent Mass Hunter Qualitative Analysis B.07.00.
Statistical analysis
Data were analyzed statistically using the Statistical Package for Social Sciences (SPSS) software. The data were subjected to a one-way analysis of variance (ANOVA) for main effects followed by the Tukey post hoc test at p value < 0.05.
Results and discussion
Biodegradation of LDPE, PET, and PS
In the present study, soil and sediment samples from the two most plastic-polluted sites (mangrove forest and sanitary landfill) were screened for the degradation study. In the past 2 decades (2000–2021), 290 research articles describing plastic degradation have been published (Akinpelu and Nchu 2022), and most of them have presented partial degradation or structural damage of the plastic. It is, therefore, very important to identify and screen common microorganisms that can target more than one type of plastic to accelerate the degradation capacity. The present study showed the identification of novel isolates from plastic-polluted sites that had a broad host range and were able to target one of the most common pollutants in the environment (LDPE, PET, and PS, together constitute 37% of plastic waste generated, Fotopoulou and Karapanagioti 2017). In the present study, the three polymers LDPE, PET, and PS were pre-treated by using UV radiations. The process of pre-treatment using UV rays is a well-studied and environmentally friendly technique. In nature, biodegradation of plastic waste is only carried out by microorganisms after the physiochemical deterioration (Gewert et al. 2015). It was observed that the UV treatment reduced the hydrophobicity of the polymers and enhanced the enzyme activity for bacterial surface colonization as seen in FESEM images. By-product analysis (LC-MS) showed the formation of short-chain compounds (such as ether, ester, and alcohol). It is reported that UV or heat pre-treatment can generate oxygen free radicals within the polymeric surface, that further participate in the polymer degradation pathway to form short-chain compounds (olefins and ketones). These short-chain compounds are easily attacked by exoenzymes secreted by microorganisms for further degradation (Gewert et al. 2015; Yamada-Onodera et al. 2001). One of the earlier studies has shown the successful implication of UV rays (longer incubation time, shorter distance, and a dose of 7.02 × 1012 μW cm−2 s) resulted in higher roughness, hydrophilicity, biofilm formation, and surface degradation of PS and LDPE after 45 days of incubation (Taghavi et al. 2021). Alternatively, Montazer et al. (2018) showed that the pre-treatment with UV radiation (artificial and natural) modified the LDPE surface and accelerated the biodegradation.
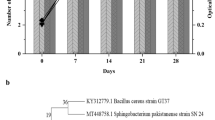
Identification of the bacterial isolates was performed using partial 16S rRNA gene sequencing, phylogenetic analysis, and sequence similarity is presented with the top match from the EzBioCloud database (Fig. 1). The isolates (MR5, MR10, BS5, and TL1) were screened qualitatively for lipolytic activity (Fig. S1) and further selected for biodegradation analysis of LDPE, PET, and PS. The 16S rRNA gene sequencing of the isolates obtained from mangrove sediment i.e., MR5 showed 99.72% sequence similarity with Dermacoccus nishinomiyaensis strain DSM 20448(T). Dermacoccus sp. MR5 is an aerobic Gram-positive coccus bacterium and it belongs to the Dermacoccaceae family that has been studied in the degradation of polyethylene (Bolo et al. 2015) but the mechanism and the enzymes involved in the degradation were not explored. Also, Dermacoccus sp. MR5 has not been explored in the degradation of any other polymers. Similarly, MR10 showed 100% 16S rRNA gene sequence similarity with Corynebacterium sanguinis CCUG 58655(T). Corynebacterium sp. MR10 is an aerobic Gram-positive short rod-shaped bacterium, and it belongs to the Corynebacteriaceae family. The common habitats for the bacterial family are soil, water, and plants, and the members have been earlier explored in the degradation of hydrocarbons (Zhang et al. 2016), but have not been studied in plastic degradation. Alternatively, 16S rRNA gene sequencing of the isolates obtained from the sanitary landfill i.e., BS5 showed 99.86% sequence similarity with Bacillus siamensis KCTC 13613 (T) and has been explored in the degradation of LDPE (Maroof et al. 2021), however the enzymatic mechanism of action was unexplored. Similarly, TL1 showed 99.71% 16S rRNA gene sequence similarity with Priestia megaterium NBRC 15308 (T). Priestia sp. TL1 is a Gram-positive rod-shaped bacterium that belongs to the family Bacillaceae. Priestia sp. TL1 has been explored in the bioremediation of salinized soil and metal removal (Biedendieck et al. 2021). The relationship between isolates and the type of members of their respective genera is presented in the phylogenetic tree (Fig. 1). All four isolates served as novel candidates to be studied in the degradation of LDPE, PET, and PS.
LDPE, PET, and PS were screened for biodegradation by analyzing bacterial growth (absorbance and microbial count) (Fig. 2 and Fig. 3), cell dry weight (Fig. 4), and quantification of laccase and esterase enzymes (Fig. 5 and Fig. 6). Residual polymers were collected after 4 weeks of incubation in the BHB media to analyze weight loss (Fig. 7), surface damage (Fig. 8), and structural changes (Table 2). The identification of small molecular weight compounds released during degradation assay is presented in Table 3. During the LDPE degradation, after 1 week of incubation, the four bacterial isolates Dermacoccus sp. MR5, Corynebacterium sp. MR10, Bacillus sp. BS5 and Priestia sp. TL1 showed growth which was recorded as increased microbial count and cell dry weight (Figs. 3 and 4). Out of the four isolates, BS5 was observed to be the most efficient isolate for the degradation of all three polymers and this was followed by TL1. The maximum absorbance (OD 600 nm) for LDPE with BS5 was recorded to be 0.327 ± 0.029, with a microbial viable count of 8.01 × 107 CFU/mL and dry cell biomass as 0.066 ± 0.001 mg/mL. This was followed by TL1, where maximum absorbance was recorded to be 0.166 ± 0.008, microbial viable count of 6.21 × 107 CFU/mL, and dry cell mass of 0.0572 ± 0.008 mg/mL. Further BS5 produced the highest amount of laccase (12.33 ± 0.7 U/mL) and esterase (0.13 ± 0.003 U/mL), followed by TL1 (laccase produced, 7.06 ± 0.4 U/mL and esterase, 0.09 ± 0.001 U/mL) after 4 weeks of degradation (Figs. 5 and 6). The isolate BS5 showed a weight loss of 3.83 ± 0.8% followed by TL1 (2.21 ± 0.2%) (Fig. 7). Similarly, bacterial isolates MR10 showed increased growth (0.165 ± 0.0039, microbial viable count of 6.12 × 107 CFU/mL, dry cell biomass of 0.044 ± 0.008 mg/mL) for 4 weeks. The isolate also produced laccase (6.78 ± 0.7 U/mL) and esterase (0.09 ± 0.003 U/mL) and showed a weight loss of LDPE (2.1 ± 0.1%). The growth for the three isolates was recorded to be increasing after 4 weeks of incubation. Alternatively, MR5 showed a decline in the growth after 21 days of incubation. The isolate produced laccase (4.22 ± 0.2 U/mL) and esterase (0.011 ± 0.0003 U/mL) after 3 weeks of incubation, which was recorded to be highest in the 4 weeks and showed a weight loss of 1.91 ± 0.2%. The growth, production of enzymes (laccase and esterase), and weight loss of LDPE showed the efficiency of all the isolates in utilizing LDPE as the sole carbon source during the degradation assay. LDPE has a C–C backbone which makes them recalcitrant to biodegradation. UV-pretreatment of LDPE initiated auto-oxidation that facilitated surface colonization and biofilm formation by bacterial isolates (Dey et al. 2020). The auto-oxidation resulted in the formation of low molecular weight fragments such as alcohols, aldehydes, and ketones via enzymatic activities. Laccase belongs to the oxidase group of enzymes that performs depolymerization of polymer chains by oxidative cleavage of the amorphous region. The oxidative cleavage hence provides an accessible carbonyl region within the polymer chain (here LDPE) for degradation (Kang et al. 2019). In the present study, laccase mediated the oxidation of hydro-carbon backbone of the polyethylene which initiated degradation (produces water as a by-product) (Nunes and Kunamneni 2018) as evidenced by the weight loss recorded. The most significant role of laccase was reported during the depolymerization of polyethylene via oxidation of the polymer backbone by Santo et al. (2013). The study also reported the efficiency to degrade polyethylene immediately within 2 days of incubation. Some of the genera that have been reported for LDPE degradation include Bacillus (10.7% weight loss in 28 days), Brevibacillus, Listeria, Lysinibacillus (20% weight loss in 120 days), Micrococcus, Moraxella, Proteus, Pseudomonas (20% weight loss in 120 days), Rhodococcus (2.5% weight loss in 30 days mediated by laccase enzyme), Serratia (70% weight loss in 36 days), Staphylococcus, Streptococcus, Streptomyces, and Vibrio (Singh and Ting 2022; Restrepo-Flórez et al. 2014; Arutchelvi et al. 2008). Although a lot of microorganisms have been explored for the degradation of LDPE, most of them have only been highlighted with activities that altered the structural changes. Isolates identified in the present study showed promising LDPE degradation efficiency by involving plastic degrading enzymes and forming small molecules obtained in LC-MS.
Quantitative analysis of laccase enzyme activities (U/mL) at 7 day intervals for 4 weeks during (a) LDPE; b PET, and c PS degradations with MR5, MR10, BS5, and TL1. Error bars indicate the standard error (n = 3). Values followed by the different letters are significantly different within sampling days
Quantitative analysis of esterase enzyme activities (U/mL) at 7 day intervals for 4 weeks during (a) LDPE; b PET, and c PS degradations with MR5, MR10, BS5, and TL1. Error bars indicate the standard error (n = 3). Values followed by the different letters are significantly different within sampling days
FESEM micrographs showing the surface appearance of (a) control LDPE; b control PET; c control PS; d MR5-treated LDPE; e MR5-treated PET; f MR5-treated PS; g MR10-treated LDPE; h MR10-treated PET; i MR10-treated PS; j BS5-treated LDPE; k BS5-treated PET; l BS5-treated PS; m TL1-treated LDPE; n TL1-treated PET and o TL1-treated PS. Yellow arrows show the surface erosion and biofilm formation by bacterial isolates
Similarly, during the biodegradation of PET, at the end of 4 weeks, BS5 showed the highest absorbance (0.332 ± 0.008) (Fig. 2), microbial viable count (7.85 × 107 CFU/mL) (Fig. 3), and cell dry biomass (0.056 ± 0.007 mg/mL) (Fig. 4). This was followed by TL1 (absorbance 0.22 ± 0.02, microbial viable count of 5.89 × 107 and cell dry biomass of 0.045 ± 0.009) and MR10 (absorbance 0.22 ± 0.04, microbial viable count of 5.89 × 107 CFU/mL and cell dry biomass of 0.037 ± 0.001 mg/mL). The three isolates BS5, TL1, and MR10 showed a gradual increase in the growth and biomass. In contrast, MR5 showed the highest growth in the 3 week (absorbance 0.14 ± 0.023, microbial viable count of 3.58 × 107 CFU/mL, and cell dry biomass of 0.025 ± 0.0003 mg/mL) and the growth declined at the end of the fourth week compared to the three isolates. All the isolates produced laccase and esterase enzyme which subsequently increased for 4 weeks, where BS5 produced the highest laccase (17.11 ± 0.5 U/mL) (Fig. 5) and esterase (1.1 ± 0.02 U/mL) (Fig. 6) and showed a weight loss of (4.44 ± 1.12%) (Fig. 7). This was followed by TL1 (11.69 ± 0.1 U/mL of laccase, 0.45 ± 0.02 U/mL of esterase production and 4.03 ± 0.5% of weight loss) and MR10 (8.16 ± 0.1 U/mL of laccase, 0.13 ± 0.003 U/mL of esterase production and weight loss of 3.43 ± 0.7%). Whereas, MR5 produced maximum laccase (5.07 ± 0.25 U/mL) after 3 weeks, however, maximum esterase (0.09 ± 0.003 U/mL) was recorded on the fourth week of degradation and exhibited a weight loss of 2.42 ± 0.3%. The results highlighted the efficiency of bacterial isolates to utilize PET as a sole carbon source for their growth. Unlike, PE and PS, PET has carbon and hetero atoms (ester or amide bonds) in the main chain, has better thermal stability (can be molded by melting), and is easily hydrolyzed compared to polymers with only carbon backbone (Olabisi et al. 1997; Wei and Zimmermann 2017). The monomers of PET are linked together with ester bonds which can be hydrolyzed by hydrolytic enzymes (which target ester bonds) found in nature (Hiraga et al. 2019). The hygroscopic nature (i.e., it absorbs water from its surroundings and when heated, the water hydrolyzes the polymer), contributed to decreasing the resilience nature of PET which would have added to the biodegradation (Auta et al. 2022). It is therefore evident from the present study that, UV pretreatment, modified the surface by decreasing the crystallinity (dense structure) of PET which would have led to fragmentation (Gong et al. 2018). The surface modification and hydrolysis initiated the bacterial colonization (as observed in Fig. 8) and biofilm formation. Bond cleavage of PET was mediated by the esterase enzyme which resulted in the formation of smaller molecules (such as esters, amides, and heteroarenes) (Table 3) to be assimilated by bacterial isolates. The activity of laccase was observed with the weight loss of PET and the formation of ester and amides (Table 3). Earlier reports have shown efficient and complete degradation of PET after incubation of about 6 weeks at 30 °C, with the involvement of PETase and MHETase (Yoshida et al. 2016). However, only a handful of studies have shown the biodegradation of PET by bacterial isolates, namely, Streptomyces scabies (Jabloune et al. 2020), Thermobifida alba AHK119 (54.2% weight loss in 21 days by hydrolase enzymes) (Hu et al. 2010), Arthrobacter sulfonivorans, and Serratia plymuthica (Janczak et al. 2020), Saccharomonospora viridis (27% weight loss in 3 days at 63 °C, Kawai et al. 2019). Our result showed increased laccase activity by BS5, TL1, and MR10 over a period of 28 days. Also, the isolates were able to depolymerize the PET as evidenced by the formation of small molecules (ester and amides) as observed in LC-MS analysis.
