Abstract
Laccases or laccase-like multicopper oxidases have great potential in bioremediation to oxidase phenolic or non-phenolic substrates. However, their inability to maintain stability in harsh environmental conditions and against non-substrate compounds is one of the main reasons for their limited use. The gene (mco) encoding multicopper oxidase from Bacillus mojavensis TH309 were cloned into pET14b( +), expressed in Escherichia coli, and purified as histidine tagged enzyme (BmLMCO). The molecular weight of the enzyme was about 60 kDa. The enzyme exhibited laccase-like activity toward 2,6-dimethoxyphenol (2,6-DMP), syringaldazine (SGZ), and 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid) (ABTS). The highest enzyme activity was recorded at 80 °C and pH 8. BmLMCO showed a half-life of ~ 305, 99, 50, 46, 36, and 20 min at 40, 50, 60, 70, 80, and 90 °C, respectively. It retained more than 60% of its activity after pre-incubation in the range of pH 5–12 for 60 min. The enzyme activity significantly increased in the presence of 1 mM of Cu2+. Moreover, BmLMCO tolerated various chemicals and showed excellent compatibility with organic solvents. The Michaelis constant (Km) and the maximum velocity (Vmax) values of BmLMCO were 0.98 mM and 93.45 µmol/min, respectively, with 2,6-DMP as the substrate. BmLMCO reduced the antibacterial activity of cefprozil, gentamycin, and erythromycin by 72.3 ± 1.5%, 79.6 ± 6.4%, and 19.7 ± 4.1%, respectively. This is the first revealing shows the recombinant production of laccase-like multicopper oxidase from any B. mojavensis strains, its biochemical properties, and potential for use in bioremediation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Laccases and laccase-like multicopper oxidases’ (LMCOs) enzymes catalyze the reduction of four electrons reducing oxygen to water by oxidation of one electron from the substrate, and exhibit broad substrate specificity. They can catalyze the oxidation of aromatics, polyphenols, terphenyls, nitriles, and various non-phenolic compounds. The fact that laccases have an unusually wide range of activities allows them to be used effectively in many industrial applications. They are used to clarify fruit juices and wines, bio-discoloration of denim (Sarafpour et al. 2022) and bio-bleaching of pulp, improve yogurt texture, enhance bread half-life, modify lignin, and produce energy from fuel cells. LMCOs or laccases can be incorporated as formulations of healthcare products, such as hair dye, deodorant, perfume, hand-face cream, mouthwash, and toothpaste, to reduce their toxic effect (Gigli et al. 2022; Mojtabavi et al. 2022). Furthermore, they are effective bio-agents in environmental remediation applications due to their ability to degrade synthetic dyes, plasticizers, plastics, polycyclic aromatic hydrocarbons (PAHs), pesticides, fertilizers, and pharmaceuticals (Dong et al. 2023).
Antibiotics are listed as emerging contaminants that threaten water resources. Even in low concentration ranges, they can be toxic to organisms and cause microorganisms to become antibiotic-resistant. Conventional treatment techniques contain filtration, sedimentation, coagulation, and flocculation, are insufficient for antibiotic removal (Al-sareji et al 2023). On the other hand, advanced oxidation processes, including ozonation, Fenton oxidation, and photocatalytic oxidation, can cause additional health and environmental problems due to toxic chemicals used or harmful by-products (Ekeoma et al 2023). It is also costly to adapt these methods to an industrial scale (Al-sareji et al 2023). Therefore, studies regarding the use of laccases for antibiotic removal as an environmentally friendly and easily applicable alternative have increased recently. Such studies have been performed with laccases from a limited number of sources, mostly fungi such as Myceliophthora thermophila (García‐Delgado et al. 2018), Trametes versicolor (Wen et al. 2019), Trametes hirsuta (Navada and Kulal 2019), and Aspergillus spp. (Lou et al. 2022). However, their use in antibiotic removal or other bioremediation applications is quite limited due to rapid activity loss in wastewaters, including alkali ingredients and other contaminants, such as heavy metals, inhibitors, and organic solvents (Mandic et al. 2019; Li et al. 2020; Cheng et al. 2021; Chopra and Sondhi 2022). Fungal laccases are also extremely sensitive to temperature and pH changes during application. Moreover, the heterologous expression of genes in fungi by genetic engineering is quite difficult compared to that of bacterial counterparts.
Bacillus species, which are Gram-positive, endospore-forming, string aerobic/facultative aerobic bacteria with rod shapes, are widely distributed in environments (Roberts et al. 1994). They can also survive in extreme habitats, including deserts, hot springs, salterns, and alkaline lakes, due to the longevity of their endospores (Mandic-Mulec et al. 2016). In time, they may develop mechanisms that allow them to adapt to these environments and thus produce active and stable bioactive compounds resistant to relatively harsh conditions (Parrilli et al. 2021). Bacillus mojavensis TH309, previously isolated from plastic waste, is a halotolerant endophyte (Adıgüzel 2020). The strain, which has growth ability over broad temperature (30–60 °C) and pH (5–11) ranges, is a potential source of laccase with desired properties for bioremediation applications. Further, laccases from various Bacillus species, such as Bacillus sphaericus (Claus and Filip 1997), Bacillus halodurans (Ruijssenaars and Hartmans 2004), Bacillus vallismortis (Zhang et al 2013), Bacillus clausii (Brander et al. 2014), Bacillus tequilensis (Sondhi et al. 2014), Bacillus amyloliquefaciens (Wang et al. 2020b), Bacillus velezensis (Li et al. 2020), Bacillus subtilis (Cheng et al. 2021), Bacillus licheniformis (Sun et al. 2023), Bacillus pumilus (Liu et al. 2023), and Bacillus cereus (Shafana Farveen et al. 2023). However, there are no reports describing the biochemical and structural features of laccase or LMCO from any B. mojavensis strains.
LMCOs are a component of the endospore coat in Bacillus species (Martins et al. 2002). Its extraction and purification necessitate using techniques that require high cost and extensive effort. In addition, these techniques applied sequentially cause significant losses in the product. Recombinant DNA technology eliminates such problems by expressing the target gene in high amounts with a tag that facilitates purification (Sinirlioglu et al. 2013). Therefore, in the present study, the LMCO gene of B. mojavensis TH309 was cloned into a pET14b( +) vector that carries an N-terminal Histidine taq sequence and expressed in Escherichia coli BL21 (DE3) under the control of T7 promoter. The expressed enzyme (BmLMCO) was purified and then characterized. In addition, the antibiotic removal potential of BmLMCO was determined by agar well-diffusion experiments (Zhang et al. 2020).
Materials and methods
Chemicals and kits
Yeast extract, tryptone, NaCl, ethylenediaminetetraacetic acid (EDTA), centrifugal ultrafiltration tubes, and buffer components were purchased from Merck (Darmstadt, Germany). Solvents, phenylmethylsulfonyl fluoride (PMSF), 1,4-dithiothreitol (DTT), sodium dodecyl sulfate (SDS), β-mercaptoethanol (βME), 2,6-dimethoxyphenol (2,6-DMP), syringaldazine (SGZ), 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid) (ABTS), agarose, acrylamide, bis-acrylamide, bromophenol blue, Coomassie Brilliant Blue R-250, and chromatography column were obtained from Sigma-Aldrich (Darmstadt, Germany). Ampicillin was procured from Fisher Bioreagent (Fair Lawn, NJ). Genomic DNA isolation, plasmid extraction, gel extraction, and DNA clean up kits were purchased from Favorgen (Pintung, Taiwan). Ni–NTA Rezin was purchased from Thermo Fisher Scientific (Waltham, MA).
