Abstract
A bacterial isolate PM1 obtained from the rhizosphere of healthy plants was identified as Pseudomonas aeruginosa by biochemical characteristics and 16S rRNA gene sequence (GenBank ID OL321133.1). It induced resistance in Nicotiana tabacum cv. Xanthi-nc and Cyamopsis tetragonoloba, against Tobacco mosaic virus (TMV) and Sunn-hemp rosette virus (SRV), respectively. Foliar treatment with isolate PM1 curbed TMV accumulation in susceptible N. tabacum cv. White Burley. PM1 was more effective as a foliar than a root/soil drench treatment, evident through a comparative decrease in ELISA values, and reduced viral RNA accumulation. Foliar and soil drench treatment with PM1 resulted in a disease index of 48 and 86 per cent, and a control rate of 48.9 and 8.5 per cent, respectively. PM1 exhibited phosphate solubilization, produced siderophores, auxins, HCN, and ammonia, all important plant growth-promoting traits. Foliar treatment with PM1 enhanced growth in tobacco, while its volatiles significantly promoted seedling growth in C. tetragonoloba. Of the several metabolites produced by the isolate, many are known contributors to induction of systemic resistance, antibiosis, and growth promotion in plants. Soluble metabolites of PM1 were less effective in inducing antiviral resistance in N. tabacum cv. Xanthi-nc in comparison with its broth culture. PM1 and its metabolites were antagonistic to Gram-positive Bacillus spizizenii and Staphylococcus aureus, and fungi Fusarium oxysporum, Aspergillus niger, and Rhizopus stolonifer. Its volatiles were inhibitory to F. oxysporum and R. stolonifer. Thus, PM1 exhibited considerable potential for further evaluation in plant virus control and production of diverse metabolites of use in agriculture and medicine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Controlling plant viruses through deployment of defence responses of the host is an attractive means of protecting crops. Incompatibility between the host and virus, governed through resistance genes, offers excellent protection, and has formed the basis of breeding programmes for virus control and enhancing productivity (Ronde et al. 2014). However, this strategy often fails due to the co-evolution of the pathogen, and subsequent breakdown of R-gene-dependent resistance (Merrick et al. 2021). Other forms of defence responses however are known that can efficiently control virus infection in susceptible hosts by inducing antiviral resistance. Triggered by various agents, these include systemic acquired resistance (SAR), the onset of which mainly involves infection by necrotizing pathogens and chemicals such as salicylic acid (Conrath 2006; Oostendorp et al. 2001), induced systemic resistance (ISR) that develops following application of plant growth-promoting rhizobacteria (PGPR) (Pieterse et al. 2014), and induced antiviral resistance with plant proteins such as CAP-34, CIP-29, and BDP-30 (Prasad and Srivastava 2017). While SAR and ISR depend on signalling pathways largely mediated by salicylic acid, and ethylene/jasmonic acid, respectively (Pieterse et al. 1996), resistance induced by plant proteins has thus far always been associated with accumulation of virus inhibitory agents in the resistant plants (Prasad and Srivastava 2017). Hence, pathways of resistance induction may differ, involving different sets of fingerprints, but they converge eventually, providing long-lasting protection against diverse biotic and abiotic stresses.
Isolates of PGPR induce a broad-spectrum resistance against pathogens and promote plant health and growth (Backer et al. 2018). This is achieved through direct, or indirect mechanisms, including production of auxin, siderophore, antibiosis, ISR, or competition for nutrients and niches; and the underlying molecular mechanisms for growth promotion and ISR have been dissected to a large extent (Pieterse et al. 1998; van Loon et al. 1998). Several isolates of Pseudomonas and Bacillus species have proven valuable in biological control and growth promotion (Bakker et al. 2007; Kloepper et al. 2004). Studies on PGPR strains associated with resistance induction against plant viruses are relatively few in comparison with fungal and bacterial pathogens. For instance, resistance could be demonstrated against Tobacco necrosis virus (TNV) in tobacco (Maurhofer et al. 1994), Cucumber mosaic virus (CMV), Tomato spotted wilt virus (TSWV), and Potato virus Y (PVY) in tomato and cucumber (Beris et al. 2018; Raupach et al. 1996; Zehnder et al. 2000) with PGPR strains P. fluorescens, Serratia marcescens and B. amyloliquefaciens. P. aeruginosa strain 7NSK2 was reported as an efficient biocontrol agent that protected tomato from Pythium splendens, and bean from Botrytis cinerea (Buysens et al. 1996; De Meyer and Hofte 1997), but its ability to elicit antiviral resistance in susceptible plants has not been adequately studied. Nevertheless, the mechanism and determinants of resistance induction have been explored by using mutants of strain 7NSK2, and TMV/tobacco model system based on lesion development (De Meyer et al.1999a; 1999b). In other studies, formulations of PGPR consortia that included P. aeruginosa strain NML2212, could reduce sunflower necrosis disease incidence caused by Sunflower necrosis virus by 51.4%, with an increase in plant height and yield (Srinivasan and Mathivanan 2009), while a PGPR mixture comprising isolates of P. aeruginosa and Stenotrophomonas rhizophilia, and CMV-KU1, a strain of CMV associated with a benign satellite RNA, were used to control infection on tomato caused by CMV-16, a virulent strain of CMV causing severe stunting and fruit malformation (Dashti et al. 2012). A reduction in disease severity and reduced virus accumulation was observed in sets treated with a combination of PGPR and CMV-KU1, but not in those that were treated with the PGPR isolates alone (Dashti et al. 2012). There are not many reports of plant virus control utilizing a single strain of P. aeruginosa that could effectively lower the virus titre/accumulation in planta.