In the present study, all the bacterial isolates (MR5, MR10, BS5, and TL1) showed biodegradation of PS efficiently. BS5 showed the highest growth for 4 weeks (absorbance of 0.59 ± 0.5, microbial viable count of 7.04 × 107 and cell dry biomass of 0.061 ± 0.006 mg/mL) (Figs. 2, 3 and 4) followed by TL1 (absorbance of 0.41 ± 0.4, microbial viable count of 6.91 × 107 and cell dry biomass of 0.059 ± 0.001 mg/mL) and MR10 (absorbance of 0.45 ± 0.01, microbial viable count of 6.06 × 107 and cell dry biomass of 0.045 ± 0.001 mg/mL). MR5 showed the highest growth and cell dry biomass after 3 weeks of incubation and declined by the end of the fourth week. PS has the same C–C backbone as PE, which makes PS recalcitrant to biodegradation in natural conditions (Zhang et al. 2022). UV pretreatment irradiated the PS surface, cleaving the nearest hydrogen in the C-H bond which would have resulted in chain scission and formed amines, alcohols, and olefins (Table 2). The surface erosion, initiated colonization and biofilm formation by the bacterial isolates, which thereby used PS as the sole carbon source for their growth during 4 weeks of biodegradation. As PS contains a bulky styrene group on the side chain, it hinders the binding of PS polymers within the active site of enzymes (Zhang et al. 2022). Therefore, PS undergoes the initial depolymerization into oligomers or monomers (as seen in the LC-MS result in Table 3). The depolymerization in the present study was led by extracellular laccase and esterase by the bacterial isolates (Fig. 5 and Fig. 6), which resulted in the weight loss of PS (Fig. 7). The highest biodegradability for PS was observed with BS5 that produced the highest laccase (15.35 ± 0.19 U/mL) and esterase (1.04 ± 0.02 U/mL) and exhibited a weight loss of 4.84 ± 0.6% compared to the other isolates. Similarly, TL1 showed growth until fourth week and produced extracellular laccase (12.15 ± 1.0 U/mL) and esterase (0.44 ± 0.02 U/mL) and exhibited a residual loss of PS (3.43 ± 0.5%), followed by MR10 (6.26 ± 0.2 U/mL of laccase, 0.22 ± 0.003 U/mL of esterase produced and exhibited a weight loss of 3.23 ± 0.34%). It was observed that MR5 showed growth until 3 weeks and produced extracellular laccase (maximum laccase produced on the 4 week 6.57 ± 0.14 U/mL) and exhibited a weight loss of 2.42 ± 0.3%. However, the growth and enzyme production decreased after the 3 week. Overall, PS has a hydrophobic property, and the addition of antioxidants, stabilizers, and additives makes the polymer less interactive with extracellular enzymes (Jadaun et al. 2022; Zhang et al. 2022). However, the isolates obtained from sanitary landfill (BS5 and TL1) and mangrove sediment (MR5 and MR10) successfully colonized the PS surface and initiated biodegradation with the help of laccase and esterase enzymes. Recent reports have shown the involvement of various genera in PS degradation including Bacillus (9.9% weight loss in 28 days), Pseudomonas (10% weight loss in 28 days), Enterobacter (0.8% weight loss in 60 days), Klebsielaa, Micrococcus, Exiguobacterium, Lysinibacillus (29.5% weight loss in 126 days), Serratia, and Acinetobacter (13 and 25% weight loss in 60 days) (Zhang et al. 2022). The isolates reported in the present study are novel and have not been reported in the biodegradation of PS. While BS5 and TL1 showed maximum laccase activity on the 28th day of degradation activity, the weight loss was observed to be 4.84 ± 0.6%. Also, the depolymerization of PS generated smaller molecules including amide, ester, ether, acid, alcohol, and amines.
Although MR5 showed increased growth for 3 weeks when incubated with LDPE, PET, and PS, the growth and enzyme production declined after 3 weeks. The decline in growth could be due to cell lysis, nutrient depletion, and the formation of inhibitory products (Auta et al. 2018; Montazer et al. 2018) (not analyzed in this study) and needs further investigation. The experimental control sets containing only bacterial isolates showed no growth and the control set with only polymers showed no changes in the absorbance value for 4 weeks. It was also observed that although the four isolates showed laccase and esterase production, the concentration of enzymes varied significantly. This could be due to various factors, such as the molecular weight of the polymers, the functional groups present, chemical bonds, hydrophilic or hydrophobicity with the surface, and crystallinity (Chamas et al. 2020; Singh and Ting 2022). Considering the complex structure of polymers, the pre-treatment by UV radiation played a significant role in surface modification, thereby accelerating the biodegradation. The time of incubation and the nutrient media also play a significant role in the degradation. As the present study was carried out for 4 weeks (BS5, TL1, and MR10 showed a gradual increase in the microbial viable count and enzyme production), increasing the time of incubation and change of the nutrient media can help to increase the efficiency of the bacterial isolates.
Structural analysis of LDPE, PET, and PS during biodegradation
The surface morphology of the degraded polymers was analyzed using FESEM (Fig. 8). The image showed the rough and damaged surface of the three polymers LDPE, PET, and PS. The change in the surface morphology was due to UV irradiation, which can be further seen in FTIR images (Fig. S2–S7). The images showed that MR5 (Fig. 8d–f), MR10 (Fig. 8g–i), BS5 (Fig. 8j–l), and TL1 (Fig. 8m–o), colonized the polymer surface and formed biofilm. The colonization of the bacterial isolates on the surface was mediated by the auto-oxidation which was initiated by the UV pre-treatment. Further, the isolates produced extracellular laccase and esterase to oxidize and hydrolyze the polymer backbone. This can be observed after 4 weeks of incubation of LDPE, PET, and PS where surface damage was visible through numerous pores/holes, cracks, erosions, and grooves. However, the uninoculated polymer remained smooth, and no changes in the surface morphology were observed (Fig. 8a–c).