Strains, cultivation, and vector
The strain Bacillus mojavensis TH309, previously isolated from waste plastic in our laboratory, was routinely cultured in Luria–Bertani (LB) broth at 45 °C for 24 h under 200 rpm shaking condition (Adıgüzel 2020). LB broth consists of (in g/L): 10 tryptone, 5 yeast extract, and 10 NaCl. E. coli DH5α and E. coli BL21 (DE3) were used for the amplification of plasmid and heterologous expression of the mco gene, respectively (Liu et al. 2017). The strain Bacillus subtilis ATCC6633 was used in well-diffusion experiments (Vehapi and Özçimen 2021). All strains of E. coli were grown in LB broth at 37 °C with 175 rpm agitation. Transformants were cultivated in LB broth supplemented with 50 µg/mL ampicillin under the same conditions. pET-14b (Novagen, Madison, WI, USA) was used for both cloning and expression vector (Carrillo and Borthakur 2022).
Amplification of mco gene
The genomic DNA of B. mojavensis TH309 was extracted using a FavorPrep™ Tissue Genomic DNA Extraction Mini Kit according to the manufacturer’s protocol (Esmkhani and Shams 2022). Then, it was used as a template for amplifying the mco gene. The genomic DNA of B. mojavensis TH309, extracted using a bacterial DNA isolation kit according to the manufacturer’s protocol, was used as a template for amplifying the mco gene. Forward (5'-AACGCTCGAGTTATTTATGGGGGTCAATCAC-3’) and reverse (5'-AACGCTCGAGATGACACTTGAAAAATTTGC-3') primers with XhoI restriction sites (italic) were designed based on sequence deposited with the accession number of NZ_CP051464.1 in NCBI database. The PCR reaction mixture (100 µL) was prepared with 1 µL of Q5® High-Fidelity DNA polymerase (New England Biolabs, Ipswich, MA, USA), 20 µL of 5X Q5® Reaction Buffer (5X), 20 µL of 5X Q5® High GC Enhancer, 2 µL of genomic DNA (≌ 40 ng), 5 µL of forward primer, 5 µL of reverse primer, 2 µL of dNTP (10 mM), and 45 µL nuclease-free water. The PCR was performed for 30 cycles in a thermal cycler (Biorad-T100, Hercules, CA) under the following order: 98 °C-30 s, 98 °C-30 s, 55 °C-30 s, 72 °C-47 s, and 72 °C-300 s. The amplicons were recovered from 0.8% (w/v) agarose gel using a gel extraction kit after visualization.
Cloning of the mco gene
pET14b( +) isolated using plasmid extraction kit and amplified mco gene were digested in reaction mixture containing 20 µL of DNA, 20 µL of nuclease-free water, 6 µL of 10X buffer (10 mM Tris–HCl, 10 mM MgCl2, 100 mM KCl, 0.1 mg/mL BSA, pH 8.5), and 3 µL of XhoI restriction enzyme (10 U/µL). Digestion was performed at 37 °C for 2 h. Digestion products were extracted from 0.8% (w/v) agarose gel and purified (Mathews et al. 2016). Mixtures of 7 µL Linearized pET14b( +), 1 µL digested amplicon, 1 µL buffer (400 mM Tris–HCl containing 100 mM MgCl2, 100 mM DTT, 5 mM ATP; pH 7.8), and 1 µL T4 DNA ligase (10 U/µL) were incubated overnight at 25 °C (Tabor 1989). The ligation mixture (3.5 µL) was introduced to CaCl2-competent cells of E. coli DH5α (50 µL) to heat-shock transformation (Chung and Miller 1993). Transformants were grown on LB Agar supplemented with ampicillin and then screened to detect clones containing the recombinant vector (pET14b( +)-mco). Subsequently, pET14b( +)-mco subcloned to E. coli BL21(DE3) using the same transformation method.
Sequencing of mco and sequence analysis of BmLMCO
Plasmid pET14b( +)-mco was sequenced (BM Labosis, Ankara, Turkey) using T7 forward (5’-TAATACGACTCACTATAGGG-3’) and T7 reverse (5’-GCTAGTTATTGCTCAGCGG-3’) primers. The DNA sequences of the mco from B. mojavensis TH3069 were converted to amino acid sequences to assess sequence similarity with laccases deposited in the NCBI database. Amino acid sequences were aligned using Clustal W (https://www.genome.jp/tools-bin/clustalw). Multiple sequence alignment was visualized using Jalview 2.11.2.0 (Waterhouse et al. 2009). The phylogenetic tree of BmLMCO and various laccases was then built by the neighbor-joining method through MEGA X software (Kumar et al. 2022). The secondary and tertiary structures of BmLMCO predicted with “Sequence Annotated by Structure (SAS)” (Noby et al. 2020) PSIPRED (http://bioinf.cs.ucl.ac.uk/), and SWISS-MODEL (Waterhouse et al. 2018) web tools, respectively.
Heterologous expression of BmLMCO
E. coli BL21(DE3) carrying pET14b( +)-mco was cultured at 37 °C for 18 h in LB broth supplemented with ampicillin at a concentration of 50 μg/mL. Thereafter, 500 µL of the culture were inoculated into 250 mL Erlenmeyer flasks containing 50 mL fresh medium. After the flask was incubated at 18 °C and 175 rpm until the OD600 of the culture reaches 0.4, IPTG was added to the culture at a final concentration of 0.3 mM. Then, the culture was incubated for an additional 5 h under the same conditions. At the end of incubation, the cells were retrieved by centrifugation (8000 × g, 20 min) at + 4 °C and disrupted through sonication (Noby et al. 2020). The cell lysate was centrifuged at 8000 × g for 20 min at + 4 °C and the supernatant was used as crude enzyme solution.
Purification of BmLMCO
BmLMCO was purified using HisPur™ Ni–NTA Resin equilibrated with 50 mM phosphate buffer (pH 7.4) (Cordas et al. 2022). The crude enzyme solution was loaded onto the resin and incubated at + 4 °C for 1 h. Unbound proteins were eluted from the resin by passing the equilibration buffer through the column. The bounded proteins were eluted with 50 mM phosphate buffer (pH 7.4) containing imidazole at a final concentration of 200 mM. Fractions (1 mL) in which enzyme activity was detected were pooled, concentrated, and dialyzed for further studies.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and zymogram analysis
Protein solutions were mixed with 2X loading buffer (62.5 mM Tris–HCl, pH 6.8, 2% SDS, 25% glycerol, and 0.01% bromophenol blue) with βME (5%) for SDS-PAGE analysis. Mixtures were incubated at 97 °C for 7 min and then loaded into the wells of stacking gel (5%). Separation was achieved in 10% polyacrylamide gel using Tris–glycine–SDS buffer under a constant voltage of 120 V using the Mini-Protean Tetra Cell Electrophoresis System (Bio-Rad Laboratories, Hercules, CA) (Laemmli 1970). Proteins were stained by Coomassie Brilliant Blue R-250 according to Zehr et al. (1989). The mixture of purified BmLMCO and βME-free loading buffer was subjected to electrophoresis without incubation at 97 °C. The gel was used for zymogram analysis after performing the electrophoresis as described above. First, the gel was kept in potassium phosphate buffer containing Triton X-100 at a final concentration of 10 mM for 60 min to remove SDS. Afterward, it was washed with distilled water three times. Finally, the gel was incubated in 100 mM potassium phosphate buffer containing 0.2 mM CuCl2 and 0.02 mM 2,6-DMP until a band corresponding to the laccase activity appeared (Yang et al. 2012).
Laccase assay and total protein assays
The laccase activity of BmLMCO was measured spectrophotometrically using 2,6-DMP as substrate. Briefly, the reaction was performed in 50 mM potassium phosphate buffer (pH 7.0) containing 2.96 mM 2,6-DMP and 0.27 mM CuCl2 at 40 °C for 10 min. The activity was calculated based on the increase in absorbance of the reaction mixture at 470 nm (\({\mathrm{\Delta G}}_{470}\)) using the following formula [Eq. (1)] (Baltierra-Trejo et al. 2015; Abdelgalil et al. 2022):
where \({V}_{t}\),\({ D}_{f}\), \(\mathrm{t}\), \(\upvarepsilon\), and \({V}_{e}\) are volume of the reaction mixture (mL), dilution factor, reaction time (min), absorption coefficient incorporated with correction factor (0.035645 µM−1 cm−1), and volume of enzyme solution (mL), respectively.