PGPR strains also release volatile organic compounds (VOCs) that are known to promote growth in seedlings and initiate defence responses in plants against pathogens. In particular, VOCs like 2,3-butanediol and its precursor acetoin have been reported to afford high levels of protection and growth promotion in Arabidopsis seedlings (Ryu et al. 2003; Sharifi and Ryu 2018). Bacterial volatiles exert varied effects on plants such as enhancing seed germination and biomass, increasing yield, and often increasing rhizosphere volume, leading to improved availability of nutrients to plants (Sharifi and Ryu 2018). Some reduce biotic stress on plants by being toxic to their pathogens. It is generally believed that the major mechanism of disease suppression by volatiles is through ISR (Sharifi and Ryu 2016).
This paper reports a Pseudomonas aeruginosa isolate (PM1) that demonstrates resistance towards viral infections possibly through suppression of virus replication, as well as growth promotion in tobacco, and seedling growth promotion in Cyamopsis tetragonoloba induced by its volatiles. This paper also describes the inhibitory effects of PM1 on select bacterial and fungal species. The repertoire of volatile organic compounds and soluble metabolites produced by the isolate have also been studied to relate its ability to directly inhibit microbes, promote growth and induce resistance in plants.
Materials and methods
Isolation of rhizobacterial strain PM1
Strain PM1 was isolated from the rhizosphere soil according to Barillot et al. (2013), with modifications. A healthy Parthenium plant was uprooted, and the bulk soil loosely adhering to the roots was removed by gentle shaking. The roots were immersed in 100 mL of sterile distilled water, with intermittent shaking for 10 min, and left undisturbed for an additional 30 min. The soil suspension was filtered using sterile Whatman number 1 filter paper, and a tenfold dilution series of the sample was prepared. Each dilution was spread in triplicate on nutrient agar (NA) plates. On an average, 3–4 distinct colonies were picked up from each plate. A loopful from each colony was spread on a fresh NA plate by streak-plate technique, to ensure formation of individual, well-isolated colonies. Following incubation of the plates at 37 οC, the process was repeated, to ensure purity of the isolate obtained, and a single colony was then transferred to a NA slant and maintained as a pure culture. A total of 12 independent isolates thus collected, were independently screened for resistance induction against virus infection. We named the most promising isolate as PM1. All investigations were carried out using this isolate. The broth culture of isolate PM1 was used at a cell density of 1.5 × 108 CFU mL−1 in the experiments designed to evaluate antiviral resistance induction, estimated using McFarland standards (HiMedia, India). The opacity of the culture was adjusted with normal saline to an OD of 0.5 at 600 nm, for the desired cell density.
Morphological and physiological characteristics of isolate PM1
A morphological identification of the isolate involved light microscopy after the Gram-stain procedure. Preliminary identification of the isolate was performed using KB002 HiAssorted Biochemical Test Kit (HiMedia Labs, India). An overnight broth culture of the PM1 isolate (50 µL) was used to inoculate each well. Identity of the isolate was based on the identification index provided by the manufacturer along with the test kit. All other tests were performed with an overnight culture of PM1 on nutrient agar. Catalase enzyme activity was determined through addition of 4–5 drops of hydrogen peroxide on the colony surface and development of effervescence in samples that were positive (Schaad et al. 2001). Cellulase, lipase, amylase, and protease activities were detected by streaking the isolate onto agar medium supplemented with 0.2% carboxymethyl (CM) cellulose, 2% Tween 20 and methyl red, starch, and skimmed milk, respectively. To detect cellulase activity, CM cellulose hydrolysis was visualized by flooding the plates with 0.1% Congo red and rinsing with 1 M NaCl (Teather and Wood 1982). Lipase activity was observed as a colour change from red to pale around the streak (Samad et al. 1989), while amylase activity was detected by flooding the starch agar plates with iodine and observing a zone of hydrolysis (Collins 1995). Protease activity was observed as a zone of hydrolysis on milk agar (Chaiharn 2008).
