Abstract
Aim of this study was to optimize the production of Ligninolytic enzyme for the degradation of complex pollutants present in pulp paper industrial effluent (PPIE). Two ligninolytic enzyme-producing bacterial strains were isolated from PPIE and identified as Bacillus paramycoides strain BL2 (MZ676667) and Micrococcus luteus strains BL3 (MZ676668). The identified bacterial strain Bacillus paramycoides strain BL2 showed optimum production of LiP (4.30 U/ml), MnP (3.38 U/ml) at 72 h of incubation, while laccase (4.43 U/ml) at 96 h of incubation. While, Micrococcus luteus strains BL3 produced maximum LiP (3.98) and MnP (3.85 U/ml) at 96 h of incubation and maximum laccase (3.85 U/ml) at 72 h of incubation, pH 7-8, and temperatures of 30–35 °C. Furthermore, in the presence of glucose (1.0%) and peptone (0.5%) as nutrient sources, the enzyme activity of consortium leads to reduction of lignin (70%), colour (63%) along with COD (71%) and BOD (58%). The pollutants detected in control i.e. 3.6-Dioxa-2,7-disilaoctane, 2-Heptnoic acid,trimethylsilyl ester, 7-Methyldinaphtho [2,1-b,1’,2’-d] silole, Hexadeconoic acid, trimethylysilyl ester, Methyl1(Z)-3,3-dipheny.1-4-hexenoale, 2,6,10,14,18,22-Tetracosahexane,2,2-dimethylpropyl(2Z,6E)-10,11epoxy5,6 Dihyrostigmasterol, acetate were completely diminished. The toxicity of PPIE was reduced up to 75%. Hence, knowledge of this study will be very useful for industrial sector for treatment of complex wastewater.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The pulp paper industry's discharged wastewater comprises around 700 organic and inorganic contaminants that are directly responsible for global warming. following biological pollution, soil and water pollution as well as chemical treatment (Sharma et al. 2017). The pulp paper Industrial effluent is one of the largest freshwater consumers and generates huge amounts of wastewater worldwide (CPCB 2015). Aerobic bacteria were transformations of the lignin-derived aromatic compounds. To depolymerize and mineralize lignin, several bacteria and white-rot fungi have developed an oxidative and unspecific mechanism including extracellular enzymes, low molecular weight metabolites, and activated oxygen species (Janusz et al. 2017). In recent years, microbial techniques for wastewater degrading process optimization under laboratory circumstances have piqued the interest of many academics around the world, prompting them to investigate practical, cost-effective, and long-term wastewater treatment technologies (Kaushik et al. 2010). Lignin degradation: microorganisms, enzymes involved genomes analysis and evolution. Microorganisms and their enzymes are being studied for their possible use in the breakdown of aromatic pollutants that cause environmental problems owing to the system's lack of selectivity in lignin depolmerization (Janusz et al. 2017). Extracellular enzymatic systems that produce extracellular hydrogen peroxide (H2O2) include Lignin peroxidases, laccases, and other oxidases (Dashtban et al. 2010). Dependent on the species, strains, and culture situations, these enzyme systems have varied properties (Perez-Garcia et al. 2011). Pulp paper Industrial effluent, pH values are generally alkaline (pH 7.0–9.0) and the requirement to reduce the pH before fungal inoculation maintains the conditions there is an additional cost. Unlike fungi, bacteria can play an important role in the bioremediation of PPIE without requiring pH adjustment because they survive in both acidic and alkaline condition. Bacterial laccases can use a wide variety of substrates and are active at high pH and temperatures (Kumar and Chandra 2020). Lignin peroxidase (LiP) may oxidise non-phenolic components directly, whereas manganese peroxidase (MnP) and laccase oxidise phenolic units preferentially but moreover act on non-phenolic units in the existence of mediators (Hammel et al. 1989; Long et al. 2018). Recently, ligninolytic enzyme activity has been discovered in several bacteria that have catalytic properties and can oxidize lignocellulosic substrates, mineralize 14C-milled wood lignin and other organic pollutants. Pulp paper Industrial effluent is substantial contributors to the environment's Persistent Organic Pollutants (POPs). More than 200 organic and 700 inorganic chemicals have been discovered now pulp paper effluents by several studies (Yadav and Chandra 2018; Singh and Chandra 2019). POPs are compounds that have a high resistance to chemical/biological breakdown, are mobile in the environment, bioaccumulate in human and animal tissues, and create an adverse effect on human health and the environment even at extremely low concentrations. The POPs have also been proven genotoxic or DNA damaging, carcinogenic, endocrine disrupting chemicals (Marlatt et al. 2022). The discharge of untreated effluents from pulp and paper mills into water bodies degrades the water quality. POPs present in industrial waste could cause chronic toxic effects and endocrine disruptions. As a result, USEPA considers POPs to be priority pollutants, and their discharge is strictly banned (Boguniewicz and Kłosok 2020). Several physical, biological and enzymatic techniques have been developed for the degradation of POPs. Biological degradation of these types of recalcitrant pollutants was found most effective which is might be due to the presence of ligninolytic enzyme activity. Degradation of POPs was also easier for biofilm-forming bacteria which gives extra protection to ligninolytic enzymes responsible for the breakdown of recalcitrant pollutants (Tripathi et al. 2021a, b). Several studies showed that the involvement of ligninolytic enzyme activity, but the effect of different environmental conditions on enzyme activity is not reported so far. Hence, in this study we isolate the bacterial strains from the soil contaminated with PPIE and examined ligninolytic enzyme activity. Further, the effect of sources and concentration of carbon and nitrogen on ligninolytic enzyme activity was evaluated. Furthermore, the effect of pH, temperature and metal concentration were also evaluated on ligninolytic enzyme activity. Individual and consortium of potential bacterial strains were also evaluated for PPIE degradation. Toxicity of PPIE before and after bacterial treatment was also evaluated. Various persistent organic pollutants and metals are a major component of industrial effluent, hence finding of this study is very useful to know the mechanism of degradation of complex wastewater. The new knowledge regarding the effect of environmental factor on ligninolytic enzyme activity will be helpful for the development of a better management and treatment regime for the industries in India. Finding of this study fulfill the gap between ligninolytic enzyme-producing bacteria and optimized environmental and nutritional conditions for the degradation and detoxification of PPIE. The presence of harmful pollutants in wastewater, known as ROPs, is significant for a variety of reasons. In this study, we have tried to address this gap by enzyme-producing bacteria isolated from pulp paper industrial effluent as well as optimized environmental and nutritional conditions for degradation and detoxification of PPIE. The untreated and bacterial treated PPIE was analyzed by FTIR and GC–MS analysis has been performed to identify the degraded and transformed residual organic pollutants (ROPs). The toxicity reduction of bacterial-treated PPIE was also assessed using the seed germination of Phaseolus mungo L.
Results
Isolation and screening of ligninolytic enzyme-producing bacterial strains
To isolate ligninolytic enzyme-producing bacteria, kraft lignin (KL) was introduced as the only carbon source for MSM agar. Additional carbon and nitrogen sources, glucose (1%) and peptone (05) were introduced as co-substrates to enhance lignin-degrading bacterial growth and ligninolytic enzyme activity. In this experiment, 48 bacterial strains designated as BL1 to BL48 were isolated on MSM agar plates and purified using the streaking technique. All bacterial strains were collected, and screening was done using MSM agar media containing guiacol, methylene blue, and phenol red on separate plates using the plate assay technique for ligninolytic enzyme activity. All 48 bacterial strains were tested for ligninolytic enzyme production. Only 12 bacterial strains produced positive findings for various ligninolytic enzymes, among these only two strains (BL2 and BL3) produced the highest activity of LiP, MnP and laccase.