Biodegradation of LDPE, PET, and PS was further determined by FTIR spectroscopy (Table 2, Fig. S2–S7) after 4 weeks of degradation assay. Exposure to UV irradiation for a long period resulted in structural change with the addition and deletion of functional groups in the LDPE, PET, and PS which was observed in the FTIR analysis. FTIR spectra for the LDPE without bacteria (control) showed peaks at 2916.09 cm−1 and 2848.40 cm−1, which correspond to the C–H stretch, and 1462.85 cm−1, which corresponds to the C-H bond (Fig. S2–S7). Both peaks correspond to alkanes, which are the primary constituents of the LDPE and are susceptible to bacterial enzymes (Jeon and Kim 2016). Microbial colonization reduced the hydrophobicity of the LDPE and accelerated biofilm formation (as observed in Fig. 8). LDPE treated with MR5, showed two new peaks when compared to the control LDPE, i.e., 1054.89 cm−1, which corresponds to –C–O stretch for alcohol, carboxylic acid, esters, ether, whereas the other peak at 1654.14 cm−1 showed –C = O bond for carbonyl group. Similarly, LDPE treated with MR10 showed a peak at 1683.90 cm−1 corresponding to the –C = O stretch, which shows the formation of ketones and aldehydes. The formation of smaller molecules (ketones, aldehydes, ester, ethers, and many more like them) could be mineralized by the bacterial cells via β oxidation that breaks down the LDPE surface (Bitalac et al. 2023). Similarly, the FTIR spectra of LDPE with BS5 and TL1 showed one common peak at 1472.43 cm−1, which attributed to the –C–H bonds for alkanes (Mohanan et al. 2020; Yoon et al. 2012). The alkanes are the primary constituents of LDPE that are susceptible to degradation by bacterial enzymes (Jeon and Kim 2016). The current study also demonstrated enhanced levels of laccase during LDPE degradation, which may also play a role in the shifting of peaks. An earlier study reported LDPE film degradation using bacteria from marine sites and showed the FTIR spectra of LDPE with a shift in peaks with a decrease in transmittance of functional group and side-chain peak (Khandare et al. 2021).
FTIR spectra of PET treated with MR5 showed three peaks, in which 2959.82 cm−1 (–C–H stretch) and 1452.96 cm−1 (–C–H bend) both corresponded to alkanes. In addition, the formation of a peak at 1174.83 cm−1 (–C–O stretch) attributed to alcohol, carboxylic acid, esters, and ethers was observed. MR10 showed the maximum number of peaks, showing the efficiency of mangrove microbes in the surface modification of the PET. MR10 showed peaks at 2916.84 cm−1 (–C–H stretch), 2848.95 cm−1 (H–C=O:C–H stretch), 1472.68 cm−1 (–C=O stretch), 1462.72 cm−1 (–C–H bend), 1186.32 cm−1 (–C–O stretch) and 1173.60 cm−1 (–C–O stretch), which correspond to alkanes, aldehydes, inorganic carbonates, alcohol, carboxylic acid, esters and ethers. PET treated with BS5 and TL1 showed the development of new peaks at 2920.63 cm−1 and 2961.99 cm−1, which correspond to the –C–H stretch reflecting alkanes (Fig. S3). In addition, BS5-treated PET showed an additional peak at 969.91 cm−1, which is attributed to =C–H bond (alkenes). Earlier reports suggest spectra range from 1700–1500 cm−1 reflect the formation of small molecules during microbial degradation (Denaro et al. 2020). Similarly, MR5 and MR10-treated PS showed broad peaks at 3661.20 cm−1 (–O–H stretch) which confirms the insertion of the hydroxyl group in the PS, after bacterial decomposition (Kim et al. 2021). MR5 showed an additional peak at 1685.01 cm−1 (–C=O stretch) which corresponds to ketones and aldehydes and MR10 showed an additional peak at 1269.90 cm−1 (–C–O stretch) which attributed to alcohol, carboxylic acid, esters, and ethers. BS5-treated PS showed peaks at 1685.04 cm−1 (–C=O stretch), 1311.37 cm−1 (–C–O stretch), and 1199.87 cm−1 (–C–O stretch) which correspond to ketones, aldehydes, alcohol, carboxylic acid, esters, ethers while TL1 showed only one peak at 1311.37 cm−1 (–C–O stretch) which corresponded to alcohol, carboxylic acid, esters, and ethers. The FTIR spectra of PS-treated with LDPE showed two additional peaks at 3661.20 cm−1 (–O–H stretch) corresponding to alcohol and 1685.01 cm−1 –C=O stretch) corresponding to ketones and aldehydes. Similarly, MR10 showed peaks at 3661.20 cm−1 (–O–H stretch) corresponding to alcohol and 1269.90 cm−1 (–C–O stretch) corresponding to alcohol, carboxylic acid, esters, and ethers. TL1-treated PS showed only one additional peak at 1311.37 cm−1 (–C–O stretch) corresponding to alcohol, carboxylic acid, esters, and ethers. Whereas, BS5-treated PS showed peaks at 1685.04 cm−1 (–C=O stretch) which attributed to ketones and aldehydes, and 1311.37 cm−1 (–C–O stretch), and 1199.87 cm−1 (–C–O stretch) corresponding to alcohol, carboxylic acid, esters, and ethers.
Overall, by comparing all the spectra obtained (Table 2), it can be observed that different strains might be targeting different moieties of the LDPE, PET, and PS backbones (Bitalac et al. 2023). In the present study, the identified strains successfully modified the polymer surface by forming carbonyl groups (C=O) (range 1730–1650 cm−1) and alcohol groups (O–H). Similarly, the transmittance at 1100–1150 cm−1 (C–O stretch) represented the formation of small molecules such as alcohol, carboxylic acid, esters, and ether by all the isolates during biodegradation. This could be due to the aerobic degradation process, which incorporated oxygen atoms in the damaged structure of the polymers (Kowalczyk et al. 2016). The oxidation products would have been then transformed into functional groups, possibly by the Norrish type I and II mechanisms, as stated by Dey et al. (2020). The addition of functional groups by bacterial isolates reduced the hydrophobicity of the polymers and facilitated bacterial colonization on the surface. The activity of esterase and laccase thereby converted the complex polymers to small molecules that can be taken up by the isolates for their growth. This was further confirmed by the weight loss of the residual polymer.