Plant growth-promoting traits of isolate PM1
Production of ammonia, indole acetic acid, HCN, and siderophore
A loopful of an overnight culture was inoculated in peptone water (10 mL) and incubated for 48–72 h at 37 οC. Addition of Nessler’s reagent (0.5 ml) to the tube and development of brown to rust colour indicated a positive result for ammonia production (Cappucino et al. 1992). Production of auxin was determined according to Bric et al. (1991). The King’s B broth (50 ml) containing tryptophan (500 µgmL−1) was inoculated with 100 μL of the overnight broth culture and incubated at 28 οC for 48 h in a shaker incubator. 1 ml of the cell-free supernatant was added to 50 µL of 10 mM orthophosphoric acid and 2 ml of Salkowski’s reagent. Development of red colour confirmed production of IAA. Quantification of the IAA was based on the colorimetric assay developed by Gordon and Weber (1951). For detection of HCN, PM1 isolate was inoculated on a nutrient agar medium containing glycine (4.4 gL−1). Picric acid-soaked Whatman filter paper No.1 was placed inside the petri lid and plates sealed with parafilm. After incubation of plates for 4 days at 30 οC, a change in the colour of filter paper from yellow to orange/brown confirmed the production of HCN (Bakker and Schippers 1987). Production of siderophore was detected on Chrome-Azurol S (CAS) agar medium (Schwyn and Neilands 1987). A disc soaked in the overnight broth culture of the isolate PM1 was placed on the surface of the medium and, following a 72 h incubation, siderophore production was indicated by the development of orange colour around the disc.
Phosphate solubilization
The phosphate solubilization test was performed with Pikovskaya agar medium containing an insoluble form of calcium phosphate (Pikovskaya 1948). A sterile disc was soaked in 20 μL of an overnight broth culture of PM1was placed in the centre of the petri plate, followed by an incubation at 37 οC for 72 h. Formation of a clear zone around the disc indicated phosphate solubilization by the bacterial isolate.
Growth promotion of seedlings through volatiles produced by the PM1 isolate
The effect of volatiles on growth promotion of seedlings was studied using two sets of petri plates, the smaller placed within the larger one (Cordovez et al. 2018). C. tetragonoloba seeds were sterilized in 0.2% HgCl2 for 5 min and placed in the larger plate that contained Murashige and Skoog (MS) medium in 0.8% agar. The smaller plate with nutrient agar medium was inoculated with an overnight culture of PM1 isolate (treated set) or sterile broth (control set) and left uncovered within the larger petri plate. The larger plate was covered with the lid, sealed with Parafilm, and placed in an incubator at 37 οC for 72 h in the dark. After the initial incubation, the petri plates were moved to a glass house and observed for seedling growth.
Amplification of 16S rRNA gene, PCR product purification, sequencing, and BLAST analysis
Genomic DNA was isolated from PM1 using DNA Mini Prep kit (Chromous Biotech, India) and the universal primer pair of 27f and 1492r (F-5′AGAGTTTGATCCTGGCTCAG3′/R-5′TACCTTGTTACGACTT3′) was used for amplification of the 16S rRNA gene through PCR. The PCR was performed in 50 µL containing 2X Dream Taq PCR Master Mix (25 µL, Thermo Fisher Scientific, USA) along with primers (8 µL, 10 µM each), DNA template (4 µL, 25 ng), and RNase-free water (13 μL). The reaction was set up in a T100 Thermal cycler (Bio-Rad, USA) that was programmed as follows: Initial denaturation (95 οC, 5 min), 30 cycles of amplification involving denaturation (94 οC, 45 s), annealing (52 οC, 45 s), and extension (72 οC, 45 s), followed by final extension (72 οC, 10 min). The resultant amplicon of 1494 bp was separated on a 1% agarose gel in Tris–borate-EDTA buffer, 0.5 M, pH-8.0, stained with ethidium bromide, and visualized on a UV trans-illuminator. The PCR product was purified from gel using GeneJET PCR purification kit (Thermo Fisher Scientific, USA), and submitted to Chromous Biotech, India, for sequencing by the Sanger di-deoxy chain termination method using ABI 3500 Genetic Analyzer (Thermo Fisher Scientific, USA). The partial 16S rRNA sequence was subjected to nucleotide BLAST analysis (NCBI) and the molecular identity of the PM1 isolate established. The sequence was duly submitted to GenBank. The submitted sequence was also verified using the SILVA rRNA database (https://www.arb-silva.de), which contains curated sequences of bacterial isolates with previously removed chimeric sequences, if any. Using ten nucleotide sequences from NCBI GenBank database, the phylogenetic tree was constructed using software MEGA X, and the phylogeny inferred using the Neighbour-Joining method. The distances were calculated using Maximum Composite Likelihood method.