16S RNA identification of potential ligninolytic bacterial strains
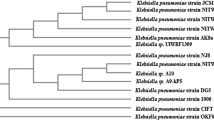
Using 16S rRNA sequencing, the selected prospective bacterial strains BL2 and BL3 with the highest ligninolytic enzyme production were found Bacillus paramycoides strain BL2 and Micrococcus luteus strain BL3 are related genera with 97% to 100% similarity. Phylogenetic tree showing relation with potential bacterial with other bacterial strains is shown in Fig. 1. The degree to which various species are related was determined by comparing their sequences. The strains' respective nucleotide sequences were deposited in the gene bank under accession numbers MZ676667 and MZ676668.
Effect of different environmental conditions on ligninolytic enzymes activity
Optimization of different substrate and incubation period
Result of the effect of a different substrate during ligninolytic activity analysis showed that Methyl blue (40 mM) for LiP, Phenol red 80 mM) for MnP and guaiacol (20 mM) for laccase in both isolates of this study (Fig. 2a–f). Because the isolated bacterial strains in this study are capable of producing all main ligninolytic enzymes (LiP, MnP, and Lac), the results are encouraging. Bacillus paramycoides strain BL2 produces LiP (2.174 and 1.815U/ml), MnP (1.275 and 0.823 U/ml), and laccase (1.528 and 2.483 U/ml), whereas Micrococcus luteus strain BL3 produces LiP (1.075 and 2.121 U/ml), MnP(1.621and 0.268 U/ml), and laccase (1.47 and 2.942 U/ml) after 72 and 96 h, respectively. Bacillus paramycoidesstrain BL2 showed maximum LiP (2.174 U/ml) and MnP (1.275 U/ml) activity at 72 h of incubation, while, laccase activity was found maximum (2.483 U/ml) at 96 h incubation (Fig. 3a b). In opposite, Micrococcus luteus strain BL3 showed LiP and MnP activity at 96 h and laccase activity at 72 h of incubation. Several researchers found a peak in ligninolytic enzyme synthesis in 96 to 120 h, which then vanished quickly when MnP and LiP activity increased (Yadav et al. 2011).
Effects of different pH
pH has been demonstrated to have a significant impact on Bacillus paramycoides strain BL2 and Micrococcus luteus extracellular ligninolytic activities. B. paramycoides produces LiP (2.215U), MnP (1.822U), and lac (2.783U) at pH 8 at the optimal incubation period, whereas M. luteus strain BL3 produces LiP (2.246 U), MnP (2.147 U), and lac (1.245 U) in pH 7 at the optimum incubation period (Fig. 3c–d). The pH of the culture media affects growth, ligninolytic enzyme production, and breakdown. According to various bacteria in acidic to alkaline circumstances, laccase production is greatest when the pH is between 7.0 and 9.0. In this work, pH 8.0 and 7.0 were shown to be favourable for B. paramycoides and M. luteus growth, respectively. In this study, the optimal pH for both enzymes was 7–8, beyond which both enzymes lost their activity (Fig. 3c–d). This might be due to a shift in the concentration of hydrogen ions in the medium.
Effects of different temperature
The extracellular ligninolytic enzyme activity of Bacillus paramycoides strain BL2 and Micrococcus luteus strain BL3 was clearly affected by temperature. The temperature of the culture medium is crucial for the production of ligninolytic enzymes. The optimal temperature for the synthesis of ligninolytic enzymes, as found in several bacteria, is greater than that of fungus (Debnath and Saha 2020). Very interesting finding of this study showed that the greatest LiP, MnP and laccase production was noted at 30–35 °C (Fig. 3e–f). At temperatures over 35 °C, very little ligninolytic activity was found. Optimal temperature for LiP and MnP was found 30 °C to 35 °C by several researchers (Zeng et al. 2015).
Effect of different carbon source
Laccase synthesis in B. paramycoides strain BL2 and M. yunnanensis strain BL3 was evaluated at 1% using five different carbon sources: sucrose, glucose, fructose, lactose, and starch. In the presence of glucose (1%), B. paramycoides strain BL2 generated LiP (3.915U), MnP (3.742U), and lac (3.545U), whereas M. luteus strain BL3 produces LiP (3.845U), MnP (3.478U), and lac (2.845U). Pattern of carbon sources on enzyme secreted by B. paramycoides strain BL2 was found glucose > sucrose > fructose > lactose > starch (Fig. 4a). While Pattern of carbon sources on enzyme secreted by B. paramycoides strain BL3 were found glucose > fructose > lactose > starch > sucrose (Fig. 4b).
Effect of different nitrogen source
In microorganisms, nitrogen is mostly processed to make amino acids, proteins, nucleic acids, and cell wall components. Extracellular laccase synthesis in B. paramycoides strain BL2 and M. luteus strain BL3 was evaluated using five different nitrogen sources: yeast extract, peptone, ammonium chloride, and sodium nitrate. In the presence of peptone, B. paramycoides strain BL2 generated maximum LiP (4.3 U/ml), MnP (3.8 U/ml), and lac (3.9 U/ml), whereas M. luteus strain BL3 produces maximum LiP (4.6 U/ml), MnP (4.2 U/ml), and lac (3.5 U/ml), respectively (Fig. 4c,d). Furthermore, 1% concentration of glucose and 0.5% was favorable for growth and Ligninolytic enzyme activity of both strains, beyond this concentration inhibits the enzyme activity. This study showed the 1% glucose, 0.5% peptone, 30–35 °C temperature, 7 to 8 pH favourable for the growth of bacterial strains and the highest LiP (43.12 U/ml), laccase (44.54 U/ml) and MnP (122.152 U/ml).
Trace elements
Microbes produce extracellular enzymes in large quantities that are influenced not only by carbon and nitrogen sources but also by trace elements (Myszograj et al. 2018). Hence, this study also focused on the effect of trace element on ligninolytic enzyme activity. In comparison to the control, the inclusion of BaCl2, KCl, ZnSO4, MgSO4, CaCl2, and FeSO4 in the medium appeared to boost bacterial growth. In the presence of FeSO4-containing media, the highest LiP and MnP generated were 5.121 and 4.724 U/ml, respectively, for B. paramycoides strain BL2 and 4.621 and 4.824 U/ml for M. luteus strain BL3 (Fig. 5a,b). On addition, in a CuSO4-containing medium, the highest laccase production was observed 4.368 and 3.668 U/ml for B. paramycoides strain BL2 and M. luteus strain BL3, respectively. While CuSO4 not much affected the activity of other tested enzyme.
The physico-chemical analysis
The physicochemical characterization of PPIE was shown in Table 1. The pulp paper industrial effluent was dark brown and alkaline in nature, which turned into light brown colour after bacterial treatment with a nearly neutral pH (7–8). The analysis of the industrial effluent sample showed several physico-chemical parameters beyond the permissible limits along with heavy metals. Several parameters of untreated industrial effluent in (mg/l) i.e. total solid (612 ± 115), total dissolved solid (555 ± 11.12), COD (14,959 ± 101.5), BOD (5800 ± 124), lignin (832 ± 20.20), sulphate (1589 ± 10.98), phosphorus(178 ± 5.40), total phenol (411 ± 17.25), Cl− (1.98 ± 0.92), Chlorophenol (198 ± 18.24), total suspended solid (53 ± 1.12) and heavy metals such as Fe (65.02 ± 0.03), Ni (3.10 ± 0.75) and Zn (11.08 ± 0.15), Cu (2.01 ± 0.95), Cr (2.11 ± 0.82), Cd (0.215 ± 0.75), Mn (10.01 ± 0.11). However, a sharp reduction in pH (7.1 ± 0.10), total solid (122 ± 1.24), total dissolved solid (102 ± 1.42), COD (4235 ± 15.64), BOD (2431 ± 13.05), lignin (246 ± 1.57), sulphate (1211 ± 3.24), phosphorus(164 ± 1.35), total phenol (317 ± 16.45), Cl− (1.132 ± 0.10), chlorophenol (185 ± 0.15), total suspended solid (18 ± 1.06) and heavy metals such as Fe (13.01 ± 0.65), Ni (0.96 ± 0.45) and Zn (2.7 ± 0.67), Cr (1.01 ± 0.34),Cd (0.11 ± 0.75), Mn (6.06 ± 0.31) was noted as shown in Table 1. After bacterial treatment lignin 70%, colour 63%, COD 71%, BOD 58% reduction were observed.