Total protein was determined by the Bradford method. Briefly, 1 mL of Bradford solution (10 mg of Coomassie Blue G250, 5 mL of 95% ethanol, 10 mL of 85% ortho-phosphoric acid, and 85 mL of distilled water) were added to 100 µL of protein solution. The mixture was vortexed for 5 s and incubated at room temperature for 5 min. Then, the absorbance of the mixture at 595 nm was determined spectrophotometrically (Thermo Scientific Multiskan GO). Total protein was quantified with the help of a standard curve drawn with different concentrations of BSA solution.
Biochemical characterization of BmLMCO
Lyophilized BmLMCO was used in the characterization studies. Lyophilization was carried out in the Karadeniz Advanced Technology Research and Application Center (KİTAM) using Labconco FreeZone 12 Plus (Kansas City, Missouri, USA) for 48 after the purified enzyme was frozen at -80 °C.
Effects of temperature and pH on the activity of BmLMCO
The influence of temperature on the activity of BmLMCO was investigated by assaying laccase activity in potassium phosphate buffer (pH 7.0) at different temperatures ranging from 20 to 90 °C as described earlier (laccase assay) (Birge et al. 2022). The optimum pH of BmLMCO was determined by assaying the laccase activity at different pHs ranging from 4 to 12 using sodium citrate (pH 4.0–6.0), potassium phosphate (pH 6.0–8.0), and Tris–HCl (pH 8.0–12.0) buffer systems (Olmeda et al.2021; Cilmeli et al. 2022). The final concentration of BmLMCO in reaction mixtures was 0.25 mg/mL.
Effects of temperature and pH on the stability of BmLMCO
The thermostability of BmLMCO was examined by pre-incubation of the enzyme solution (1 mg/mL, pH 7.0) at 20–90 °C for 480 min following the calculation of residual activity at optimum temperature. The pH stability of BmLMCO was evaluated by pre-incubation of the enzyme at 0.25 mg/mL of final concentration in the above-stated buffer systems over the pH 4.0–12.0 for 60 min at room temperature. The initial laccase activity of the BmLMCO was accepted as 100%.
Effects of metal ions and chemicals on the activity and stability of BmLMCO
The laccase activity of BmLMCO (0.25 mg/mL) was separately assessed in the presence of various metal ions (Ag2+, Ni2+, Fe2+, Co2+, Cu2+, K+, Mn2+, Mg2+, Ca2+, and Zn2+) and chemicals (EDTA, BME, Triton X-100, Tween 20, PMSF, and DTT) at a final concentration of 1 mM under optimum conditions. Relative activities were calculated by comparison with the enzyme activity determined without reagents and presented as percentages (Ghatge et al. 2018). To determine the effect of metal ions and reagents on the stability, relative laccase activity (%) was calculated after BmLMCO (1 mg/mL) was kept with the reagents individually at room temperature for 60 min. The initial laccase activity of the BmLMCO was accepted as 100% (Jeon and Park 2020).
Effects of solvents on activity and stability of BmLMCO
The laccase activity of BmLMCO (0.25 mg/mL) was assayed in the presence of various organic solvents at a final concentration of 10% or 30% to assess their effect on the activity. Reactions were carried out at an optimum temperature and pH of BmLMCO. To investigate the influence of organic solvent on activity, a buffered BmLMCO solution (1 mg/mL) containing organic solvents (10% or 30%) was incubated at room temperature for 60 min before enzymatic assay. Afterward, their activities were detected at optimum pH and temperature. Results were expressed as percentages after calculating by comparing the activity of solvent-free buffered BmLMCO solutions incubated in the same conditions (100%) for the same time (Cilmeli et al. 2022).
Kinetic properties of BmLMCO
The kinetic parameters (Km and Vmax) of BmLMCO were calculated via the Lineweaver–Burk plot constructed by testing the activity of the enzyme toward different concentrations of 2,6-DMP (0.2–20 mM). Assays were performed at 80 °C in Tris–HCl buffer system (pH 8.0). Data from triplicate experiments were fitted to the Michaelis–Menten equation by linear regression (Olmeda et al. 2021).
Storage stability of BmLMCO
The laccase activity was assayed after BmLMCO solution (1 mg/mL), prepared in 50 mm potassium phosphate buffer (pH 8), and was kept at -20 and + 4 °C for 0.5, 1, 2, 4, 8, 16, 32, and 48 days. The remaining activity was presented as a % relative to the initial activity.
Antibiotic removal potential of BmLMCO
The reaction was carried out in 50 mM potassium phosphate buffer containing 20 µg/mL antibiotic and 20 µg/mL lyophilized BmLMCO at 40 °C for 4 h. BmLMCO was removed by ultrafiltration after the reaction was completed. The enzyme-free reaction mixture was incubated under the same conditions for reaction control. On the other hand, 20 µg/mL of the antibiotic solution was prepared in the same buffer to control antibiotic stability. All solutions were subjected to an agar well-diffusion experiment after passing through a 10 kDa molecular weight cut-off (MWCO) ultrafiltration membrane and sterilizing by a syringe filter. The agar well-diffusion experiment was done as the following procedure (Mazmancı et al. 2022). First, approximately 5 × 107 B. subtilis ATCC6633 cells were spread on Mueller–Hinton Agar (MHA). Wells with a diameter of 8 mm were created in MHA and then filled 150 µL of suspensions. The plate was incubated at 35 °C for 16 h. The diameters of the inhibitory zone (the area where bacteria cannot grow) surrounding the wells were determined. The ability of BmLMCO to reduce the antimicrobial activity of the antibiotic was calculated according to the formula Eq. (2)
where \({T}_{0}\), \(E\), and \({T}_{4}\) are inhibition zone areas of the antibiotic solution, reaction mixture after enzymatic treatment, and enzyme-free reaction mixture, respectively. The area of the well is calculated as 50.24.
Statistical analysis
Standard deviations and standard errors of results from each experiment in triplicate were calculated using Microsoft Excel Software. The statistical meaningfulness of the results was assessed by a one-way ANOVA test using XL Toolbox NG (https://www.xltoolbox.net/) tool integrated with the Microsoft Excel Software.
Results and discussion
The gene mco was successfully amplified from the genomic DNA of B. mojavensis TH309 using the PCR technique. Electrophoretic analysis showed that the PCR product was about 1500 bp in length, as predicted. The mco gene and pet14b( +), digested with XhoI restriction enzyme, hybridized by applying the ligation process. Transformation, selection, and subcloning were performed as described in Materials and methods. Afterward, the plasmid was extracted from the selected colony grown overnight at 37 °C in the presence of ampicillin. Orientation of the gene was checked and approved by sequencing and restriction digestion with HindIII and XhoI. Thus, the recombinant plasmid was named pet14b( +)-mco. Genomic DNA, PCR product, plasmids, and digested DNA fragments, which were obtained as a result of the processes applied during cloning, are shown in Fig. 1.