Profiling of PM1 metabolite content by GC–MS
A loopful of isolate PM1 in 5 mL nutrient broth was incubated in a shaker incubator at 37 °C for 72 h. Broth culture was centrifuged at 10,000 rpm for 10 min and the supernatant was filter sterilized. The cell-free supernatant was submitted to Chromous Biotech (India), for GC–MS analysis. Briefly, 1 ml of the supernatant was mixed with Chloroform and water (1:3:1), mixed well, and centrifuged at 4000 rpm for 10 min. The upper aqueous phase was collected. The process of solvent extraction was repeated thrice, after which the aqueous phase was dried (Eppendorf SpeedVac, Germany). BF3-methanol solution was added, and the mixture was incubated at 60 °C for 20 min, followed by addition of ice-cold water and chloroform, and subjected to another round of centrifugation at 4000 rpm for 5 min. The supernatant was dried on SpeedVac, dissolved in 100 μL chloroform, and injected into Shimadzu GCMS-QP2010 Plus analyser (Japan), to determine secondary metabolites and volatiles.
Assay for antibacterial activity of PM1 and its metabolites
Antibacterial activity was tested by disc diffusion method (Asghar et al. 2020). The overnight broth culture of PM1, and the secondary metabolites obtained from its 72 h culture filtrate were used in a disc diffusion assay to determine their antibacterial activity against the Gram-positive Bacillus spizizenii (ATCC 6633) and Staphylococcus aureus (ATCC 33862), and the Gram-negative Escherichia coli (ATCC 11775) and Pseudomonas mosselii (ATCC 49838). The reference strains were procured through HiMedia (India). The cell-free supernatant (culture filtrate) from a 72 h culture, was mixed with ethyl acetate (1:1) and incubated overnight at RT. The secondary metabolites (SM) were extracted in ethyl acetate, and after the evaporation of the organic solvent, metabolites were obtained in a powdered form and dissolved in DMSO. Sterile discs impregnated with 20 μL of PM1 broth culture, or its metabolites, were placed on a lawn culture of the reference strains growing in a petri dish, along with the control discs saturated with either sterile broth or DMSO. The plates were incubated for 24 h and the zone of inhibition around the discs, representing antibacterial activity, was noted.
Evaluation of antifungal activity of PM1 and its soluble and volatile metabolites
Isolate PM1 was checked for potential antifungal activity against Aspergillus niger (MTCC 281), Rhizopus stolonifer (MTCC 162), and Fusarium oxysporum (MTCC 284). The fungal cultures were obtained from Microbial Type Culture Collection, IMTECH (India), and maintained in the laboratory on PDA medium. The overnight culture of PM1 was streaked on one side, while a plug of fungal hyphae was placed on the other side of the PDA plate. The plates were incubated at 25 °C for 24–72 h to observe the inhibition in fungal growth. PM1 and its metabolites were evaluated for antifungal activity against the selected fungi by a disc diffusion method. Diameter of the mycelial growth was checked periodically and the per cent inhibition in growth over the controls was calculated. Inhibition of fungal growth by volatiles produced by the bacterial isolate PM1 was determined by using a partitioned petri plate, with a space between the lid and the partition for movement of the volatiles. PDA contained in one half was inoculated with a fungal plug, while nutrient agar in the other half was streaked with PM1. Controls comprised of plates inoculated with respective fungi alone.
Maintenance of viral inoculum and test hosts
Tobacco mosaic virus (TMV) and Sunn-hemp rosette virus (SRV) were maintained on their systemic hosts, Nicotiana tabacum cv. White Burley, and Crotalaria juncea, respectively. The infected leaves were harvested and homogenized in 10 mM phosphate buffer, pH 7.0 (1:1, w/v), passed through two layers of muslin cloth, and subsequently centrifuged at 5000 × g for 15 min. The supernatant served as the inoculum. The inoculum dilution was suitably adjusted to yield 200–300 lesions on N. tabacum cv. Xanthi-nc and Cyamopsis tetragonoloba that exhibit a hypersensitive response to infection by TMV and SRV, respectively, and were hence used in the bioassays to determine induction of resistance to virus infection. Assay hosts were grown in an insect-free glass house and used at a six- or a three-leaf stage, respectively. N. tabacum cv. White Burley, a cultivar of tobacco susceptible to TMV infection, was employed to study the effect of PM1 isolate on symptom development, and viral replication.
Bioassay for induction of resistance to virus infection by the rhizobacterial isolate PM1 and its secondary metabolites
An overnight broth culture (1.5 × 108 CFU mL−1) was mixed with CM cellulose (0.5% w/v) and spread onto the basal leaf of the test hosts, N. tabacum cv. Xanthi-nc and C. tetragonoloba, with the help of a sterile cotton swab. Secondary metabolites (SM) produced by isolate PM1 were similarly applied. Plants that received a foliar treatment with sterile broth or DMSO solution constituted the control set. After 24 h of treatment, the plants were challenge-inoculated on treated (site) and upper non-treated (remote site) leaves with TMV and SRV, respectively. A decrease in the number of local lesions compared to the control constituted induced resistance and the per cent reduction in lesion number was calculated as follows: C-T/C × 100, where C = average lesion number in control set, and T = average lesion number in treated set.