Detection of functional groups analyzed through FT-IR analysis present in treated and untreated pulp paper industrial effluent
FT-IR spectroscopy is used to identify the functional groups and chemical bonds exhibiting chemical compounds present in untreated and bacterial pulp paper mill industrial effluent showed Table 2. FTIR spectrum of Pulp Paper industrial effluent (a) untreated (Control) (b) bacterial treated showed in Fig. 6a,b. The complex bond structure of wastewater in both spectra revealed that constitutes complicated organic and inorganic compounds. Wide absorption peaks between 3600 and 3200 cm−1 give O–H stretching appeared due to the phenolic group in alcohol phenols and acids (Chandra and Kumar 2017). Before and after Bacterial treatment wide absorption at 3414.8 cm−1 to 3396.7 cm−1, respectively, and Weak absorption at 2128.1 cm−1 to 2118.1 cm−1, respectively, were present in wastewater. Similar findings were previously reported by Chandra and Kumar (2017) and Chandra et al. (2018). After bacterial treatment, the sharpness of peak at 3414.8 cm−1 (Stretching O–H asymmetric grp) and 2128.4 1 cm−1 (A combination of hindered rotation and O–H bonding grp) observed in control slightly reduced up to 3396.7 1 cm−1 (Stretching O–H asymmetric grp) and 2118.1 1 cm−1 (A combination of hindered rotation and O–H bonding grp (Fig. b). The peak at 2938.6 1 cm−1 (C–H stretching bands grp) observed in Bacterial treated was slightly reduced and shifted to 2128.4 cm −1 (O–H bending grp) in Control to Bacterial treated Further, absorption at 1653.4 and 1636.9 cm−1 observed in Control to treated. represent the presence of C = O grp and β-sheet structure of amide I grp, respectively, were maximally reduced and slightly shifted into 1636.1 cm −1 (β-sheet structure of amide I grp) after bacterial treatment due to the overall decrease in organic compounds. Absorption at 13,320.1 cm −1 present in control was completely disappeared and 1295.1 cm−1 shifted to 1137.5 cm −1 due to the complete degradation of the compound showing the presence of Stretching PO2 2 symmetric (phosphate II) group. After treatment, 611.9 cm−1 shifted to 599.3 cm−1 might be due to the formation of compounds as intermediated compounds. Shifting, vanishing and the advent of new peaks indicated the conversion of complex toxic compounds into simple compounds (Kadam et al. 2016).
Pollutants detected through GC–MS analysis
GC–MS is a unique method used to detect trace levels of organic compounds found in pulp paper mill industrial effluent. Figure 7a,b depicted GC–MS chromatogram of compounds extracted from untreated and bacterial treated PPIE. Major peaks in untreated pulp paper industrial effluent showed the presence of persistent organic compounds present in PPIE. After bacterial treatment, the peak intensity reduction compared to the control and the appearance of new peaks can be observed in the treated sample. The major pollutants identified in untreated sample at different RT were 3.6-Dioxa-2,7-disilaoctane(RT: 6.07), Hexanoic acid,trimethylsilyl ester(RT:7.78), 2-iodo-3- (p-tolyl) prop-2-ene / 7,8- Dimethyl1-4(RT:8.74), Talaromycin B(RT:9.23), 2-Heptnoic acid, trimethylsilyl ester(RT: 10.78), Trimethylsilyl ether of glycerol(RT:12.40), Benzenepropanoic acid,trimethylsilyl ester(RT:15.58), Tetradecane,1-iodo/Docosane (CAS)(RT: 17.07), Silane, (dodecyloxy( trimethyl- (CAS)(RT: 18.68), 7-Methyldinaphtho [2,1-b,1’,2’-d] silole(RT: 20.01), 2,2’- Methly 1enebis (4-t- butyl phenol)(RT:21.68), 1-Monolinoleoylglycerol trimethylsilyl ester(RT:20.60), 1-Monolinoleoylglycerol trimethylsilyl ester(RT:27.07), Octadecanoic acid, trimethylysilyl ester(RT: 29.94), 3,5-Dihydroxybenzoic acid 3 TMS(RT:31.08), Methyl1(Z)-3,3-dipheny.1–4-hexenoale (RT:33.16), 2,6,10,14,18,22-Tetracosahexane(RT: 35.54), Docasane(CAS) (RT: 36.27), 1,3, Bis (t-Bytylthio) benzene-4,6-bis (methyleneox)(RT: 38.71), 5,6-Dihyrostigmasterol,acetate (RT:39.36), a-D-Galactopyranoside (RT:41.45),2,2-dimethylpropyl (2Z,6E)—10,11-epoxy(RT:45.09), Pentamethyl 1 pentaphenyl 1cyclopentasiloxane(RT: 45.37), respectively (Table 3). In the untreated (control) sample, these pollutants were completely absent due to the complete degradation of PPIE by potential isolated strains and some new metabolites were formed. However, the analysis of bacteria treated pulp paper mill industrial effluent sample has showed the existence of various ROPs such as Ethyl 2-fluro-1-trifluro methyl lphyrrolo [2,1-a] isoquinlin-3 carboxylate(RT:6.48), Ethyl1[4’-acetylphenycarbomate](RT: 10.12), Trimethylsily ether of glycerol(RT: 12.42), 3-Choloro-5-(dichloromehtyl)-5-methoxy-2-fluranone(RT: 13.68), 3-Nanonone, 2-methyl(RT: 17.33), Silane,(dodecyloxy) trimethyl-(CAS)(RT:18.69), Tricholoroacetic acid, hexadecyl ester(RT:21.12), 3-[4’-(t-butyl) phenyl] furan-2,5-dione(RT: 24.00), Docasane (CAS) (RT: 24.82), (2R,4S,5R,6S,8S,10E,12R)-12-(tert-Butyldimethylsily)-4,6-dimethoxy-2,8,10-trimethyl-10,14-pentadecadiene-1,5diol(RT:28.77),Octadecanoicacid, trimethylsilyl estermethyl (RT: 29.96), 5- Diphenylphosphinoyl-5 (1-hyroxycyloxyl)pentan-3-one ethylene acetal (RT:35.25), Mauritamide A(RT:37.76), Octasiloxane (RT: 39.39), 1,8-Diphenyl 1–3,4,10,11-tertahydro[1,4] dioxino [2,3–9:5,6–9’] diisoquinoline(RT:43.21), 2-(2,4-Dicholoro-6-Nitrophenoxy) ethanol (RT:45.49), Dimethyl exo-6-(dibromomethyl)-6-methyl 1–5 oxobicyclo[2,2.2]octa-2,7-diene-2,3-dicarboxylate dimethyl(RT:47.62), at different RT. While, some compounds Docasane (CAS), Hexadeconoic acid, trimethylysilyl ester, Silane,(dodecyloxy) trimethyl-(CAS), Trimethylsilyl ether of glycerol, Octadecanoic acid, trimethylysilyl ester, Octadecanoic acid, trimethylysilyl ester were pulp paper mill industrial effluent still persistent in the pulp paper industrial effluent even after bacterial treatment. GC–MS data showed that various organic pollutants are degraded and biologically transformed into simpler compounds after bacterial treatment. These POPs might be used as a primary energy source for bacteria which leads to the degradation and decolorization of PPIE. The result indicates that BL2 and BL3 strains transformed or degraded chlorinated, phenolic, non-phenolic as well as lignin derivatives compounds from the PPIE.