Analysis of degraded products using liquid chromatography–mass spectrometry
The BHB broth used during the degradation of LDPE, PET, and PS inoculated with MR5, MR10, BS5, and TL1 was analyzed for the degraded product by LC–MS analysis. The control sets (broth without bacterial isolates) were compared with the experiment sets (all three polymers with bacterial isolates). The extraction method used in the study has limitations, and the extracts collected in our experiment might not include all of the intermediates formed during the biodegradation of LDPE, PET, and PS. The LC–MS data showed that the eluted compounds (putative identification) contained carbon, hydrogen, and oxygen elements, and significant differences were recorded in abundance, which suggested unique compounds were obtained among the experiment sets (Table 3). In total, 27 eluted products were obtained from the experiment sets (polymers and bacterial culture) that were absent in the control set (experiment set with only LDPE, PET, and PS). The ion chromatogram obtained showed peaks that depicted individual products generated during degradation. LDPE degradation by the four isolates showed the formation of ester (eplerenone), fatty acids (1-monopalmitin), fatty acid amide (oleamide), hydrocarbon (tetralin), conjugate acid (pyropheophorbide a), and a few aromatic compounds containing benzene (3-butylidene-7-hydroxyphthalide, N-alkylindole and 4,5-di-O-methyl-8-prenylafzelechin-4-beta-ol) (Table 3). LC-MS also identified some unknown compounds such as pyran (avermectin B1b aglycone) which is a topic to be further explored. The present study showed oxidation of the long carbon–carbon chain of LDPE which could have been depolymerized through enzymatic hydrolysis (laccase) leading to the formation of small molecules (Fujisawa et al. 2001). The degraded products were then possibly consumed by the bacterial isolates via β-oxidation and TCA cycle metabolism (Ji et al. 2013). However, the associated gene regulation studies can substantiate the process of complete degradation. It can also be estimated that all the isolates commonly oxidized the LDPE by targeting –C–O and –C=O bonds during biodegradation assay (Ji et al. 2013). The results from the current study are in line with the earlier reports published by Ren et al. (2019) and Kyaw et al. (2012). The studies showed enzymatic hydrolysis of polyethylene indicating oxidation reaction on the surface of the film by bacterial degradation, and formation of small molecules mainly ester, acids, and hydrocarbons.
PET degradation showed the formation of ester, amide, hydrocarbon, and fatty acids which were commonly observed with all the isolates (MR5, MR10, BS5, and TL1). MR5-treated PET showed two unique peaks at retention times of 14.8 and 19.59 min, that were putatively identified as eplerenone (steroid acid ester) and oleamide (a fatty acid ester). MR10, BS5, and TL1-treated PET showed one chromatogram peak, at a retention time of 20.01, 0.93, and 19.50 min, and the compounds were identified as 2-pentadecylfuran (heteroarene, M/z value of 296.29), hypoxanthine (purine, M/z value of 137.0455) and 1-Monopalmitin (fatty acid, M/z value of 353.2662). The degradation of PET was mediated by a PET-hydrolyzing enzyme (esterase) which was produced by all the bacterial isolates. The esterase enzyme is a member of α/β hydrolase superfamily with a special function to convert the PET into monomers of terephthalic acid (TPA) and mono (2-hydroxyethyl terephthalate (MHET as reported by Yoshida et al. (2016). The current study identified smaller molecules which shows the surface damage of PET by the action of esterase enzymes. It can also be predicted that all the isolates initiated the biodegradation of PET commonly by hydrolyzing the –C–H bond during the biodegradation assay (Jadaun et al. 2022). Earlier studies have identified hydrocarbons such as phenanthrene, naphthalene, tetradecane, and diesel during the biodegradation of PET (Denaro et al. 2020). Similarly, PET degradation by Streptomyces species identified xylene and ethyl benzene as the main metabolites (Farzi et al. 2019).
PS degradation showed the formation of amide, ester, ether, acid, alcohol, and amines, which were commonly observed with all the isolates (MR5, MR10, BS5, and TL1). MR5-treated PS was identified with pyropheophorbide a (conjugate acid) and MR10 and BS5-treated PS were identified with 2-pentadecylfuran (heteroarene). TL1-treated PS identified 11S-hydroxy-tetradecanoic acid, 1-monopalmitin (fatty acid), xestoaminol C (fatty alcohol), eplerenone (steroid acid ester), and dodemorph (water-soluble morpholine) and some unknown compounds. Elution of smaller molecules suggested the metabolism of bacterial isolates to break the carbon–carbon backbones and aromatic rings in the PS. Similar to LPDE degradation, laccase, lipases, and oxidoreductases have been proposed to be involved in PS degradation (Zhang et al. 2022). Although the main C–C bond could be cleaved by the enzymes, the side chain of the styrene group interferes with degradation. Therefore, the enzymes first attack either the β-carbon of the main chain (chain cleavage) or the aromatic ring (side-chain cleavage) to break the PS into smaller molecules (Hou and Majumder 2021). The aromatic ring is further cleaved by hydrolases into short-chain hydrocarbons, which are utilized as carbon sources through the TCA (tricarboxylic acid) cycle (Zhang et al. 2022; Kim et al. 2021). Although studies have highlighted the degradation of PS, by-product analysis is limited, and detailed determination of the enzymatic pathway can further enhance the degradation mechanism (Kim et al. 2021).
Conclusion
This study demonstrated the microbe-assisted degradation of UV-pretreated LDPE, PET, and PS using bacterial isolates Dermacoccus sp. MR5, Corynebacterium sp. MR10 isolated from a Malaysian mangrove, and Bacillus sp. BS5, and Priestia sp. TL1 from a sanitary landfill. The study identified novel microbial isolates that showed a broad host range and played a critical role in the degradation of three different polymers LDPE, PET, and PS. The degradation was mediated by esterase and laccase enzymes which played crucial roles in the breakdown of the polymers. Further, the growth of bacterial isolates showed significant changes in the physical and chemical structure of the polymers. These changes were confirmed through FTIR, FESEM, and LC–MS. FTIR analysis showed the formation of new functional group peaks resulting from oxidation and hydrolysis during polymer degradation. The depolymerization process further led to the fragmentation of the polymers into small molecules (esters, amides, acids) as evidenced by LC-quidMS analysis. FESEM analysis provided visual evidence of microbial attachment, biofilm formation, and surface erosion, which indicated that polymers served as a carbon source for the bacterial isolates and provided energy for their metabolic activities. Overall, the microbial degradation with the involvement of laccase and esterase showed weight reduction of the LDPE, PET, and PS, where the highest weight reduction was observed for BS5-treated PS (4.84 ± 0.6%) and BS5-treated PET (4.44 ± 1.12%) in a very short incubation time (4 weeks). The results will serve as the basis for future studies to look into potential bacterial isolates that target simultaneous biodegradation of multiple polymers. Future studies should focus on increasing higher degradation rates by engineering enzymes and microorganisms, biodegradation strategies using multiple enzymes, and construction of metabolic pathways to upcycle the monomers generated into value-added products.
Data availability
The data presented in this study are available in the manuscript and are available from the corresponding author on reasonable request.