Evaluation of induction of resistance to virus infection and plant growth promotion in Nicotiana tabacum cv. White Burley
The overnight broth culture of bacterial isolate PM1 (1.5 × 108 CFU/mL) was used either as a root/soil drench (10 mL per plant), or as a foliar application on all leaves of tobacco plants (5–6 leaf stage). Three treatments were given, on alternate days, in sets of plants that were either used to measure growth promotion or to study induction of resistance against TMV. Each set comprised 10 test plants. The plants that received foliar treatment with sterile broth alone constituted the control set.
Induction of resistance to virus infections and evaluation of disease severity index
The top three leaves of individual plants were dusted with carborundum powder (600 mesh) and mechanically inoculated with TMV, after 24 h of the third treatment. The disease index, infection and control rates were compared between sets that received foliar and soil drench treatments. Plants were observed over four weeks for disease onset and severity of mosaic symptoms. The symptoms were rated on a scale of 1–5, where 0 = no symptoms; 1 = mild mosaic; 2 = severe mosaic covering 50% leaf area; 3 = severe mosaic covering entire leaf surface; 4 = severe mosaic accompanied with leaf distortion; 5 = severe mosaic accompanied with leaf distortion and stunting.
Detection of TMV by dot blot
Dot blot was performed according to Hibi and Saito (1985). Leaf tissue was analysed for presence of TMV at 14 dpi, when the typical mosaic was clearly visible in the DW-treated control set. Leaf extract from plants in each set was spotted onto nitrocellulose membrane and probed using anti-TMV antibody (1:1000). The serological reaction was detected using alkaline phosphatase (ALP) conjugated goat anti-rabbit antibodies (1:2000). Nitro-blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) were used as substrates for ALP. All reagents were sourced from Sigma-Aldrich (USA).
Determination of viral load by indirect ELISA
DAC-ELISA was performed as described by Clark and Adams (1977) for detecting viral load in the control and treated sets of plants. The top two leaves from three randomly selected plants in each set were harvested at 28 dpi, and homogenized in 0.1 M phosphate buffer, pH 7.2. Total protein was quantified according to Bradford (1976). The protein content was equalized in all sets of leaf extracts and a 100-fold dilution prepared in coating buffer. The primary and ALP-conjugated secondary antibodies were diluted in PBS-T and used at 1:1000 and 1:2000 (v/v), respectively. The reaction was developed by addition of the substrate p-nitro phenyl phosphate and stopped by adding 3 M NaOH. ELISA plate reader (Bio-Rad Laboratories, USA) was used to read the absorbance at 405 nm.
RT-PCR of viral RNA
Primer BLAST on an isolate (GenBank MT108232.1) was used to design TMV coat protein-specific primers VP-02-F1 (5′-CAAGCTCGAACTGTCGCTCA-3′) and VP-02-R1 (5′-ACCGTTGCGTCGTCTACTCT-3′), that fetched an amplicon of 242 bp. Total RNA was extracted from 100 mg leaf tissue obtained from three representative plants in each set at 28 dpi and subjected to RT-PCR using a one-step RT-PCR kit (Qiagen, Germany). Reaction mix was prepared as per manufacturer’s instructions, and PCR performed in T100 Thermal cycler (Bio-Rad, USA) as follows: Reverse transcription (50 °C, 30 min); initial PCR activation (95 °C, 15 min); denaturation (94 °C, 45 s), annealing (58 °C, 45 s) and extension (72 °C, 1 min) × 30 cycles; final extension (72 °C, 10 min). 18S rRNA gene-specific primer pair (F-5′-CCTGCGGCTTAATTGACTC-3′, R-5′-GTTAGCAGGCTGAGGTCTCG -3′) were used to fetch an amplicon of 174 bp and included as an internal reference.
Evaluation of growth promotion in tobacco
Plants were evaluated for growth promotion at the end of four weeks by measuring the leaf area using a Systronics Leaf Area Meter 211 (India), and determining the plant height. At the end of eight weeks, the number of fruits and flowers was noted to compare the effect of treatment with isolate PM1.
Statistical analysis
All experiments were performed in triplicate, and three biological replicates were used per treatment for assessing induction of resistance in hypersensitive hosts, while ten biological replicates were used in determining seedling growth promotion, resistance induction, and growth promotion in tobacco cultivar White Burley. Data were analysed using one-way analysis of variance (ANOVA One-way) test, a-nd independent t test with Least Significant Difference (LSD) at P ≤ 0.05, using SPSS software (IBM SPSS statistics version 20).