Agarose gel (0.8%) DNA electrophoresis blot showing the different molecular structures used during this work: (L) ladder; (1) genomic DNA of B. mojavensis TH309; (2) amplified PCR products after amplification of mco; (3) PCR product digested by XhoI restriction enzyme; (4) pET14b( +); (5) pET14b( +) digested by XhoI restriction enzyme; (6) pET14b( +) after ligation (pET14b( +)-mco); (7) pET14b( +)-mco digested by HindIII restriction enzyme; (8) pET14b( +)-mco digested by XhoI restriction enzyme
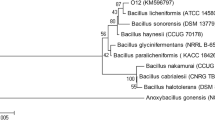
Sequences analysis showed that mco gene contains a 1584 bp length open reading frame (ORF) and encodes a protein of 527 amino acid residues. Analysis with Signal P 6.0 (https://services.healthtech.dtu.dk/service.php?SignalP) elicited that the protein did not contain any signal peptide sequences. BLASTp analysis of the protein revealed that 78.72%, 74.67%, 73.72%, 73.53%, 61.93% 53.51%, and 51.04% identity with those of laccases from Bacillus subtilis (NCBI, AEK80414.1), Lysinibacillus fusiformis (NCBI, AYW03784.1), Bacillus vallismortis (NCBI, AGR50961.1), Pseudomonas stutzeri (NCBI, AND62506.1), Bacillus amyloliquefaciens (NCBI, QHT73050.1), Bacillus pumilus (NCBI, QKI37671.1), and Bacillus licheniformis (NCBI, QGX86460.1), respectively. Multiple sequence alignment revealed that BmLMCO, like other aligned laccases, has four conserved histidine-rich sequence motifs (105–107 HGH, 153–155 HDH, 428–433 HPIHLH, 503–510 HF CHSLEH) associated with the binding of coppers to the enzyme's T1, T2, and T3 centers (Solano et al 2001) (Fig. 2). Moreover, the phylogenetic neighbor-joining tree generated from the multiple sequence alignment is shown in Fig. 3. It showed that BmLMCO is in a different evolutionary branch from fungal laccases as well as some bacterial counterparts, including Sinorhizobium meliloti, Pseudomonas stutzeri, Acetobacter ascendens, and Paenibacillus glucanolyticus.
Alignment of copper-binding motifs in BmLMCO and laccases from Bacillus subtilis, Bacillus stratosphericus, Bacillus pumilus, Bacillus amyloliquefaciens, Bacillus licheniformis, Bacillus vallismortis, Acetobacter ascendens, Paenibacillus glucanolyticus, Pseudomonas stutzeri, Thermus thermophilus, Sinorhizobium meliloti, Lysinibacillus fusiformis, Fusarium oxysporum f. sp. raphani, Phanerochaete flavidoalba. Acc. Number imply the accession number of the enzymes in NCBI database. The asterisk symbol (*) shows histidine residues associated with copper ions. The black line separates prokaryotic laccases from eukaryotic
The secondary structure prediction of BmLMCO revealed that the enzyme consists of a high number of β-sheets (≌27) and three α-helix (Supplementary material 1). It was observed that 3% and 30% of the amino acid residues of BmLMCO are located in the alpha helix and the beta layers, respectively. PSIPRED analysis also showed that BmLMCO includes three domains with boundary locations at 181th and 336th amino acids. The three-dimensional structure of BmLMCO was plotted using a template (crystal structure of the CotA native enzyme) deposited in PDB with 4989.2.A code through SWISS-MODEL (Fig. 4). BmLMCO shared an identity 76.17% with the template and had a 0.79 of GMQE value. Constructed structure of BmLMCO was highly compatible with its predicted secondary structure. Furthermore, it clearly showed that the enzyme has three cupredoxin domain with a β-barrel folding motif formed by the binding of antiparallel/parallel β-strands with β-turn and relatively large loops. Considering that laccases (except for a few archaeal and bacterial laccases) and ascorbate oxidases (found in plants and fungi) contain three cupredoxin domains, unlike ferroxidases, nitrite oxidases, and ceruloplasmin found in MCO, structural analysis shows that the enzyme is a laccase-like multicopper oxidase (Nakamura and Go 2005). According to ProtParam analysis, the computed isoelectric point (pI) of BmLMCO was 6.80. The half-life of BmLMCO was estimated to be 30 h, ˃20 h, and ˃10 h in mammalian, yeast, and E. coli cells, respectively. The aliphatic index and grand average of hydropathicity (GRAVY) values were 72.63 and -0.468, respectively, which indicated that the enzyme was thermally stable and hydrophilic.
First, lysates of E. coli BL21(DE3), E. coli BL21(DE3) with pET14b( +), and E. coli BL21(DE3) with pET14B( +)-mco cells grown in the absence and presence of IPTG were analyzed by SDS-PAGE (Fig. 4a). A distinct protein band with a molecular weight of about 65 kDa in lysates of E. coli BL21(DE3) cells harboring pET14b( +)-mco, unlike lysates of E. coli BL21(DE3) cells with and without pet14b( +) indicated that BmLMCO was expressed heterologously. Furthermore, laccase activity was only observed in the lysate of E. coli cells harboring pET14b( +)-mco.
Crude enzyme solution with 121.71 ± 6.71 U/mg of specific laccase activity subjected to Ni–NTA chromatography and thus BmLMCO was purified 3.25 ± 0.09 –fold with 32.86 ± 0.91 of the final recovery yield. BmLMCO was confirmed to have a molecular weight of ≌65 kDa by SDS-page analysis performed after purification (Fig. 5a). This value was similar to that of laccases and LMCOs from Lactobacillus plantarum (Callejón et al. 2016), B. licheniformis (Koschorreck et al. 2008; Chopra and Sondhi 2022), B. subtilis (Cheng et al. 2021), Bacillus aquimaris (Kumar et al. 2022), and B. pumilus (Reiss et al. 2011; Yan et al. 2022). Zymogram analysis resulted in the appearance of an orange band resulting from the laccase activity of BmLMCO (Fig. 5b).
SDS-PAGE showing the protein cell lysate of the normal and recombinant bacteria. (M) Molecular weight protein ladder; (1) E. coli BL21(DE3) protein cell lysate; (2) E. coli BL21(DE3)—pET14b( +) protein cell lysate; (3) E. coli BL21(DE3) pET14b( +)-mco protein cell lysate; (4) SDS-PAGE of the purified BmLMCO protein. (5) Zymography of BmLMCO after purification revealed with 2,6-DMP
The relative laccase activity of BmLMCO was above 80% in the temperature range of 50–70 °C, and still above 73% at 90 °C and 40 °C. The relative activity reached its optimum level at 80 °C (Fig. 6a). However, its activity significantly dropped off when the reaction temperature was decreased to 20 °C and 30 °C. The same temperature optima has been reported for laccases from Geobacillus sp. JS12 (Jeon and Park 2020), B. velezensis (Li et al. 2020), and Enterococcus faecium A2 (Birge et al. 2022). The temperature optima of BmLMCO was higher than those of thermostable/thermo-tolerant laccases from Thioalkalivibrio sp. ALRh (Ausec et al. 2015), Meiothermus ruber (Kalyani et al. 2016), Klebsiella pneumoniae (Liu et al. 2017), Caldalkalibacillus thermarum (Ghatge et al. 2018), Bacillus sp. PC-3 (Sharma et al 2019), Achromobacter xylosoxidans (Unuofin et al. 2019), Anoxybacillus ayderensis (Wang et al. 2020a, b), Geobacillus sp. ID17 (Atalah and Blamey 2022), Methylobacterium extorquens (Ainiwaer et al. 2022), and B. licheniformis VNQ (Sharma et al. 2022). On the other hand, the laccase obtained from Thermus thermophilus (Stevens et al. 2020), B. vallismortis fmb-103 (Zhang et al. 2013), Bacillus tequilensis (Sondhi et al. 2014), and Anoxybacillus sp. UARK-01 (Al-kahem Al-balawi et al. 2017) showed optimal activity at above 80 °C.
BmLMCO exhibited oxidizing activity against 2,6-DMP at all pH values (4–12) (Fig. 6b). Relative activity was over 70% between pH 7 and 12, and maximum activity was measured at pH 8.0 in both potassium phosphate and tris–HCl buffers. As with many bacterial laccases, the relative activity was found to be over 50% in low pH environments (pH 5–7) (Mandic et al. 2019; Cheng et al. 2021; Birge et al. 2022). Our study findings showed that BmLMCO could be used effectively in biotechnological applications in alkaline, neutral, and moderately acidic conditions.