References
Akinpelu EA, Nchu F (2022) A bibliometric analysis of research trends in biodegradation of plastics. Polymers 14:2642. https://doi.org/10.3390/polym14132642
Albertsson AC, Karlsson S (1990) The influence of biotic and abiotic environments on the degradation of polyethylene. Prog Polym Sci 15:177–192. https://doi.org/10.1016/0079-6700(90)90027-X
Arutchelvi J, Sudhakar M, Arkatkar A, Doble M, Bhaduri S, Uppara PV (2008) Biodegradation of polyethylene and polypropylene. Ind J Biotechnol 0975–0967:9–22
Auta HS, Emenike CU, Jayanthi B, Fauziah SH (2018) Growth kinetics and biodeterioration of polypropylene microplastics by Bacillus sp. and Rhodococcus sp. isolated from mangrove sediment. Mar Pollut Bull 127:15–21. https://doi.org/10.1016/j.marpolbul.2017.11.036
Auta HS, Abioye OP, Aransiola SA, Bala JD, Chukwuemeka VI, Hassan A, Aziz A, Fauziah SH (2022) Enhanced microbial degradation of PET and PS microplastics under natural conditions in mangrove environment. J Environ Manag 15:114273. https://doi.org/10.1016/j.jenvman.2021.114273
Banerjee U, Chisti Y, Moo-Young M (1993) Spectrophotometric determination of mycelial biomass. Biotechnol Tech 7:313–316. https://doi.org/10.1007/BF00150905
Biedendieck R, Knuuti T, Moore SJ, Jahn D (2021) The “beauty in the beast”—the multiple uses of Priestia megaterium in biotechnology. Appl Microbiol Biotechnol 105:5719–5737. https://doi.org/10.1007/s00253-021-11424-6
Binnerup SJ, Højberg O, Sørensen J (1998) Gram characteristics determined on single cells and at the microcolony level of bacteria immobilized on polycarbonate membrane filters. J Microbiol Method 31:185–192. https://doi.org/10.1016/S0167-7012(97)00102-4
Bitalac JMS, Lantican NB, Gomez NCF, Onda DFL (2023) Attachment of potential cultivable primo-colonizing bacteria and its implications on the fate of low-density polyethylene (LDPE) plastics in the marine environment. J Hazard Mater 451:131124. https://doi.org/10.1016/j.jhazmat.2023.131124
Bolo NR, Diamos MAJC, Sia S, Ocampo MAB, Suyom LM (2015) Isolation, identification, and evaluation of polyethylene glycol and low density polyethylene-degrading bacteria from Payatas dumpsite, Quezon city, Philippines. Phil J Health Res Dev 19:50–59
Bond T, Ferrandiz-Mas V, Felipe-Sotelo M, Van Sebille E (2018) The occurrence and degradation of aquatic plastic litter based on polymer physicochemical properties: a review. Crit Rev Environ Sci Technol 48:685–722. https://doi.org/10.1080/10643389.2018.1483155
Cai L, Wang J, Peng J, Wu Z, Tan X (2018) Observation of the degradation of three types of plastic pellets exposed to UV irradiation in three different environments. Sci Total Environ 628:740–747. https://doi.org/10.1016/j.scitotenv.2018.02.079
Campanale C, Savino I, Massarelli C, Uricchio VF (2023) Fourier transform infrared spectroscopy to assess the degree of alteration of artificially aged and environmentally weathered microplastics. Polymers 15:1–16. https://doi.org/10.3390/polym15040911
Chamas A, Moon H, Zheng J, Qiu Y, Tabassum T, Jang JH, Abu-Omar M, Scott SL, Suh S (2020) Degradation rates of plastics in the environment. ACS Sust Chem Eng 8:3494–3511. https://doi.org/10.1016/j.envpol.2018.05.008
Coates J (2000) Interpretation of infrared spectra, a practical approach. In: Meyers RA (ed) Encyclopedia of analytical chemistry. John Wiley & Sons Ltd, Chichester, pp 10815–10837
Danso D, Chow J, Streit WR (2019) Plastics: environmental and biotechnological perspectives on microbial degradation. Appl Environ Microbiol 85:e01095–e01019. https://doi.org/10.1128/AEM.01095-19
Denaro R, Aulenta F, Crisafi F, Di Pippo F, Viggi CC, Matturro B, Tomei P, Smedile F, Martinelli A, Di Lisio V, Venezia C (2020) Marine hydrocarbon-degrading bacteria breakdown poly (ethylene terephthalate) (PET). Sci Total Environ 749:141608. https://doi.org/10.1016/j.scitotenv.2020.141608
Deng H, He J, Feng D, Zhao Y, Sun W, Yu H, Ge C (2021) Microplastics pollution in mangrove ecosystems: a critical review of current knowledge and future directions. Sci Total Environ 753:142041. https://doi.org/10.1016/j.scitotenv.2020.142041
Dey AS, Bose H, Mohapatra B, Sar P (2020) Biodegradation of unpretreated low-density polyethylene (LDPE) by Stenotrophomonas sp. and Achromobacter sp., isolated from waste dumpsite and drilling fluid. Front Microbiol 11:603210. https://doi.org/10.3389/fmicb.2020.603210
Farzi A, Dehnad A, Fotouhi AF (2019) Biodegradation of polyethylene terephthalate waste using streptomyces species and kinetic modeling of the process. Biocatal Agric Biotechnol 17:25–31. https://doi.org/10.1016/j.bcab.2018.11.002
Fotopoulou KN, Karapanagioti HK (2017) Degradation of various plastics in the environment. In: Takada H, Karapanagioti HK (eds) Hazardous chemicals associated with plastics in the marine environment. The Handbook of Environmental Chemistry, Springer Cham New York
Fritz M, Lauschke T, Schlebrowski T, Beucher L, Schweyen P, Alenezi B, Hahn B, Dierkes G, Ternes T, Fischer CB (2022) Photoaging phenomena of biodegradable polybutylene succinate and conventional low density polyethylene by artificial weathering–a comparative surface study. Appl Surf Sci 590:153058. https://doi.org/10.3390/polym15040911
Fujisawa M, Hirai H, Nishida T (2001) Degradation of polyethylene and nylon-66 by the laccase-mediator system. J Polym Environ 9:103–108. https://doi.org/10.1023/A:1020472426516
Gewert B, Plassmann MM, MacLeod M (2015) Pathways for degradation of plastic polymers floating in the marine environment. Environ Sci Process Impacts 17:1513–1521. https://doi.org/10.1039/C5EM00207A
Geyer R, Jambeck JR, Law KL (2017) Production, use, and fate of all plastics ever made. Sci Adv 3:e1700782. https://doi.org/10.1126/sciadv.1700782
Gong J, Kong T, Li Y, Li Q, Li Z, Zhang J (2018) Biodegradation of microplastic derived from poly (ethylene terephthalate) with bacterial whole-cell biocatalysts. Polymers 10:1326. https://doi.org/10.3390/polym10121326
Harshvardhan K, Jha B (2013) Biodegradation of low-density polyethylene by marine bacteria from pelagic waters, Arabian sea, India. Mar Pollut Bull 77:100–106. https://doi.org/10.1016/j.marpolbul.2013.10.025
Hiraga K, Taniguchi I, Yoshida S, Kimura Y, Oda K (2019) Biodegradation of waste PET: A sustainable solution for dealing with plastic pollution. EMBO Rep 20:e49365. https://doi.org/10.15252/embr.201949365