Results
Biochemical and molecular characterization of isolate PM1
PM1 isolate grew as a greenish and viscid colony on nutrient agar medium. Microscopic examination revealed Gram-negative short but slender rods. A few biochemical characteristics of the isolate were studied using the HiMedia kit, KB002, and the results are included in Table 1, and indicated by a superscript ‘b’. Out of the twelve tests, reaction of the isolate to two tests differed from the listed attributes of P. aeruginosa. Isolate PM1 produced H2S, though not conclusively in the kit-based test, although H2S production was observed on a separate SIM agar medium. Hence, this strain did possess the ability to produce H2S. PM1 utilized citrate, glucose, and arabinose as substrates, while it was unable to use lysine, ornithine, adonitol, and sorbitol. It reduced nitrate, but lacked urease, and phenylalanine deamination ability. Amongst the growth promotion traits studied, isolate PM1 solubilized phosphate, produced siderophore, ammonia, auxin, and HCN. It exhibited enzymic activities that included catalase, lipase, and protease, but not amylase and cellulase (Table 1, Fig. 1).
Biochemical characteristics and PGPR traits of isolate PM1. Pure culture of PM1(a), KG002 HiMedia kit-based biochemical tests as indicated in Table 1 (b), Phosphate solubilisation (c), production of siderophore (d), auxin (e), ammonia (f), H2S (g), HCN (h), cellulase (i), Lipase (j), caseinase (protease) (k), amylase (l), and catalase (m). Con = control
Amplification of the 16S rRNA gene with universal primer set yielded a product of 1494 bp, and a 1375 bp sequence was obtained from the product purified out of the gel. The sequence was submitted to NCBI GenBank and assigned GenBank ID OL321133.1. A sequence identity of 99.78% was observed with several strains of P. aeruginosa when subjected to NCBI nucleotide BLAST analysis. Figure 2 represents the phylogenetic position of the isolate PM1 based on 16S rRNA gene sequences of a few bacterial strains.
Effect of volatiles on growth of seedlings of Cyamopsis tetrogonoloba
Volatiles produced by the bacterial isolate PM1 introduced significant differences in the affected set of seedlings as compared to the control set (Fig. 3a and b). The treated seedlings produced longer shoots and roots. The primary roots appeared stouter, with well-developed secondary roots. An increase of nearly 50% was observed in the root length and the number of secondary roots in the seedlings (Fig. 3c, d, and e).
Growth promotion of Cyamopsis tetragonoloba seedlings by volatiles produced by the Pseudomonas aeruginosa isolate PM1. The rhizobacterial isolate PM1 was physically separated from the seedlings by inoculating PM1 on nutrient agar medium contained in a small petridish (PM1), or leaving it uninoculated (control), and placing it within a larger petridish containing Murashige and Skoog medium with 0.8% agar, along with the seedlings (a). The effect of the volatiles on the growth of seedlings was observed after 7 days of exposure (b). Figures c, d and e represent the effects on shoot and root length, and the number of secondary roots, respectively. Error bars indicate ± SEM, and the P values calculated statistically at 0.003, 0.00 and 0.004 for shoot length, root length and number of secondary roots, respectively, using Leven’s Independent T test
Analysis of secondary metabolite content by GC–MS
A total of 96 compounds were identified in the chloroform extract of PM1 isolate through gas chromatography–mass spectroscopy (GC–MS) analysis. A variety of alkanes, ketones, alkenes, esters, fatty acids, alcohols, amines, and aromatic compounds were identified. The relative abundance of different metabolites was calculated on a per cent basis. While some metabolites were produced in significantly higher amounts, such as 13-Docosenamide (18.25%), 2,4-Di-t-butylphenol (6.08%), Quinolone (4.13%), n-Pentatriacontane (3.98%), Eicosane (3.8%) and 1-triacontanol (2.67%); others formed a minor component. Compounds with a similarity index greater than 90% in reference database search are considered in Table 2 and Fig. 4.
Antagonism of PM1 and its secondary metabolites against bacterial species
PM1 isolate and its secondary metabolite (SM) were tested for antibacterial activity against the Gram-positive B. spizizenii (ATCC 6633) and S. aureus (ATCC 33862), and against the Gram-negative E. coli (ATCC 11775) and P. mosselii (ATCC 49838). PM1 and its SM inhibited the Gram-positive species only, evident as a zone of inhibition. The PM1 broth culture had a greater inhibitory effect as compared to its metabolites (Fig. 5).