The thermal stability of BmLMCO assessed by measuring the residual enzyme activity against 2–4 DMP after pre-incubation at temperatures ranging from 20 to 90 °C for 480 min is shown in Fig. 7a. BmLMCO was pretty stable up to 40 °C. It exhibited moderate stability at 50–70 °C. The calculated half-life of BmLMCO was 98.66 (R2:0,9268), 49.90 (R2:0,9103), and 45.97 (R2:0,8522) at 50, 60, and 70 °C, respectively. However, its activity dropped to below 50% after incubation at 80 and 90 °C for 30 min. It is well known that fungal laccases have lower thermal stability than their bacterial counterparts. On the other hand, it has been reported that some bacterial laccases can maintain their stability at high temperatures. It was reported that the half-life of laccase from B. amyloliquefaciens was 87 min at 70 °C and 42 min at 80 °C, respectively (Wang et al 2020b). Lončar et al. (2016) reported that a thermostable and organic solvent-tolerant laccase from B. licheniformis ATCC 9945a lost half of its activity after pre-incubation at 60 °C for 100 min and at 70 °C for 59 min. In another study, it was reported that the temperature value at which the Pp4816 laccase from Pediococcus pentosaceus could maintain half of its activity after 5 min of pre-incubation (T50) was 83.7 °C (Olmeda et al. 2021). The thermo-tolerant property of BmLMCO may be due to the tight packing of the native protein around the copper centers (Fan et al 2015).
a The effect of temperature on BmLMCO stability at 20 °C (△), 30 °C (○), 40 °C (◇), 50 °C (□), 60 °C (▲), 70 °C (●), 80 °C (◆), and 90 °C (■). b The effect of pH on BmLMCO stability. Sodium citrate (pH 4–6, △), potassium phosphate (pH 6–9, ○), and Tris–HCl (pH 9–12, ◇) buffers were used in experiments. Initial laccase activity of BmLMCO assessed as 100%
Results plotted in Fig. 7b showed that BmLMCO was stable over a broad pH range (pH 5–12). Even more, the activity of BmLMCO increased by 5% from that of the initial after pre-incubated at pH 8 for 1 h. The relative activity of BmLMCO, pre-incubated at pH 10.0 in Tris HCl buffer, was 97.18%. Moreover, in the same buffer, it retained more than 79% of its activity against 2,4-DMP after pre-incubation at pH 12.0. The alkaline tolerant capacity of BmLMCO was superior to that of many laccases or LMCOs from bacteria (Chauhan and Jha 2018; Ainiwaer et al. 2022).
BmLMCO activity was tested in the presence of metal ions at a final concentration of 1 mM (Fig. 8). Results showed that Ag2+ and Cu2+ enhanced the activity of BmLMCO by 1.73% and 28.53%, respectively. However, the metal ions Fe2+, Co2+, K+, Mg2+, and Zn2+ inhibited the enzyme activity significantly. Among metal ions, Ni2+, Mn2+, and Ca2+ had little influence on BmLMCO activity. The effect of metal ions on the stability of BmLMCO was also tested (data was not shown). Pre-incubation of BmLMCO with Cu2+ increased its catalytic activity toward 2,4-DMP. On the other hand, the enzyme showed residual activity ranging from 78.28% to 99.32% after pre-incubation with Ag2+, Ni2+, Fe2+, Co2+, and Ca2+ for 60 min at room temperature. It is well known that copper accelerates the activity of many laccases (Sondhi et al. 2014; Chauhan and Jha 2018). However, the effects of other metal ions on the activities of laccases cannot be generalized. Chauhan and Jha (2018) reported that 1 mM Na+, K+, Pb+2, Ca+2, Cu+2, and Co+2 promoted the activity of laccase from Pseudomonas sp. S2. In another study, Zhang et al. (2022) found that K+, Co2+, Ni2+, and Zn2+ stimulated the activity of laccase obtained from Psychrobacter sp. NJ228.
The effect of various chemicals on the activity of BmLMCO is depicted in Fig. 9. Accordingly, no noticeable reduction in BmLMCO activity was seen in the presence of DTT, tween 20, triton X-100, and SDS. Adding PMSF, βME, and EDTA to the reaction mixture decreased BmLMCO activity by 20.07%, 35.00%, and 22.58%, respectively. Pre-incubation of the enzyme with DTT and SDS for 60 min increased the activity by 9.15% and 6.13%, respectively (data were not shown). The remaining enzyme activity was still higher than 88% after pre-incubation with the other chemicals except for, βME. Similarly, SDS-activated laccases from B. licheniformis (Lu et al. 2013), B. tequilensis SN4 (Sondhi et al. 2014), and Alcaligenes faecalis (Mehandia et al. 2020) have also been reported previously. Results also showed that BmLMCO was more stable than that of many previously reported laccases or LMCOs (Neelkant et al 2020; Wang et al. 2020b; Olmeda et al. 2021).
The influences of various organic solvents on BmLMCO activity are illustrated in Fig. 10. The presence of acetone, chloroform, or isopropanol at a final concentration of 30% in the reaction mixture caused an increase in the activity up to 123.83%. A moderate increase in laccase activity was also detected by pre-incubation of BmLMCO with chloroform at a final concentration of 10% (data was not shown). On the other hand, ethanol affected adversely the activity and stability. In addition, BmLMCO was pretty much compatible with other tested solvents.
The results of the storage stability study are illustrated in Fig. 11a. The relative activity of lyophilized BmLMCO was 57.53% and 49.86% after the enzyme was stored for 32 days at – 20 °C and + 4 °C, respectively. In addition, BmLMCO maintained 37.98% and 30.47% of its activity at the end of the 48-day storage period at -20 °C and + 4 °C, respectively. Xu et al. (2015) observed a more than about 60% reduction in the activity of laccase from T. versicolor after 10 days of storage at + 4 °C. Qiu et al. (2021) reported an activity loss in laccase from Aspergillus oryzae was over 80% after a storage period of 15 days at + 4 °C.
Km and Vmax values of BmLMCO were 0.98 mM and 93.45 µmol/min, respectively, with 2,6-DMP as the substrate (Fig. 11b). The Km value of BmLMCO was lower than those of laccases from B. tequilensis SN4 (Sondhi et al. 2014), Streptomyces viridochromogenes (Trubitsina et al. 2015), Thioalkalivibrio sp. ALRh (Ausec et al. 2015), Lactobacillus plantarum (Callejón et al. 2016), Paenibacillus glucanolyticus (Mathews et al. 2016), K. pneumoniae (Liu et al. 2017), Geobacter metallireducens (Berini et al 2018), and P. pentosaceus (Olmeda et al. 2021), while higher than those of laccases from M. ruber (Kalyani et al. 2016), Aquisalibacillus elongatus (Rezaei et al. 2017), A. faecalis (Mehandia et al. 2020), and B. licheniformis VNQ (Sharma et al. 2022) for the same substrate.
Antibiotic removal potential of BmLMCO
The potential for the use of BmLMCO in antibiotic removal was evaluated based on the loss in antimicrobial activity of antibiotics. Cefprozil, gentamicin, and erythromycin, classified as cephalosporin, aminoglycoside, and macrolide antibiotics, were used and treated with BmLMCO in a potassium phosphate buffer system at 40 °C for 4 h. A decrease in their antimicrobial effect was observed with well-diffusion experiment is shown in Fig. 12(d-f). BmLMCO caused the 72.3 ± 1.5%, 79.6 ± 6.4%, and 19.7 ± 4.1% decreases in the antimicrobial activity of cefprozil, gentamycin, and erythromycin, respectively. The excellent effect of BmLMCO on gentamicin and cefprozil is most likely due to the opening of the beta-lactam ring, which causes the antimicrobial effect in antibiotics as a result of catalytic activity. Their chemical structures are shown in Supplementary material 2. To sum up, results showed that BmLMCO can be used for removal of antibiotics.
a Antimicrobial activity of cefprozil solution (T0), the enzyme-free reaction mixture (T4), and the reaction mixture after enzymatic treatment. b Antimicrobial activity of gentamicin solution (T0), the enzyme-free reaction mixture (T4), and reaction mixture after enzymatic treatment. c Antimicrobial activity of erythromycin solution (T0), the enzyme-free reaction mixture (T4), and reaction mixture after enzymatic treatment
Conclusion
The mco gene from B. mojavensis is cloned and expressed in E. coli, successfully. The enzyme called BmLMCO showed excellent activity and moderate stability at elevated temperatures. It was active and stable over a broad pH range. BmLMCO tolerated various metal ions, chemicals, and solvents considerably. In addition, it diminished the antimicrobial activity of cefprozil, gentamycin, and erythromycin. BmLMCO, with the aforementioned remarkable properties, is a promising candidate to meet the needs of biotechnological applications like the degradation of emerging pollutants.