Toxicity assessment of PPIE for environmental safety
Toxicity assay of untreated and bacterial treated PPIE on seeds germination of P. mungo L. in 3 days observations.. The various parameters of seed germination were observed with untreated and bacterial-treated PPIE (Table 4). The test indicated the removal of toxicity based on germination or suppression of seed and early growth of seeds. The toxicity test of P. mungo L. seeds with different concentrations of untreated PPIE (concentration 0, 25, 50, 75, 100%) was more toxic showing only 84, 60, 40, and 20% germination, respectively. While in bacterial treated PPIE toxicity was significantly 75% reduced. The presence of more toxic pollutants and dissolved solids that are absorbed by the seeds before germination and the effect of the different physicochemical and biochemical parameters of seeds due to germination suppression at high concentrations of pulp paper industrial effluent (Sonkar et al. 2019a, b). Increased germination percent in treated PPIE may be due to decreased and detoxified ROPs that have formed suitable environmental conditions for seed germinations and utilization as nutrients present in industrial effluent. Recently, reported toxicity analysis of untreated pulp paper industrial effluent through seed germination tests and results were correlated with several previous studies (Kumar et al. 2020; Sharma and Singh 2021; Singh et al. 2021)
Discussions
All 48 bacterial strains were collected on MSM agar because it provides a minimal requirement for bacterial growth. Among these two bacterial strains oxidized guaiacol and produced a reddish-brown hue, whereas they decolorized other substrates such as methylene blue and phenol red dye i.e. these two strains showed Ligninolytic enzyme activity. Similar results were collaborated with the findings of Bendary et al. (2021). Bacillus paramycoides strain BL2 showed MnP and LiP activity higher in the initial stage followed by laccase activity. Finding of this study corroborates with the findings of several other researchers (Bandounas et al. 2011; Chandra et al. 2012; Yadav and Chandra 2015; Lai et al. 2016; Noman et al. 2020). Most of the lignin-degrading bacteria only generated one or two enzymes among ligninolytic enzymes (Chen et al. 2012; Shi et al. 2013; Buraimoh et al. 2015). The presence of three main ligninolytic enzymes in this study is critical because these enzymes worked together to degrade lignin which is the main component of pulp-paper mills. Only a few bacteria have been shown to be capable of generating all three enzymes, including B. subtilis, Klebsiella pneumonia and Bacillus sp (Yadav and Chandra 2015). Further, pH has been demonstrated to have a significant impact on Bacillus paramycoides strain BL2 and Micrococcus luteus BL3 extracellular ligninolytic activities. This might be due to the pH significantly influence the ligninolytic activity. Every organism has its optimum condition in which they perform a best metabolic activity. The pH of the culture has a significant impact on several enzymatic activities might be due to the increase in the transport of many components through the cell membrane (Guan et al. 2020). Veloz Villavicencio et al. (2020) showed that most of the fungal cultures favoured growth and enzyme activity at a slightly acidic pH of medium. Lowe et al. (2020) found that enzyme production increased steadily as the starting pH increased, peaking at pH 7 (0.472 U/ml) and subsequently declining at higher pH values such as pH 9 (0.005 U/ml).Temperature also effects the enzyme production and high temperature reduced the enzyme activity most likely because increasing the temperature hindered the growth of microorganisms, hence lowered enzyme activity (Edae and Alemu, 2017). Temperature affects the enzyme activity because at higher temperature denaturation of protein takes place and accordingly effect the enzyme activity (Feller et al. 2010). Hence, every activity has an optimal condition on which activity of enzymes was found best. Glucose was one of the carbon sources that promoted excellent growth and laccase synthesis. Ligninolytic enzyme activity was maximal in glucose-containing medium because enzymes are substrate selective (Elisashvili et al. 2017.) According to Kumar et al. (2020), a 1% carbon supply is required for maximal laccase synthesis, while Lip and MnP were shown to have the highest enzyme activity at low-nutrient media. MnP production was suppressed in the presence of a high-carbon substrate. Lactose, fructose and starch on the other hand, greatly inhibited laccase development in P. ostreatus, which was consistent with efficient enzyme synthesis (AdoB et al. 2021). MnP synthesis is a secondary metabolic process induced by N and C deficiency, as previous research has shown (Starnes et al. 2016; Rahman et al. 2018). Nutrient concentration also affects the Ligninolytic enzyme activity and it has been found that lower concentrations and higher concentrations of nutrients inhibit bacterial growth and enzyme activity. This might be due to a higher concentration increase the osmotic stress resulting in the water efflux from the cell, loss of the turgor pressure and inhibiting the bacterial growth. LiP and MnP activity was found maximum in presence of FeSO4. This might be due to Fe work as a co-substrate for MnP. The function of CuSO4and FeSO4 might be maintaining the protein structure lead to stabilize the enzyme activity. Extracellular Ligninolytic enzymes are generated in tiny amounts by nature; nevertheless, the presence of inducers, primarily aromatic or phenolics chemicals linked to lignin or lignin derivatives, can significantly increase their synthesis (Olajuyigbe et al. 2018). Copper sulphate enhanced growth to a higher amount than the other inducers when compared to the control. This is might be due to the filling of type-2 copper binding sites with copper ions may be responsible for the activation of laccase by Cu ions (Estevinho et al. 2021). Laccase activation was induced by copper ions, while laccase inhibition was caused by Fe and Mn ions, according to Yang et al. (2020). In most situations, adding Cu and Mn ions increases the activity of manganese peroxidase enzymes (Chauhan and Choudhury 2020).The findings revealed that the effect of metal ions on laccase activity was highly reliant on the source and kind of metals utilized, both of which had a significant impact on the enzyme's catalytic activity. The present study concludes that the ligninolytic enzyme is thought to be an optimal green catalyst. They are used in a variety of sectors, including food, textiles, pulp and paper mills, distilleries, tanneries, medicine, cosmetics, etc. hence, they can also be utilized as a biocatalyst for the treatment of their wastewater. Several reports showed the use of ligninolytic enzyme for the treatment of recalcitrant pollutants (Kumar and Chandra 2018). But, due to the lack of optimum conditions for maximum enzyme production, there is a need for the enhanced protein production of these enzymes for the betterment of the ecosystem. Hence, the purpose of this study is to evaluate the effect of several factors on the ligninolytic enzyme activity of bacterial isolates isolated from pulp paper industrial effluent. Two bacterial strains B. paramycoides strain BL2 and M. luteus strain BL3 were isolated from pulp-paper mills. Result showed that glucose (1%) and nitrogen (0.5%) was found optimum for bacterial growth and Ligninolytic enzyme activity. Moreover, pH (7–8), temperature (30–35 °C), 96 h of incubation, CuSO4, FeSO4were enhances the ligninolytic enzyme activity. Most of the studies reported the effect of environmental conditions on individual enzyme activity, but this study showed effects on LiP, MnP and laccase together. Physico-chemical analysis of effluent showed its complexity. Phenols and lignin components of PPIE are recalcitrant in nature and major plant constituents of PPIE. Dark colour of pulp paper mill industrial effluent might be due to lignin which is a major component of raw material used during paper production. Presence of high value of COD and phenols are toxic for the aquatic and terrestrial environment, even at relatively low levels in discharged pulp paper mill industrial effluent lead to toxic nature of effluent (Yadav and Chandra 2018; Gupta et al. 2017). Phenol at a higher concentration blocked the photosynthesis of blue algae, diatom, and even higher concentration range of 100–400 g/mL also induced full inhibition of photosynthesis (Duan et al. 2017). Moreover, sodium sulfite, which is used during the pulping process, may be the source of sulfate ions in wastewater, and the nitrate found in the industrial effluent might be generated from lignin (Yadav and Chandra 2015). The metal content in industrial effluent samples can be attributed to the bioaccumulated metals by plants, and these plants are used as raw materials. Furthermore, the source of heavy metals in the effluent might be due to the chemicals and pipe lines used during the processing of paper production. The isolated bacterial strains had ability to destroy and decolorize PPIE with a good additional nutritional source, (glucose: 1.0 percent, peptone: 0.5 percent) as well as ambient factors (pH: 7.0–8.0) temperature: 30–35 °C, agitation: 120 rpm) during the optimal incubation period 96 h. The breakdown and decolorization of contaminants existing in the environment takes 120 h. FTIR and GC–MS analysis showed that the majority of pollutants are derived from the fatty acids, organic acids generated during the pulping process (Kumari et al. 2016; Chandra and Kumar 2017). This finding is corroborated by earlier data from Chandra and Kumar 2017; Lowe and Groger 2020; Kumar et al. 2021. Presence of various ROPs, phenolics, chlorinated, and aliphatic compounds in wastewater was previously reported by several researchers for the degradation through bacterium (Sonkar et al. 2019a, b). Table 2 showed the compounds detected in untreated and treated effluent. Various POPs of PPIE transformed or degraded phenolic, non-phenol halide derivatives as well as lignin derivatives from wastewater have been identified in by (Raj et al. 2014). Degradation of complex compounds was possible only due to the enzyme activity (MnP, Laccase, Lip) shown by potential bacterial strains.