Hou L, Majumder ELW (2021) Potential for and distribution of enzymatic biodegradation of polystyrene by environmental microorganisms. Materials 14:503. https://doi.org/10.3390/ma14030503
Hu X, Thumarat U, Zhang X, Tang M, Kawai F (2010) Diversity of polyester-degrading bacteria in compost and molecular analysis of a thermoactive esterase from Thermobifida alba AHK119. Appl Microbiol Biotechnol 87:771–779. https://doi.org/10.1007/s00253-010-2555-x
Jabloune R, Khalil M, Moussa IEB, Simao-Beaunoir AM, Lerat S, Brzezinski R, Beaulieu C (2020) Enzymatic degradation of p-nitrophenyl esters, polyethylene terephthalate, cutin, and suberin by Sub1, a suberinase encoded by the plant pathogen Streptomyces scabies. Microb Environ 35:ME19086. https://doi.org/10.1264/jsme2.ME19086
Jadaun JS, Bansal S, Sonthalia A, Rai AK, Singh SP (2022) Biodegradation of plastics for sustainable environment. Bioresour Technol 347:126697. https://doi.org/10.1016/j.biortech.2022.126697
Jaiganesh R, Jaganathan MK (2018) Isolation, purification, and characterization of lipase from Bacillus sp. from kitchen grease. Asian J Pharm Clin Res 11:224–227. https://doi.org/10.22159/ajpcr.2018.v11i6.24955
Janczak K, Dąbrowska GB, Raszkowska-Kaczor A, Kaczor D, Hrynkiewicz K, Richert A (2020) Biodegradation of the plastics PLA and PET in cultivated soil with the participation of microorganisms and plants. Int Biodeterior Biodegrad 155:105087. https://doi.org/10.1016/j.ibiod.2020.105087
Jeon HJ, Kim MN (2016) Comparison of the functional characterization between alkane monooxygenases for low-molecular-weight polyethylene biodegradation. Int Biodeterior Biodegrad 114:202–208. https://doi.org/10.1016/j.ibiod.2016.06.012
Ji Y, Mao G, Wang Y, Bartlam M (2013) Structural insights into diversity and n-alkane biodegradation mechanisms of alkane hydroxylases. Front Microbiol. https://doi.org/10.3389/fmicb.2013.00058
Kang J, Zhou L, Duan X, Sun H, Ao Z, Wang S (2019) Degradation of cosmetic microplastics via functionalized carbon nanosprings. Matter 1:745–758. https://doi.org/10.1016/j.matt.2019.06.004
Kawai F, Kawabata T, Oda M (2019) Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl Microbiol Biotechnol 103:4253–4268. https://doi.org/10.1007/s00253-019-09717-y
Kay M, McCabe R, Morton L (1993) Chemical and physical changes occurring in polyester polyurethane during biodegradation. Int Biodeterior Biodegrad 31:209–225. https://doi.org/10.1016/0964-8305(93)90006-N
Khandare SD, Chaudhary DR, Jha B (2021) Marine bacterial biodegradation of low-density polyethylene (LDPE) plastic. Biodegradation 32:127–143. https://doi.org/10.1007/s10532-021-09927-0
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. https://doi.org/10.1099/ijs.0.038075-0
Kim HW, Jo JH, Kim YB, Le TK, Cho CW, Yun CH, Chi WS, Yeom SJ (2021) Biodegradation of polystyrene by bacteria from the soil in common environments. J Hazard Mater 416:126239. https://doi.org/10.1016/j.jhazmat.2021.126239
Kim HR, Lee C, Shin H, Kim J, Jeong M, Choi D (2023) Isolation of a polyethylene-degrading bacterium, Acinetobacter guillouiae, using a novel screening method based on a redox indicator. Heliyon 9:1–12. https://doi.org/10.1016/j.heliyon.2023.e15731
Kowalczyk A, Chyc M, Ryszka P, Latowski D (2016) Achromobacter xylosoxidans as a new microorganism strain colonizing high density polyethylene as a key step to its biodegradation. Environ Sci Pollut Res 23:11349–11356. https://doi.org/10.1007/s11356-016-6563-y
Kyaw BM, Champakalakshmi R, Sakharkar MK, Lim CS, Sakharkar KR (2012) Biodegradation of low-density polythene (LDPE) by Pseudomonas species. Ind J Microbiol 52:411–419. https://doi.org/10.1007/s12088-012-0250-6
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Lebreton L, Slat B, Ferrari F, Sainte-Rose B, Aitken J, Marthouse R, Hajbane S, Cunsolo S, Schwarz A, Levivier A, Noble K (2018) Evidence that the great Pacific garbage patch is rapidly accumulating plastic. Sci Rep 8:1–15. https://doi.org/10.1038/s41598-018-22939-w
Li Y, Li S, Sun J (2021) Degradable poly (vinyl alcohol)-based supramolecular plastics with high mechanical strength in a watery environment. Adv Mater 33:2007371. https://doi.org/10.1002/adma.202007371
Maroof L, Khan I, Yoo HS, Kim S, Park HT, Ahmad B, Azam S (2021) Identification and characterization of low-density polyethylene-degrading bacteria isolated from soils of waste disposal sites. Environ Eng Res 26:3. https://doi.org/10.4491/eer.2020.167
Meyer-Dombard DAR, Bogner JE, Malas J (2020) A review of landfill microbiology and ecology: a call for modernization with ‘next generation’ technology. Front Microbiol 11:1127. https://doi.org/10.3389/fmicb.2020.01127
Mohanan N, Montazer Z, Sharma PK, Levin DB (2020) Microbial and enzymatic degradation of synthetic plastics. Front Microbiol 11:580709. https://doi.org/10.3389/fmicb.2020.580709
Montazer Z, Habibi-Najafi MB, Mohebbi M, Oromiehei A (2018) Microbial degradation of UV—pretreated low-density polyethylene films by novel polyethylene-degrading bacteria isolated from plastic-dump soil. J Polym Environ 26:3613–3625. https://doi.org/10.1007/s10924-018-1245-0
Mor R, Sivan A (2008) Biofilm formation and partial biodegradation of polystyrene by the actinomycete Rhodococcus ruber. Biodegradation 19:851–858. https://doi.org/10.1007/s10532-008-9188-0
Nunes CS, Kunamneni A (2018) Laccases—properties and applications. In: Nunes CS, NunesKumar V (eds) Enzymes in human and animal nutrition. Academic Press, Cambridge
Olabisi O (1997) Conventional polyolefins. In: Olabisi O (ed) Plastics engineering. New York
Papinutti L, Martínez MJ (2006) Production and characterization of laccase and manganese peroxidase from the ligninolytic fungus Fomes sclerodermus. J Chem Technol Biotechnol 81:1064–1070. https://doi.org/10.1002/jctb.1537
Quero GM, Luna GM (2017) Surfing and dining on the “plastisphere”: Microbial life on plastic marine debris. Adv Oceanogr Limnol 8:2. https://doi.org/10.4081/aiol.2017.7211
Ren L, Men L, Zhang Z, Guan F, Tian J, Wang B, Wang J, Zhang Y, Zhang W (2019) Biodegradation of polyethylene by Enterobacter sp. D1 from the guts of wax moth Galleria mellonella. Int J Environ Res Pub Health 16:1941. https://doi.org/10.3390/ijerph16111941