Availability of data and materials
Data will be made available on reasonable request.
References
Abdelgalil SA, Soliman NA, Abo-Zaid GA, Abdel-Fattah YR (2022) Bioprocessing strategies for cost-effective large-scale production of bacterial laccase from Lysinibacillus macroides LSO using bio-waste. Int J Environ Sci Technol 19:1633–1652. https://doi.org/10.1007/s13762-021-03231-3
Adıgüzel AO (2020) Production and characterization of thermo-, halo-and solvent-stable esterase from Bacillus mojavensis TH309. Biocatal Biotransform 38(3):210–226. https://doi.org/10.1080/10242422.2020.1715370
Ainiwaer A, Liang Y, Ye X, Gao R (2022) Characterization of a novel Fe2+ activated non-blue laccase from Methylobacterium extorquens. Int J Mol Sci 23(17):9804. https://doi.org/10.3390/ijms23179804
Al-kahem Al-balawi TH, Wood AL, Solis A, Cooper T, Barabote RD (2017) Anoxybacillus sp. strain UARK-01, a new thermophilic soil bacterium with hyperthermostable alkaline laccase activity. Curr Microbiol 74:762–771. https://doi.org/10.1007/s00284-017-1239-5
Al-sareji OJ, Meiczinger M, Somogyi V, Al-Juboori RA, Grmasha RA, Stenger-Kovács C, Jakab M, Hashim KS (2023) Removal of emerging pollutants from water using enzyme-immobilized activated carbon from coconut shell. J Environ Chem Eng 11(3):109803. https://doi.org/10.1016/j.jece.2023.109803
Atalah J, Blamey JM (2022) Isolation and characterization of a novel laccase from an Antarctic thermophilic Geobacillus. Antarct Sci 34(4):289–297. https://doi.org/10.1017/S0954102022000074
Ausec L, Črnigoj M, Šnajder M, Ulrih NP, Mandic-Mulec I (2015) Characterization of a novel high-pH-tolerant laccase-like multicopper oxidase and its sequence diversity in Thioalkalivibrio sp. Appl Microbiol Biotechnol 99:9987–9999. https://doi.org/10.1007/s00253-015-6843-3
Baltierra-Trejo E, Márquez-Benavides L, Sánchez-Yáñez JM (2015) Inconsistencies and ambiguities in calculating enzyme activity: the case of laccase. J Microbiol Methods 119:126–131. https://doi.org/10.1016/j.mimet.2015.10.007
Berini F, Verce M, Ausec L, Rosini E, Tonin F, Pollegioni L, Mandić-Mulec I (2018) Isolation and characterization of a heterologously expressed bacterial laccase from the anaerobe Geobacter metallireducens. Appl Microbiol Biotechnol 102:2425–2439. https://doi.org/10.1007/s00253-018-8785-z
Birge A, Alcicek EA, Baltaci MO, Sisecioglu M, Adiguzel A (2022) Purification and biochemical characterization of a new thermostable laccase from Enterococcus faecium A2 by a three-phase partitioning method and investigation of its decolorization potential. Arch Microbiol 204(8):533. https://doi.org/10.1007/s00203-022-03054-x
Brander S, Mikkelsen JD, Kepp KP (2014) Characterization of an alkali-and halide-resistant laccase expressed in E. coli: CotA from Bacillus clausii. PLoS ONE 9(6):e99402. https://doi.org/10.1371/journal.pone.0099402
Callejón S, Sendra R, Ferrer S, Pardo I (2016) Cloning and characterization of a new laccase from Lactobacillus plantarum J16 CECT 8944 catalyzing biogenic amines degradation. Appl Microbiol Biotechnol 100(7):3113–3124. https://doi.org/10.1007/s00253-015-7158-0
Carrillo JT, Borthakur D (2022) Heterologous expression and characterization of a thermoalkaliphilic SAM-synthetase from giant leucaena (Leucaena leucocephala subsp glabrata). Plant Physiol Biochem 181:42–49. https://doi.org/10.1016/j.plaphy.2022.04.009
Chauhan PS, Jha B (2018) Pilot scale production of extracellular thermo-alkali stable laccase from Pseudomonas sp. S2 using agro waste and its application in organophosphorous pesticides degradation. J Chem Technol Biotechnol 93(4):1022–1030. https://doi.org/10.1002/jctb.5454
Cheng CM, Patel AK, Singhania RR, Tsai CH, Chen SY, Chen CW, Di Dong C (2021) Heterologous expression of bacterial CotA-laccase, characterization and its application for biodegradation of malachite green. Bioresour Technol 340:125708. https://doi.org/10.1016/j.biortech.2021.125708
Chopra NK, Sondhi S (2022) Cloning, expression and characterization of laccase from Bacillus licheniformis NS2324 in E. coli application in dye decolorization. Int J Biol Macromol 206:1003–1011
Chung CT, Miller RH (1993) Preparation and storage of competent Escherichia coli cells. Meth Enzymol 218:621–627. https://doi.org/10.1016/0076-6879(93)18045-E
Cilmeli S, Doruk T, Könen-Adıgüzel S, Adıgüzel AO (2022) A thermostable and acidophilic mannanase from Bacillus mojavensis: its sustainable production using spent coffee grounds, characterization, and application in grape juice processing. Biomass Convers Biorefin. https://doi.org/10.1007/s13399-022-02602-1
Claus H, Filip Z (1997) The evidence of a laccase-like enzyme activity in a Bacillus sphaericus strain. Microbiol Res 152(2):209–216. https://doi.org/10.1016/S0944-5013(97)80014-6
Cordas CM, Nguyen GS, Valério GN, Jønsson M, Söllner K, Aune IH, Wentzel A, Moura JJ (2022) Discovery and characterization of a novel Dyp-type peroxidase from a marine actinobacterium isolated from Trondheim fjord. Norway. J Inorg Biochem 226:111651. https://doi.org/10.1016/j.jinorgbio.2021.111651
Dong CD, Tiwari A, Anisha GS, Chen CW, Singh A, Haldar D, Patel AK, Singhania RR (2023) Laccase: a potential biocatalyst for pollutant degradation. Environ Pollut. https://doi.org/10.1016/j.envpol.2023.120999
Ekeoma BC, Ekeoma LN, Yusuf M, Haruna A, Ikeogu CK, Merican ZMA, Kamyab H, Pham CQ, Vo DVN, Chelliapan S (2023) Recent advances in the biocatalytic mitigation of emerging pollutants: a comprehensive review. J Biotechnol 369:14–34. https://doi.org/10.1016/j.jbiotec.2023.05.003
Esmkhani M, Shams S (2022) Cutaneous infection due to Bacillus cereus: a case report. BMC Infect Dis 22(1):393. https://doi.org/10.1186/s12879-022-07372-9
Fan L, Zhao M, Wang Y (2015) Expression of CotA laccase in Pichia pastoris and its electrocatalytic sensing application for hydrogen peroxide. Appl Microbiol Biotechnol 99:9483–9493. https://doi.org/10.1007/s00253-015-6720-0
García-Delgado C, Eymar E, Camacho-Arévalo R, Petruccioli M, Crognale S, D’Annibale A (2018) Degradation of tetracyclines and sulfonamides by stevensite-and biochar-immobilized laccase systems and impact on residual antibiotic activityJ. Chem Technol Biotechnol 93(12):3394–3409. https://doi.org/10.1002/jctb.5697
Ghatge S, Yang Y, Song WY, Kim TY, Hur HG (2018) A novel laccase from thermoalkaliphilic bacterium Caldalkalibacillus thermarum strain TA2. A1 able to catalyze dimerization of a lignin model compound. Appl Microbiol Biotechnol 102:4075–4086. https://doi.org/10.1007/s00253-018-8898-4
Gigli V, Piccinino D, Avitabile D, Antiochia R, Capecchi E, Saladino R (2022) Laccase mediator cocktail system as a sustainable skin whitening agent for deep eumelanin decolorization. Int J Mol Sci 23(11):6238. https://doi.org/10.3390/ijms23116238