The Phaseolus mungo L. seeds were used to test toxicity. The toxicity decrease of wastewater after bacterial treatment also confirmed. Therapy for bacteria, as a result, it may be argued that the strains have the potential to be a useful tool in the fight against cancer for the safe disposal of ROPs found in PPIE. Hence, knowledge of this study regarding Ligninolytic enzyme activity is helpful to understand the mechanisms of enzyme and treatment of wastewater discharged from the industries.
Materials and methods
Sample collection
Sludge samples for laboratory analysis were collected from the discharge site of the M/s K. R. Pulp Papers Limited, located in Shahjahanpur, India (27º50031.800N79º51015.700E), Uttar Pradesh, India. The Sludge was composed from the surface to a depth of about 0 to 10 cm with sterile spatulas, then transferred to the research laboratory and maintained the original properties at 4 °C for microbiological investigation (Sharma et al., 2020).
Isolation and purification of bacterial strains
1 g of soil sample was mixed with 100 ml of sterile 0.9% NaCl for proper solubilization. The solution was briskly agitated before being allowed to settle. 1 ml of the solution was diluted in steps of one milliliter until it reached a dilution of 106). Potential bacterial strains were isolated using a nutrient enrichment approach. A wastewater sample (20 ml) was added to a 250 ml Erlenmeyer flask containing 80 ml of sterile mineral salt medium (MSM) with the composition described by Feng et al. (2018). The pH of the medium was set at 7.50. As a carbon source, lignin (100 mg/l) was added to the medium (L-MSM), and flasks were incubated at 30 °C under shaking conditions (120 rpm). Bacteria were isolated as the method described by Hooda et al. (2015) In a subsequent investigation, 2 ml aliquots of these cultures were frozen as stock cultures in 80% glycerol stock and kept at – 80 °C.
Identification of bacterial strains
A PowerSoil® DNA Extraction Kit was used to extract the entire genomic DNA of each strain from pure cultures (MO-BIO, USA). The DNA extraction was carried out as directed by the manufacturer. The partial 16S rRNA gene sequences were amplified using universal eubacteria primers 27F (5'- AGA GTT TGA TCC TGG CTC AG-3') and 1492R (5'- GGT TAC CTT GTT ACG ACT T-3') in a polymerase chain reaction (PCR). A total of 30 PCR cycles were completed. Each cycle consists of 5 min of denaturation at 96 °C, 5 min of primer annealing at 50 °C and 1.5 min of primer extension at 72 °C. According to the manufacturer's instructions, 1.5 bp of amplified PCR products were purified using an Agencourt AMPure XP (Beckam Coulter, USA). A sample of this amount was submitted to be sequenced. An Applied Biosystems 3730xl DNA Analyzer was used to sequence the purified PCR products of the 16S rRNA genes. The produced sequences were examined using the biological sequence alignment editor (Bioedit) (Choi et al. 2010). The partial sequences were submitted to BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis using the online option database at the National Center for Biotechnology Information (NCBI) to identity the bacterial isolates. The sequences were aligned with the Clustral W tool, and a phylogenetic tree was constructed using the MEGA 6 programme and the neighbor-joining approach (Hennell et al. 2012).
Assay of ligninolytic enzymes
The LiP assay was done by monitoring the oxidation of dye Azure B in presence of H2O2. The reaction mixture contained sodium tartrate buffer (50 mM, pH 3.0), Azure B (32 M), 500 l of culture filtrate, 500 l of H2O2 (2 M). OD was taken at 651 nm after 10 min (Chandra and Singh 2012; Wittner et al. 2021). The selected isolates were screened for LiP using Azure-B agar dye as an indicator. All two bacterial isolates were streaked on Azure B-agar plates containing MSM, agar and 0.1% Kraft lignin as substrate.
The activity of MnP was evaluated using a UV–Vis spectrophotometer set to 465 nm and the oxidation of phenol red to a coloured product (Kumar et al. 2018). The quantity of enzyme required to produce 1.0 micromole of coloured product per minute at 30 °C under the test conditions with a molar extinction coefficient of A465 = 12,100 M−1 cm−1 was defined as one unit of enzyme activity.
Laccase activity was measured using Wolfenden and Willson's technique of oxidation of 2,2'-azinobis-(3-ethylbenzethiazoline-6-sulphonate) (ABTS) 0.15 ml ABTS (0.03%), 0.5 ml 0.1 M sodium acetate buffer at pH 5.0, and 0.35 ml extracellular enzyme samples made up an enzymatic combination. A UV–Vis spectrophotometer was used to quantify the production of oxidised ABTS in a 1.0 ml quartz cuvette (1 cm light path) at 30 °C and 450 nm wavelength. With a molar extinction value of A420 = 36 000 M−1 cm−1, ABTS oxidation was followed by an increase in absorbance at 420 nm (Lai et al. 2016). Different dye azure B, methylene blue Ramazol brilliant blue for LiP, Phenol red, methyl red, Vanilly acetone for MnP and Guaiacol, Tannic, Gallic for laccase substrates were also evaluated at different concentrations (5–160 mM) to know the best enzyme activity.
Optimization of ligninolytic enzyme activity
One loopful of each of the selected potential bacterial strains BL2 and BL3 was inoculated into 10 ml of LB medium at an initial concentration of 107 cells/ml for the optimization of ligninolytic enzymes producing bacterial strains. To produce a final OD600 of 1.0, samples were incubated at 30 °C for 10 h with shaking at 120 rpm (Trinh et al. 2016; Chandra et al. 2017). The inoculums were then transferred to a 250 ml Erlenmeyer flask with 90 ml LB medium with 0.9 g/l OPEFB fibres. For 7 days, the cultures were incubated at 120 rpm and 30 °C. The pH was initially fixed at 7.6. Every 24 h, samples were taken to track daily bacterial development and enzyme activity.