Antibacterial activity of Pseudomonas aeruginosa isolate PM1. The strain PM1 and its secondary metabolites (SM) were tested for antimicrobial effects against A Bacillus spizizenii (ATCC 6633), B Staphylococcus aureus (ATCC 33862), C Escherichia coli (ATCC 11775), and D Pseudomonas mosselii (ATCC 49838), through a disc diffusion assay. Discs soaked in isolate PM1 (1), sterile broth (2), secondary metabolites of isolate PM1 (SM) (3), DMSO solution (4). Both sterile broth and DMSO solution represent negative controls. Please note the lack of inhibitory effect on Gram-negative species tested. In the graph, the zones of inhibition with similar letters are statistically similar
Antifungal activity of PM1, bacterial volatiles, and metabolites
PM1 isolate exhibited antagonistic effects against Aspergillus niger (MTCC 281), Rhizopus stolonifer (MTCC 162), and Fusarium oxysporum (MTCC 284) (Fig. 6a). However, volatiles produced by the isolate were effective against F. oxysporum and R. stolonifer but showed no apparent inhibitory effect on A. niger (Fig. 6b). The antagonistic effect of PM1 and its metabolites, in terms of per cent inhibition in hyphal growth, was most obvious against F. oxysporum, inhibiting its growth radius by 88% and 75%, respectively. While both secondary metabolite (SM) and PM1 in broth culture were equally effective in inhibiting R. stolonifer, broth culture was more effective in inhibiting A. niger and F. oxysporum, compared to its SM fraction (Table 3).
Antifungal activity of Pseudomonas aeruginosa isolate PM1 and its volatiles. Antifungal effects of PM1(a) and its volatiles (b) were determined against Aspergillus niger (MTCC 281), Rhizopus stolonifer (MTCC 162) and Fusarium oxysporum (MTCC 284). The fungal inoculum was placed in one half of the petridish (a), or was partitioned in divided plates (b), while the other half was inoculated with isolate PM1(indicated by an arrow). The petri-plate on the left in each set represents the control. Note the absence of inhibitory effect of volatiles on A. niger
Induction of resistance against viruses in the assay hosts by PM1 and its secondary metabolites
A single basal leaf of the assay host was treated with an overnight culture of PM1 isolate, prior to inoculation with TMV or SRV on both treated (site) and untreated (remote site) leaves, 24 h post-treatment. Induction of antiviral resistance by isolate PM1, represented by a reduction in lesion number on both site and remote site leaves of N. tabacum cv. Xanthi-nc and C. tetragonoloba, is shown in Fig. 7a and b. Lesions due to TMV infection on N. tabacum cv. Xanthi-nc and SRV infection on C. tetragonoloba, were reduced by 97 and 96 per cent on site, and by 96 and 89 per cent on remote site leaves, respectively (Fig. 7c). Leaves similarly treated with SM produced by isolate PM1, in comparison with the broth culture, induced a moderate antiviral resistance against TMV and SRV, reducing lesions between 50 and 70% (Fig. 7c).
Induction of systemic resistance by Pseudomonas aeruginosa isolate PM1. Nicotiana tabacum cv. Xanthi-nc (a) and Cyamopsis tetragonoloba (b) plants received foliar treatments on a single lower leaf (site) with either sterile nutrient broth (control) or a suspension of PM1 isolate (1.5 × 108 CFU mL−1) in nutrient broth (treated), and were challenge inoculated with TMV and SRV, respectively, on both basal (site) and upper untreated (remote site) leaves (a and b). Figure 2c represents the comparative induction of systemic antiviral resistance by isolate PM1 and its secondary metabolites (SM), represented as a percent reduction in lesion number on both site (S) and remote site (RS) leaves on the two hosts following challenge inoculation with the viruses
Resistance induction against TMV in Nicotiana tabacum cv. White Burley and disease severity
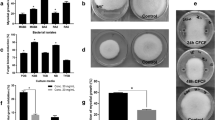
A preliminary dot blot analysis of leaf tissue was conducted for each set of tobacco plants at 28 dpi to determine the total number of plants that carried TMV. The dot blot results showed that all plants were infected with TMV, but there appeared to be a lesser accumulation of TMV in the foliar treated set as compared to sets that received root/soil treatments, and the control sets (Fig. 8). At 28 dpi, the set of tobacco plants that received a foliar application developed mild mosaic symptoms, that were difficult to observe in 2–3 plants of the set, and the disease severity rating stood between 0 and 4, whereas the set that received PM1 as a soil/root drench developed clearly visible mosaic in the majority of plants, comparable in intensity to the control set, with no delay in the symptom development in these plants vis-à-vis the control set (Fig. 9a). Plants in both sets exhibited a disease severity between 4 and 5.