Restrepo-Flórez JM, Bassi A, Thompson MR (2014) Microbial degradation and deterioration of polyethylene–a review. Int Biodeterior Biodegrad 88:83–90. https://doi.org/10.1016/j.ibiod.2013.12.014
Rillig MC, Lehmann A, de Souza Machado AA, Yang G (2019) Microplastic effects on plants. New Phytol 223:1066–1070. https://doi.org/10.1111/nph.15794
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Santo M, Weitsman R, Sivan A (2013) The role of the copper-binding enzyme–laccase–in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int Biodeterior Biodegrad 84:204–210. https://doi.org/10.1016/j.ibiod.2012.03.001
Sarker RK, Chakraborty P, Paul P, Chatterjee A, Tribedi P (2020) Degradation of low-density poly ethylene (LDPE) by Enterobacter cloacae AKS7: a potential step towards sustainable environmental remediation. Arch Microbiol 202:2117–2125. https://doi.org/10.1007/s00203-020-01926-8
Singh P, Ting ASY (2022) Plastic biodegrading microbes in the environment and their applications. In: Inamuddin TA (ed) Biodegradable materials and their applications. Scrivener Publishing LLC, Beverly. https://doi.org/10.1002/9781119905301.ch19
Singh P, Nagarajan A, Chua KO, Ting AS (2023) Biodegradation of low-density polyethylene by novel halophilic bacteria from mangrove ecosystem. Bioremediat J 19:1–2. https://doi.org/10.1080/10889868.2023.2297181
Song L, Wang Y, Tang W, Lei Y (2015) Bacterial community diversity in municipal waste landfill sites. Appl Microbiol Biotechnol 99:7745–7756. https://doi.org/10.1007/s00253-015-6633-y
Sun K, Song Y, He F, Jing M, Tang J, Liu R (2021) A review of human and animals exposure to polycyclic aromatic hydrocarbons: Health risk and adverse effects, photo-induced toxicity and regulating effect of microplastics. Sci Total Environ 773:145403. https://doi.org/10.1016/j.scitotenv.2021.145403
Taghavi N, Singhal N, Zhuang WQ, Baroutian S (2021) Degradation of plastic waste using stimulated and naturally occurring microbial strains. Chemosphere 263:127975. https://doi.org/10.1016/j.chemosphere.2020.127975
Teuten EL, Saquing JM, Knappe DR, Barlaz MA, Jonsson S, Björn A, Rowland SJ, Thompson RC, Galloway TS, Yamashita R (2009) Transport and release of chemicals from plastics to the environment and to wildlife. Philos Trans R Soc Lond B 364:2027–2045. https://doi.org/10.1098/rstb.2008.0284
Tokiwa Y, Calabia BP, Ugwu CU, Aiba S (2009) Biodegradability of plastics. Int J Mol Sci 10:3722–3742. https://doi.org/10.3390/ijms10093722
Vimala PP, Mathew L (2016) Biodegradation of polyethylene using Bacillus subtilis. Proc Technol 1:232–239
Wang K, Chen W, Tian J, Niu F, Xing Y, Wu Y, Zhang R, Zheng J, Xu L (2022) Accumulation of microplastics in greenhouse soil after long-term plastic film mulching in Beijing. China Sci Total Environ 828:154544. https://doi.org/10.1016/j.scitotenv.2022.154544
Wei R, Zimmermann W (2017) Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we? Microb Biotechnol 10:1308–1322. https://doi.org/10.1111/1751-7915.12710
Wright SL, Kelly FJ (2017) Plastic and human health: a micro issue? Environ Sci Technol 51:6634–6647. https://doi.org/10.1021/acs.est.7b00423
Yamada-Onodera K, Mukumoto H, Katsuyaya Y, Saiganji A, Tani Y (2001) Degradation of polyethylene by a fungus, Penicillium simplicissimum YK. Polym Degrad Stab 72:323–327. https://doi.org/10.1016/S0141-3910(01)00027-1
Yan F, Wei R, Cui Q, Bornscheuer UT, Liu YJ (2021) Thermophilic whole-cell degradation of polyethylene terephthalate using engineered clostridium thermocellum. Microb Biotechnol 14:374–385
Yang Y, Yang J, Wu WM, Zhao J, Song Y, Gao L, Yang R, Jiang L (2015) Biodegradation and mineralization of polystyrene by plastic-eating mealworms: part 2 role of gut microorganisms. Environ Sci Technol 49:12087–12093. https://doi.org/10.1021/acs.est.5b02663
Yoon MG, Jeon HJ, Kim MN (2012) Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell. J Bioremediat Biodegrade 3:1–8. https://doi.org/10.4172/2155-6199.1000145
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, Toyohara K, Miyamoto K, Kimura Y, Oda K (2016) A bacterium that degrades and assimilates poly (ethylene terephthalate). Science 351:1196–1199. https://doi.org/10.1126/science.aad6359
Zhang H, Tang J, Wang L, Liu J, Gurav RG, Sun K (2016) A novel bioremediation strategy for petroleum hydrocarbon pollutants using salt tolerant Corynebacterium variabile HRJ4 and biochar. J Environ Sci 47:7–13. https://doi.org/10.1016/j.biortech.2010.08.099
Zhang Y, Pedersen JN, Eser BE, Guo Z (2022) Biodegradation of polyethylene and polystyrene: from microbial deterioration to enzyme discovery. Biotechnol Adv 60:107991. https://doi.org/10.1016/j.biotechadv.2022.107991
Acknowledgements
The authors acknowledge Berjaya Sanitary Landfill, Bukit Tagar, Selangor, Malaysia, for leachate and bio-sludge samples and the Forest Department of Peninsular Malaysia, Ministry of Natural Resources and Environment for soil sample collection from mangrove forest.
Funding
This research was supported by funding from the Monash University Malaysia Post-Doctoral Fellowship Grant (STG-000058), Advancing Women’s Success Grant 2022 (CNI000054) award, and Tropical Medicine & Biology (TMB) (PLT000033) grant awarded to the first author to pursue research in microbial microplastic degradation.
Author information
Authors and Affiliations
Contributions
Pooja Singh: conceptualization, methodology, validation, data curation, and writing-original draft. Claudeen Sze Siang Lau: methodology, validation, and data curation. Sze Yin Siah: methodology, validation, and data curation. Kah Ooi Chua: bioinformatics analysis and review. Adeline Su Yien Ting: supervision and review.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest with any parties that may arise from the publication of this work. The authors do not have any other relevant financial or non-financial interest to disclose.
Additional information
Communicated by PANKAJ BHATT.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, P., Lau, C.S.S., Siah, S.Y. et al. Microbial degradation of low-density polyethylene, polyethylene terephthalate, and polystyrene by novel isolates from plastic-polluted environment. Arch Microbiol 206, 188 (2024). https://doi.org/10.1007/s00203-024-03895-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-024-03895-8