Effect of different pH and temperature on ligninolytic enzyme activity
Submerged fermentation of the chosen isolates in LB broth medium at 5 different beginning pHs was used to assess the influence of initial pH. By incubating duplicate samples of the chosen isolates at pH 5.0, 6.0, 7.0, 8.0, and 9.0 with 1 M HCl or 1 M NaOH to alter the pH, the optimal starting pH for enzyme synthesis was established. For 7 days, the tests were carried out in an incubator shaker set at 120 rpm and 30 °C. Every 24 h, samples were taken. After 7 days of development, the pH of the medium varied from 8.1 to 8.2.
To evaluate the effect of temperature on Ligninolytic enzyme activity samples were incubated at 20, 25, 30, 35, and 40 °C temperature at 120 rpm on a rotary shaker, and samples were taken every 24 h.
Effects of different nutritional conditions
Effect of various carbon sources such as sucrose, fructose, glucose, xylose, lactose, and starch, on ligninolytic activity was tested at 1.0 percent (w/v). Moreover, different organic and inorganic nitrogen sources, including peptone, beef extract, yeast extract, sodium sulphate, ammonium chloride, and urea, were introduced to the MSM medium at a concentration of 0.5% (w/v)to evaluate the effect of nitrogenous sources on enzyme activity. Furthermore, the effects of variable concentration (0.5–2 w/v) of carbon and nitrogen sources were also evaluated.
Biodegradation of pulp and paper effluent by an isolated bacterium
The degradation studies were carried out in 250 mL Erlenmeyer flasks containing 99 mL autoclaved PPIE supplemented with 1% (w/v) glucose and 0.5 percent (w/v) peptone as carbon and nitrogen sources, respectively. Inoculated overnight developed culture (1%) of BL2, BL3 and consortium with optical density (OD) 2.0, then incubated for 96 h at 35 °C and 120 rpm. As a control, flasks containing just autoclaved PPIE were employed. According to Raj et al. (2014.) an appropriate sample volume was extracted at every 24 h and examined for bacterial growth, colour decrease, and lignin content. Result showed that maximum colour and other pollution parameters reduction in PPIE treated with a consortium. Hence, the further study only analyzed effluent treated with consortium.
Physico-chemical analysis of PPIE
The usual procedures for physico-chemical examination in bacterial treated and untreated pulp paper industrial effluent were used in triplicates (Permissible limit CPCB 2015). pH, temperature, COD (open reflux method), BOD (5 days method), total dissolved solids (TDS), total suspended solids (TSS) and total solids (TS) (drying method), total nitrogen (Kjeldhal method), sulphate and phosphate (BaCl2 precipitation method and Vanadomolybdo phosphoric acid method), total nitrogen (Kjeldhal method), sulphate and phosphate (CPCB 2015). A digital pH metre (Metrohm, USA) was used to determine the pH of the industrial effluent sample after acid digestion. Moreover, heavy metals like copper, zinc, chromium, cadmium, magnesium, nickel and iron were digested and measured using the standard method described in Permissible limit CPCB (2015) Moreover, the colour was measured by UV–Vis spectrophotometer (Thermo science evolution-201) following the standard method at 465 nm (CPCB 2015), and pH was measured using a pH meter. Lignin was estimated according to Pearl and Benson (1940), and ions were also analyzed by ion meter (Orion Model 960) using the selective ion electrodes. Simultaneously, treated and untreated pulp paper industrial effluent was analyzed by atomic absorption spectrophotometry (AAS) (ZEEnit 700, Analytic Jena, Germany).
Fourier transforms infrared spectrophotometric (FT-IR) analysis
The FT-IR Spectrophotometer (NicoletTM 6700, Thermo Scientific, USA) was used to determine the functional groups present in untreated and bacteria-treated pulp paper industrial effluent. The wastewater samples from pulp and paper industrial effluent were centrifuged at 8000 rpm for 10 min at 4 °C and dried at 50 °C (Kurade et al. 2012). In a 5: 95 ratio, 1 mg of dried sample was combined with 400 mg of potassium bromide. The liquid was poured onto a translucent disc and subjected to a manual hydraulic pressure of 100 kg cm2 for 10 min before being placed in an FT-IR Spectrophotometer (NicoletTM 6700, Thermo Scientific, USA) to be analysed.
GC–MS analysis of bacterial treated and untreated industrial effluent
Dissolved organic pollutants present before and after bacterial-treated pulp paper industrial effluent were detected by gas chromatography-mass spectrometry (GC–MS) analysis. For extraction of organic pollutants, a fixed volume (10 ml) of pulp paper industrial effluent was acidified with 35% HCl up to pH < 2.0 according to Chandra and Kumar (2017). Further, an equal volume of ethyl acetate was mixed with pulp paper industrial effluent in a 100 mL separating funnel and shaken continuously for 3–5 h with intermittent rests for liquid–liquid extraction. After proper shaking, the solutions were kept constant on a separating funnel stand to separate the organic layer of solvent. The extracted organic solvent-containing organic pollutants were dried at ≤ 40 °C at low pressure. Thereafter, the dried residue was dissolved in 1 mL of ethyl acetate and filtered using a 0.22 μm syringe filter (Millipore Ltd., Carrigtwohill, Co. Cork, Ireland). Further, an aliquot (2.0 μl) of the sample was derivatives with TMS were filtered through 0.22 μm syringe filters and injected in GC–MS (PerkinElmer, UK) Thereafter, organic pollutants were identified by matching their mass spectra with those provided in the National Institute of Standards and Technology (NIST) library which was available with the instrument.
Seed germination experiment
Toxicity of effluent was evaluated by seed germination experiment, the concentration of bacteria treated and untreated PPIE used was 0, 25, 50, 75 and 100% (v/v). Subsequently, 10 seeds of Phaseolus mungo L. were placed in sterilized glass petri dishes of uniform size lined with two Whatman No. 1 filter paper discs. Treatments of seed for toxicity assessment were done as per the method followed Kumar and Chandra (2021). The germination index (GI) was calculated according to the method (Salleh et al. 2020). Further, seed germination (%), Radical length (cm), germination reduction (%), stress tolerance index (%), relative toxicity (%), phytotoxicity (%), and seedling vigor index were evaluated by the method described by David Noel and Rajan (2015) and Oleszczuk et al. (2012).
Statistical analyses for data significance
Using a Microsoft Excel Spreadsheet, the results of the replicates were aggregated and represented as mean standard deviation. The data was then submitted to a one-way analysis of variance (ANOVA) using GraphPad Prism 7.0, with the least significant difference being calculated. P 0.05 was used to determine significance (Sheikholeslami et al., 2019).