Jeon SJ, Park JH (2020) Refolding, characterization, and dye decolorization ability of a highly thermostable laccase from Geobacillus sp. Protein Expr Purif 173:105646. https://doi.org/10.1016/j.pep.2020.105646
Kalyani DC, Munk L, Mikkelsen JD, Meyer AS (2016) Molecular and biochemical characterization of a new thermostable bacterial laccase from Meiothermus ruber DSM 1279. RSC Adv 6(5):3910–3918. https://doi.org/10.1039/C5RA24374B
Koschorreck K, Richter SM, Ene AB, Roduner E, Schmid RD, Urlacher VB (2008) Cloning and characterization of a new laccase from Bacillus licheniformis catalyzing dimerization of phenolic acids. Appl Microbiol Biotechnol 79:217–224. https://doi.org/10.1007/s00253-008-1417-2
Kumar A, Singh AK, Bilal M, Chandra R (2022) Sustainable production of thermostable laccase from agro-residues waste by Bacillus aquimaris AKRC02. Catal Lett 152(6):1784–1800. https://doi.org/10.1007/s10562-021-03753-y
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680–685. https://doi.org/10.1038/227680a0
Li T, Huang L, Li Y, Xu Z, Ge X, Zhang Y, Wang N, Wang S, Yang W, Lu F, Liu Y (2020) The heterologous expression, characterization, and application of a novel laccase from Bacillus velezensis. Sci Total Environ 713:136713. https://doi.org/10.1016/j.scitotenv.2020.136713
Liu Y, Huang L, Guo W, Jia L, Fu Y, Gui S, Lu F (2017) Cloning, expression, and characterization of a thermostable and pH-stable laccase from Klebsiella pneumoniae and its application to dye decolorization. Process Biochem 53:125–134. https://doi.org/10.1016/j.procbio.2016.11.015
Liu J, Li B, Li Z, Yang F, Chen B, Chen J, Li H, Jiang Z (2023) Deciphering the alkaline stable mechanism of bacterial laccase from Bacillus pumilus by molecular dynamics simulation can improve the decolorization of textile dyes. J Hazard Mater 443:130370. https://doi.org/10.1016/j.jhazmat.2022.130370
Lončar N, Božić N, Vujčić Z (2016) Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ATCC 9945a. J Mol Catal B Enzym 134:390–395. https://doi.org/10.1016/j.molcatb.2016.06.005
Lou Q, Wu Y, Ding H, Zhang B, Zhang W, Zhang Y, Han L, Liu M, He T, Zhong J (2022) Degradation of sulfonamides in aquaculture wastewater by laccase–syringaldehyde mediator system: Response surface optimization, degradation kinetics, and degradation pathway. J Hazard Mater 432:128647. https://doi.org/10.1016/j.jhazmat.2022.128647
Lu L, Wang TN, Xu TF, Wang JY, Wang CL, Zhao M (2013) Cloning and expression of thermo-alkali-stable laccase of Bacillus licheniformis in Pichia pastoris and its characterization. Bioresour Technol 134:81–86. https://doi.org/10.1016/j.biortech.2013.02.015
Mandic M, Djokic L, Nikolaivits E, Prodanovic R, O’Connor K, Jeremic S, Topakas E, Nikodinovic-Runic J (2019) Identification and characterization of new laccase biocatalysts from Pseudomonas species suitable for degradation of synthetic textile dyes. Catalysts 9(7):629. https://doi.org/10.3390/catal9070629
Martins LO, Soares CM, Pereira MM, Teixeira M, Costa T, Jones GH, Henriques AO (2002) Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J Biol Chem 277(21):18849–18859. https://doi.org/10.1074/jbc.M200827200
Mathews SL, Smithson CE, Grunden AM (2016) Purification and characterization of a recombinant laccase-like multi-copper oxidase from Paenibacillus glucanolyticus SLM1. J Appl Microbiol 121(5):1335–1345. https://doi.org/10.1111/jam.13241
Mazmancı B, Könen Adıgüzel S, Sadak YS, Yetkin D, Ay H, Adıgüzel AO (2022) Antimicrobial, antibiofilm, and anticancer potential of silver nanoparticles synthesized using pigment-producing Micromonospora sp. SH121. Prep Biochem Biotechnol. https://doi.org/10.1080/10826068.2022.2101001
Mehandia S, Sharma SC, Arya SK (2020) Isolation and characterization of an alkali and thermostable laccase from a novel Alcaligenes faecalis and its application in decolorization of synthetic dyes. Biotechnol Rep 25:e00413. https://doi.org/10.1016/j.btre.2019.e00413
Mojtabavi S, Khoshayand MR, Torshabi M, Gilani K, Fazeli MR, Faramarzi MA, Samadi N (2022) Formulation, characterization, and bioactivity assessments of a laccase-based mouthwash. J Drug Deliv Sci Technol 69:103128. https://doi.org/10.1016/j.jddst.2022.103128
Nakamura K, Go N (2005) Function and molecular evolution of multicopper blue proteins. Cell Mol Life Sci 62:2050–2066. https://doi.org/10.1007/s00018-004-5076-x
Navada KK, Kulal A (2019) Enzymatic degradation of chloramphenicol by laccase from Trametes hirsuta and comparison among mediators. Int Biodeterior Biodegrad 138:63–69. https://doi.org/10.1016/j.ibiod.2018.12.012
Neelkant KS, Shankar K, Jayalakshmi SK, Sreeramulu K (2020) Purification, biochemical characterization, and facile immobilization of laccase from Sphingobacterium ksn-11 and its application in transformation of diclofenac. Appl Biochem Biotechnol 192:831–844. https://doi.org/10.1007/s12010-020-03371-1
Noby N, Hussein A, Saeed H, Embaby AM (2020) Recombinant cold-adapted halotolerant, organic solvent-stable esterase (estHIJ) from Bacillus halodurans. Anal Biochem 591:113554. https://doi.org/10.1016/j.ab.2019.113554
Olmeda I, Casino P, Collins RE, Sendra R, Callejon S, Huesa J, Soares AS, Ferrer S, Pardo I (2021) Structural analysis and biochemical properties of laccase enzymes from two Pediococcus species. Microb Biotechnol 14(3):1026–1043. https://doi.org/10.1111/1751-7915.13751
Parrilli E, Tedesco P, Fondi M, Tutino ML, Giudice AL, de Pascale D, Fani R (2021) The art of adapting to extreme environments: the model system Pseudoalteromonas. Phys Life Rev 36:137–161. https://doi.org/10.1016/j.plrev.2019.04.003
Qiu X, Wang S, Miao S, Suo H, Xu H, Hu Y (2021) Co-immobilization of laccase and ABTS onto amino-functionalized ionic liquid-modified magnetic chitosan nanoparticles for pollutants removal. J Hazard Mater 401:123353. https://doi.org/10.1016/j.jhazmat.2020.123353
Reiss R, Ihssen J, Thöny-Meyer L (2011) Bacillus pumilus laccase: a heat stable enzyme with a wide substrate spectrum. BMC Biotechnol 11(1):1–11. https://doi.org/10.1186/1472-6750-11-9
Rezaei S, Shahverdi AR, Faramarzi MA (2017) Isolation, one-step affinity purification, and characterization of a polyextremotolerant laccase from the halophilic bacterium Aquisalibacillus elongatus and its application in the delignification of sugar beet pulp. Bioresour Technol 230:67–75. https://doi.org/10.1016/j.biortech.2017.01.036
Roberts MS, Nakamura LK, Cohan FM (1994) Bacillus mojavensis sp. Nov., distinguishable from Bacillus subtilis by sexual isolation, divergence in DNA sequence, and differences in fatty acid composition. Int J Syst Evol Microbiol 44(2):256–264. https://doi.org/10.1099/00207713-44-2-256
Ruijssenaars HJ, Hartmans S (2004) A cloned Bacillus halodurans multicopper oxidase exhibiting alkaline laccase activity. Appl Microbiol Biotechnol 65:177–182. https://doi.org/10.1007/s00253-004-1571-0