References
AdoB V, Onilude AA, Amande TJ (2021) Optimization of total soluble protein production by Trametes sp. isolate B7 and enzymatic degradation of synthetic dyes. J Microbiol Biotech Food Sci https://doi.org/10.15414/jmbfs.2019.9.1.99-103
Bandounas L, Wierckx NJP, DeWinde JH, Ruijssenaars HJ (2011) Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol 11:1–11
Boguniewicz-Zabłocka J, Kłosok-Bazan I (2020) Sustainable processing of paper industry water and wastewater: a case study on the condition of limited freshwater resources. Polish J Env Studies 29:2063–2070
Buraimoh OM, Amund OO, Ilori MO (2015) Kraft lignin degradation by AutochtonousStrepmyces strains isolated from a tropical lagoon ecosystem. J Microbiol Biotech Food Sci. 5:248–253
Chandra R, Singh R (2012) Decolourisation and detoxification of rayon grade pulp paper mill effluent by mixed bacterial culture isolated from pulp paper mill effluent polluted site. Biochem Eng J 61:49–58
Chandra R, Kumar V (2017) Phytoextraction of heavy metals by potential native plants and their microscopic observation of root growing on stabilised distillery sludge as a prospective tool for in situ phytoremediation of industrial waste. Environ Sci Pollut Res 24(3):2605–2619
Chandra R, Kumar V, Yadav S (2017) Extremophilic ligninolytic enzymes in Extremophilic enzymatic processing of lignocellulosic feed stocks to bioenergy. Springer Cham. https://doi.org/10.1007/978-3-319-54684-1_8
Chandra R, Kumar V, Tripathi S, Sharma P (2018) Heavy metal phytoextraction potential of native weeds and grasses from endocrine-disrupting chemicals rich complex distillery sludge and their histological observations during in-situ phytoremediation. Ecol Eng 111:143–156
Chauhan AK, Choudhury B (2020) Suitability of organic solvent and cholinium based ionic liquid activated novel lignolytic enzymes of H. aswanensis for enhanced Kalson lignin degradation. Int J Biol Macromol 165:107–117
Chen YH, Chai LY, Zhu YH, Yang ZH, Zheng Y, Zhang H (2012) Biodegradation of kraft lignin by a bacterial strain Comamonas sp. B-9 isolated from eroded bamboo slips. J Appl Microbiol 112:900–906
Choi O, Das A, Yu CP, Hu Z (2010) Nitrifying bacterial growth inhibition in the presence of algae and cyanobacteria. Biotech Bioengin 107:1004–1011
CPCB Central Pollution Control Board Annexure-I Charter for Water Recycling and Pollution Prevention in Pulp & Paper Industries (Specific to Ganga River Basin States) (2015) pp 1–38
Dashtban M, Schraft H, Syed TA, Qin W (2010) Fungal biodegradation and enzymatic modification of lignin. Int J Biochem Molecul Bio 1(1):36–50
David Noel S, Rajan MR (2015) Phytotoxic effect of dyeing industry effluent on seed germination and early growth of lady’s finger. J Pollut Eff Contr 2(126):10–4172
Debnath R, Saha T (2020) An insight into the production strategies and applications of the ligninolytic enzyme laccase from bacteria and fungi. Biocatal Agricul Biotechnol 26:101645
Duan W, Meng F, Lin Y, Wang G (2017) Toxicological effects of phenol on four marine microalgae. Environ Toxicol Pharmacol 52:170–176. https://doi.org/10.1016/j.etap.2017.04.006
El-Bendary MA, Ezzat SM, Ewais EA, Al-Zalama MA (2021) Optimization of spore laccase production by Bacillus amyloliquefaciens isolated from wastewater and its potential in green biodecolorization of synthetic textile dyes. Prepar Biochem Biotech 51:16–27
Edae T, Alemu M (2017) Selection and optimization of lignocellulosic substrate for laccase production from Pleurotus species. Int J Biotechnol Mol Biol Res 8(4):38–48
Elisashvili V, Kachlishvili E, Asatiani MD, Darlington R, Kucharzyk KH (2017) Physiological peculiarities of lignin-modifying enzyme production by the white-rot basidiomycete Coriolopsis gallica strain BCC 142. Microorganisms 5(4):73
Estevinho RMG (2021) Insight into catalytic features of native and engineered laccases and synthesis of added-value compounds (Doctoral dissertation)
Feller G (2010) Protein stability and enzyme activity at extreme biological temperatures. J Phys Condens Matter 22(32):323101
Feng X, Ouyang M, Liu X, Lu L, Xia Y, He X (2018) Thermal runaway mechanism of lithium ion battery for electric vehicles: A review. Energy Storage Mater 10:246–267
Guan N, Liu L (2020) Microbial response to acid stress: mechanisms and applications. Appl Microbio Biotechnol 104(1):51–65
Gupta S, Pandey RA, Sanjay Pawar B (2017) Bioremediation of synthetic high–chemical oxygen demand wastewater using microalgal species Chlorella pyrenoidosa. Bioremediat J 21(1):38–51
Hammel KE, Tardone PJ, Moen MA, Price LA (1989) Biomimetic oxidation of nonphenoliclignin models by Mn(III): new observations on the oxidizability of guaiacyl and syringyl substructures. Arch Biochem Biophys 270(1):404–409
Hennell J R, Dagostino PM, Lee S, Khoo CS, Sucher NJ (2012) Using GenBank® for genomic authentication: a tutorial. Methods Mol Biol 862:181–200
Hooda R, Bhardwaj NK, Singh P (2015) Screening and identification of ligninolytic bacteria for the treatment of pulp and paper mill effluent. Water Air Soil Poll 226:305
Janusz G, Pawlik A, Sulej J, Swiderska-Burek U, Jarosz-Wilkołazka A, Paszczyński A (2017) Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol Rev 41(6):941–962
Kadam AA, Jang J, Lee DS (2016) Facile synthesis of pectin-stabilized magnetic graphene oxide Prussian blue nanocomposites for selective cesium removal from aqueous solution. Biores Technol 216:391–398
Kaushik G, Gopal M, Thakur IS (2010) Evaluation of performance and community dynamics of microorganisms during treatment of distillery spent wash in a three stage bioreactor. Biores Technol 101(12):4296–4305
Kumar V, Chandra R (2018) Characterisation of manganese peroxidase and laccase producing bacteria capable for degradation of sucrose glutamic acid-Maillard reaction products at different nutritional and environmental conditions. World J Microbio Biotech 34(2):1–18
Kumar P, Naik M (2020) Biotic symbiosis and plant growth regulators as a strategy against cadmium and lead stress in chickpea. Plant Arch 20(2):2495–2500
Kumar A, Chandra R (2021) Biodegradation and toxicity reduction of pulp paper mill wastewater by isolated laccase producing Bacillus cereus AKRC03. Clean Eng Technol 4:100193
Kumar A, Biswas B, Saini K, Kumar A, Kumar J, Krishna BB, Bhaskar T (2021) Py-GC/MS study of prot lignin with cobalt impregnated titania, ceria and zirconia catalysts. Renew Energy 172:121–129
Kumari S, Deori M, Elancheran R, Kotoky J, Devi R (2016) In vitro and in vivo antioxidant, anti-hyperlipidemic properties and chemical characterization of Centella asiatica (L.) extract. Front Pharmacol 7:400
Kurade MB, Waghmode TR, Kagalkar AN, Govindwar SP (2012) Decolorization of textile industry effluent containing disperse dye Scarlet RR by a newly developed bacterial-yeast consortium BL-GG. Chem Eng J 184:33–41
Lai CMT, Chua HB, Danquah MK, Saptoro A (2016) Isolation of thermophilic lignin degrading bacteria from oil-palm empty fruit bunch (EFB) compost. IOP Conf Series Mat Sci Eng 206:012016
Long LF, Couvreur J, Leriche G, Champ M, Garnier G, Allais F, Saito K (2018) Importance of mediators for lignin degradation by fungal laccase. ACS Sustain Chem Engine 6:10097–10107
Löwe J, Gröger H (2020) Fatty acid hydratases: Versatile catalysts to access hydroxy fatty acids in efficient syntheses of industrial interest. Catalysts 10(3):287
Lowe J, Dietz KJ, Groger H (2020) From a biosynthetic pathway toward a biocatalytic process and chemocatalytic modifications: three-step enzymatic cascade to the plant metabolite cis-(+)-12-OPDA and metathesis-derived products. Adv Sci 7:1902973
Marlatt VL, Bayen S, Castaneda-Cortès D, Delbès G, Grigorova P, Langlois VS, Martyniuk CJ, Metcalfe CD, Parent L, Rwigemera A, Thomson P (2022) Impacts of endocrine disrupting chemicals on reproduction in wildlife and humans. Environ Res 208:112584
Myszograj S, Stadnik A, Płuciennik-Koropczuk E (2018) The influence of trace elements on anaerobic digestion process. Civil Environm Eng Rep 28(4):105–115
Noman E, Al-Gheethi AA, Talip BA, Mohamed R, Kassim AH (2020) Oxidative enzymes from newly local strain Aspergillusiizukae EAN605 using pumpkin peels as a production substrate: Optimized production, characterization, application and techno-economic analysis. J. Hazardous Mat 121954
Olajuyigbe FM, Fatokun CO, Oyelere OM (2018) Biodelignification of some agro-residues by Stenotrophomonas sp. CFB-09 and enhanced production of ligninolytic enzymes. Biocatalysis Agricultural Biotech 15:120–130
Oleszczuk P, Rycaj M, Lehmann J, Cornelissen G (2012) Influence of activated carbon and biochar on phytotoxicity of air-dried sewage sludges to Lepidium sativum. Ecotoxicol Environ Saf 80:321–326
Pearl IA, Benson HK (1940) The determination of lignin in sulfite pulping liquor. Paper Trade J 111:35
Perez-Garcia O, Escalante FM, De-Bashan LE, Bashan Y (2011) Heterotrophic cultures of microalgae: metabolism and potential products. Water Res 45:11–36
Rahman O, Shi S, Ding J, Wang D, Ahmad S, Yu H (2018) Lignin nanoparticles: synthesis, characterization and corrosion protection performance. New J Chemis 42:3415–3425
Raj A, Kumar S, Haq I, Singh SK (2014) Bioremediation and toxicity reduction in pulp and paper mill effluent by newly isolated ligninolytic Paenibacillus sp. Ecol Eng 71:355–362
Salleh MS, Nordin MS, Puteh AB (2020) Germination performance and biochemical changes under drought stress of primed rice seeds. Seed Sci Technol 48(3):333–343
Sharma IP, Chandra S, Kumar N, Chandra D (2017) PGPR: heart of soil and their role in soil fertility. In Agriculturally important microbes for sustainable agriculture (pp. 51–67). Springer, Singapore
Sharma P, Tripathi S, Chandra R (2020) Phytoremediation potential of heavy metal accumulator plants for waste management in the pulp and paper industry. Heliyon 6:04559
Sharma P, Singh SP (2021) Pollutants characterization and toxicity assessment of pulp and paper industry sludge for safe environmental disposal. In Emerging Treatment Technologies for Waste Management 207–22
Sheikholeslami K, Ali Sher A, Lockman S, Kroft D, Ganjibakhsh M, Nejati-Koshki K, Shojaei S, Ghavami S, Rastegar M (2019) Simvastatin induces apoptosis in medulloblastoma brain tumor cells via mevalonate cascade prenylation substrates. Cancer 994
Shi Y, Chai L, Tang C, Yang Z, Zheng Y, Chen Y, Jing Q (2013) Biochemical investigation of kraft lignin degradation by Pandoraeasp. B-6 isolated from bamboo slips. Bioprocess Biosyst Eng 36:1957–1965
Singh AK, Chandra R (2019) Pollutants released from the pulp paper industry: Aquatic toxicity and their health hazards. Aquatic Toxicol 211:202–216
Singh AK, Kumar A, Bilal M, Chandra R (2021) Organometallic pollutants of paper mill wastewater and their toxicity assessment on Stinging catfish and sludge worm. Environ Technol Innov 24:101831
Sonkar G, Mall RK, Banerjee T, Singh N, Kumar TL, Chand R (2019a) Vulnerability of Indian wheat against rising temperature and aerosols. Environ Pollut 254:112946
Sonkar M, Kumar M, Dutt D, Kumar V (2019b) Treatment of pulp and paper mill effluent by a novel bacterium Bacillus sp IITRDVM-5 through a sequential batch process. Biocatalysis Agricultural Biotechnol 20:101232
Starnes DL, Lichtenberg SS, Unrine JM, Starnes CP, Oostveen EK, Lowry GV, Bertsch PM, Tsyusko OV (2016) Distinct transcriptomic responses of Caenorhabditiselegans to pristine and sulfidized silver nanoparticles. Environ Pollut 213:314–321
Trinh NTN, Masniyom P, Maneesri J (2016) Optimization of culture conditions for Acetobacteraceti TISTR 102 in coconut water with supplementary banana juice. Intern Food Res J 23:1300–1307
Tripathi S, Yadav S, Purchase D, Singh K, Al-Rashed S, Chandra R (2021a) Characterization of Persistent Organic Pollutants from Culturable and Un-culturable Bacterial Communities structure in Pulp and Paper Sludge after Secondary Treatment. Chemosphere 295:133892
Tripathi S, Chandra R, Purchase D, Bilal M, Syed A, Yadav S (2021b) Quorum sensing- a bacterial tool for robust degradation of industrial waste containing persistent organic pollutants. Environ Pollut 292:118342
Veloz Villavicencio E, Mali T, Mattila HK, Lundell T (2020) Enzyme activity profiles produced on wood and straw by four fungi of different decay strategies. Microorganisms 8(1):73
Wittner N, Broos W, Bauwelinck J, Slezsák J, Vlaeminck SE, Cornet I (2021) Enhanced fungal delignification and enzymatic digestibility of poplar wood by combined CuSO4 and MnSO4 supplementation. Process Biochem 108:129–137
Yadav S, Chandra R (2018) Detection and assessment of the phytotoxicity of residual organic pollutants in sediment contaminated with pulp-paper mill effluent. Environ Monit Ass 190(10):581
Yadav S, Chandra R (2015) Syntrophic co-culture of Bacillus subtilis and Klebsiella pneumonia for degradation of kraft lignin discharged from rayon grade pulp industry. J Environ Sci 33:229–238
Yadav S, Chandra R, Rai V (2011) Characterization of potential MnP producing bacteria and its metabolic products during decolourisation of synthetic melanoidins due to biostimulation effect of D–xylose at stationary phase. Process Biochem 46(9):1774–1784
Yang X, Wu Y, Zhang Y, Yang E, Qu Y, Xu H, Chen Y, Irbis C, Yan J (2020) A thermo-active laccase isoenzyme from Trametestrogii and its potential for dye decolorization at high temperature. Front Microbio. 241
Zeng G, Cheng M, Huang D, Lai C, Xu P, Wei Z, Li N, Zhang C, He X, He Y (2015) Study of the degradation of methylene blue by semi-solid-state fermentation of agricultural residues with Phanerochaetechrysosporium and reutilization of fermented residues. Waste Manag 38:424–430
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
Author information
Authors and Affiliations
Contributions
Prerna Verma & Sonam Tripathi: conceptualization, data curation, investigation, writing. Sangeeta yadav: writing, review and revision. Ram Chandra: supervision, conceptualization, data curation, investigation and writing -original draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
None of the authors declare a conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Verma, P., Tripathi, S., Yadav, S. et al. Degradation and decolourization potential of ligninolytic enzyme producing Bacillus paramycoides BL2 and Micrococcus luteus BL3 for pulp paper industrial effluent and its toxicity evaluation. Arch Microbiol 204, 642 (2022). https://doi.org/10.1007/s00203-022-03236-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03236-7