Sarafpour M, Alihosseini F, Bayat M (2022) New laccase-mediated system utilized for bio-discoloration of indigo-dyed denim fabrics. Appl Biochem Biotechnol 194(12):5848–5861. https://doi.org/10.1007/s12010-022-04066-5
Sharma V, Ayothiraman S, Dhakshinamoorthy V (2019) Production of highly thermo-tolerant laccase from novel thermophilic bacterium Bacillus sp. PC-3 and its application in functionalization of chitosan film. J Biosci Bioeng 127(6):672–678. https://doi.org/10.1016/j.jbiosc.2018.11.008
Sharma V, Pugazhenthi G, Vasanth D (2022) Production and characterization of a novel thermostable laccase from Bacillus licheniformis VNQ and its application in synthesis of bioactive 1, 4-naphthoquinones. J Biosci Bioeng 133(1):8–16. https://doi.org/10.1016/j.jbiosc.2021.09.008
Sinirlioglu ZA, Sinirlioglu D, Akbas F (2013) Preparation and characterization of stable cross-linked enzyme aggregates of novel laccase enzyme from Shewanella putrefaciens and using malachite green decolorization. Bioresour Technol 146:807–811. https://doi.org/10.1016/j.biortech.2013.08.032
Solano F, Lucas-Elio P, López-Serrano D, Fernández E, Sanchez-Amat A (2001) Dimethoxyphenol oxidase activity of different microbial blue multicopper proteins. FEMS Microbiol Lett 204(1):175–181. https://doi.org/10.1111/j.1574-6968.2001.tb10882.x
Sondhi S, Sharma P, Saini S, Puri N, Gupta N (2014) Purification and characterization of an extracellular, thermo-alkali-stable, metal tolerant laccase from Bacillus tequilensis SN4. PLoS ONE 9(5):e96951. https://doi.org/10.1371/journal.pone.0096951
Stevens JC, Rodgers DW, Dumon C, Shi J (2020) Characterization and enzyme engineering of a hyperthermophilic laccase toward improving its activity in ionic liquid. Front Energy Res 8:158. https://doi.org/10.3389/fenrg.2020.00158
Sun F, Yu D, Zhou H, Lin H, Yan Z, Wu A (2023) CotA laccase from Bacillus licheniformis ZOM-1 effectively degrades zearalenone, aflatoxin B1 and alternariol. Food Control 145:109472. https://doi.org/10.1016/j.foodcont.2022.109472
Tabor S (1989) DNA ligases. Curr Protoc Mol Biol 8(1):3–14. https://doi.org/10.1002/0471142727.mb0314s08
Trubitsina LI, Tishchenko SV, Gabdulkhakov AG, Lisov AV, Zakharova MV, Leontievsky AA (2015) Structural and functional characterization of two-domain laccase from Streptomyces viridochromogenes. Biochimie 112:151–159. https://doi.org/10.1016/j.biochi.2015.03.005
Unuofin JO, Moubasher HA, Okoh AI, Nwodo UU (2019) Production of polyextremotolerant laccase by Achromobacter xylosoxidans HWN16 and Citrobacter freundii LLJ16. Biotechnol Rep 22:e00337. https://doi.org/10.1016/j.btre.2019.e00337
Vehapi M, Özçimen D (2021) Antimicrobial and bacteriostatic activity of surfactants against B. subtilis for microbial cleaner formulation. Arch Microbiol 203(6):3389–3397. https://doi.org/10.1007/s00203-021-02328-0
Wang H, Huang L, Li Y, Ma J, Wang S, Zhang Y, Ge X, Wang N, Lu F, Liu Y (2020a) Characterization and application of a novel laccase derived from Bacillus amyloliquefaciens. Int J Biol Macromol 150:982–990. https://doi.org/10.1016/j.ijbiomac.2019.11.117
Wang J, Chang F, Tang X, Li W, Yin Q, Yang Y, Hu Y (2020b) Bacterial laccase of Anoxybacillus ayderensis SK3-4 from hot springs showing potential for industrial dye decolorization. Ann Microbiol 70:51. https://doi.org/10.1186/s13213-020-01593-6
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. https://doi.org/10.1093/bioinformatics/btp033
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res Spec Publ 46:W296–W303. https://doi.org/10.1093/nar/gky427
Wen X, Zeng Z, Du C, Huang D, Zeng G, Xiao R, Lai C, Xu P, Zhang C, Wan J, Hu L, Yin L, Zhou C, Deng R (2019) Immobilized laccase on bentonite-derived mesoporous materials for removal of tetracycline. Chemosphere 222:865–871. https://doi.org/10.1016/j.chemosphere.2019.02.020
Xu R, Tang R, Zhou Q, Li F, Zhang B (2015) Enhancement of catalytic activity of immobilized laccase for diclofenac biodegradation by carbon nanotubes. Chem Eng J 262:88–95. https://doi.org/10.1016/j.cej.2014.09.072
Yan N, Ma H, Yang CX, Liao XR, Guan ZB (2022) Improving the decolorization activity of Bacillus pumilus W3 CotA-laccase to Congo red by rational modification. Enzyme Microb Technol 155:1099. https://doi.org/10.1016/j.enzmictec.2021.109977
Yang L, Wenfeng Z, Yadong L, Xingguo W (2012) Cloning of multicopper oxidase gene from Ochrobactrum sp. 531 and characterization of its alkaline laccase activity towards phenolic substrates. Adv Biol Chem. 2(3):248–255. https://doi.org/10.4236/abc.2012.23031
Zehr BD, Savin TJ, Hall RE (1989) A one-step, low background Coomassie staining procedure for polyacrylamide gels. Anal Biochem 182(1):157–159. https://doi.org/10.1016/0003-2697(89)90734-3
Zhang C, Zhang S, Diao H, Zhao H, Zhu X, Lu F, Lu Z (2013) Purification and characterization of a temperature-and pH-stable laccase from the spores of Bacillus vallismortis fmb-103 and its application in the degradation of malachite green. J Agric Food Chem 61(23):5468–5473. https://doi.org/10.1021/jf4010498
Zhang C, You S, Zhang J, Qi W, Su R, He Z (2020) An effective in-situ method for laccase immobilization: excellent activity, effective antibiotic removal rate and low potential ecological risk for degradation products. Bioresour Technol 308:123271. https://doi.org/10.1016/j.biortech.2020.123271
Zhang A, Hou Y, Wang Q, Wang Y (2022) Characteristics and polyethylene biodegradation function of a novel cold-adapted bacterial laccase from Antarctic sea ice psychrophile Psychrobacter sp. NJ228. J Hazard Mater 439:129656. https://doi.org/10.1016/j.jhazmat.2022.129656
Funding
This study was supported by the Scientific and Technological Research Council of Türkiye (TUBİTAK) under Project No. 122Z601.
Author information
Authors and Affiliations
Contributions
AOA: project administration, validation, data analysis, visualization, writing, and methodology. SK-A: writing, investigation, and validation. SC: formal analysis. BM: methodology, writing, and curation. EY: data analysis, writing, and validation. SÜ-O: methodology, investigation, and validation. NGK: formal analysis. MAM: supervision, writing, and curation.
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was reported to the authors. In addition, the authors have no financial interests to disclose.
Ethical approval
This study does not involve the need for ethical approval.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Communicated by Yusuf Akhter.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Adigüzel, A.O., Könen-Adigüzel, S., Cilmeli, S. et al. Heterologous expression, purification, and characterization of thermo- and alkali-tolerant laccase-like multicopper oxidase from Bacillus mojavensis TH309 and determination of its antibiotic removal potential. Arch Microbiol 205, 287 (2023). https://doi.org/10.1007/s00203-023-03626-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-023-03626-5