Suppression of virus infection by Pseudomonas aeruginosa isolate PM1 in N. tabacum cv. White Burley against TMV. Sets of tobacco plants were treated with sterile broth (disease control), or received treatment with PM1 (1.5 × 108 CFU mL−1) as a foliar application or soil/root drench. Treatments were repeated thrice, on alternate days, and plants in each set were challenge inoculated on the top three leaves with TMV, 24 h after receiving the final treatment. The leaf extract from three randomly selected plants, at 14 dpi, were spotted on nitrocellulose membrane, and TMV detected by dot ELISA. A set of plants that received no treatment constituted a healthy control. Note the lighter colour development in the foliar treated set
Induction of resistance to virus infection by Pseudomonas aeruginosa isolate PM1 on N. tabacum cv. White burley against TMV. Sets of tobacco plants were treated with sterile broth (a1), or received treatment with PM1 (1.5 × 108 CFU mL−1) as a foliar application (a2), or root/soil drench (a3). Treatments were repeated thrice, on alternate days. Plants in each set were challenge inoculated on the leaves with TMV, 24 h after receiving the final treatment (a1–a3). Representative plants from an experimental replicate were tested for TMV load at 28 dpi by ELISA. Different lowercase letters a–c placed over error bars indicate significantly different values (P ≤ 0.05) as determined by multiple comparison LSD (b). Presence of TMV RNA was detected through RT-PCR (c1- control, c2 and c4-PM1 treated-foliar, c3- PM1 treated-soil/root drench, c5 and c6- internal control, 18S rRNA (174 bp)
The dot blot results were re-confirmed through ELISA. The mean absorbance reading at 405 nm in the foliar treated set was nearly half of the root/soil treated set. Furthermore, the control and root/soil treated samples gave statistically similar values of 2.10 and 2.09, respectively (Fig. 9b), indicating comparable viral load. Viral RNA was detectable in all three sets in a semi-quantitative RT-PCR. The viral RNA appeared as a very faint band of 242 bp in the foliar treated set compared to the DW-treated (control) sets, while a prominent amplicon of 242 bp was seen in the soil/root treated set, comparable in intensity to the control set. The 18S rRNA reference gene was amplified to a similar extent in all three sets of plants, which otherwise exhibited differences in the TMV RNA accumulation pattern (Fig. 9c).
The control and the soil drench set of plants showed an infection rate of 100%, while an infection rate of 90% was observed in the foliar treated set. A disease index and a control rate of 48 and 48.9%, respectively, were noted with the foliar treated set, and 86 and 8.5% were observed in the root/soil treatment set (Table 4).
Growth promotion in tobacco cv. White Burley by isolate PM1
Treated plants exhibited an enhanced leaf area and plant height compared to the plants treated with sterile broth alone. The effect of foliar and root/soil drench treatment was similar (Fig. 10a, c, and d). However, when evaluated for increase in the production of flowers and fruits, the plants that received foliar treatment fared better than the ones that were given a root/soil drench (Fig. 10b and e).
Effect of Pseudomonas aeruginosa isolate PM1 treatment on growth of N. tabacum cv. White Burley. Three sets of plants, with ten plants in each set were treated with sterile nutrient broth (control), or PM1 isolate (1.5 × 108 CFU mL−1). One set was given a foliar treatment, while the other set received a root/soil drench treatment with 10 mL of broth culture (a). The relative differences in leaf area (c) and plant height (d) were determined after 4 weeks of treatment, whereas the number of flowers and fruits were noted after 8 weeks of treatment (b and e). Different Lowercase letters a–b placed over error bars indicate significantly different values (P ≤ 0.05) as determined by multiple comparison LSD
Discussion
Plant growth-promoting rhizobacteria (PGPR) are relevant for their associated biocontrol potential. Our isolate, Pseudomonas aeruginosa strain PM1, not only induced antiviral resistance against TMV and SRV in N. tabacum cv. Xanthi-nc and C. tetragonoloba, respectively, but also reduced systemic virus accumulation in tobacco cv. White Burley. It lowered the disease severity rating of the virus as well. PGPR strains are usually applied as a root treatment or as soil drench for effective growth promotion and biocontrol responses (Beneduzi et al. 2012). In our studies, however, the foliar treatment of tobacco cv. White Burley with PM1 strain was effective in disease suppression while the virus accumulation in the plants that received PM1 in the form of a root drench did not differ significantly from the control set. Leaf colonizing capacity of the strain PM1 may have been better than its ability to colonize roots and hence the observed effects. Effective control through foliar applications has been reported in some earlier studies as well (Preininger et al. 2018). CMV infection on pepper could be reduced through foliar treatment with Bacillus amyloliquefaciens strain 5B6 (Lee and Ryu 2016), and a drench, foliar, or soil application of strain MB1600 reduced TSWV infection on tomato by 80% (Beris et al. 2018).
Direct inhibition of viruses by bacterial strains or their metabolites has not yet been reported hence defence priming and induction of resistance against viruses must be the most probable mode of action for virus inhibition by strain PM1. Defence priming involves accumulation of dormant kinases, particularly MPK3, a mitogen-activated protein kinase, that functions in signal transduction (Beckers et al. 2009). A challenge inoculation with a pathogen activates these latent enzymes, initiating the cascade, leading to faster and stronger activation of defence-related genes (Beckers et al. 2009). The major determinants of PGPR-mediated ISR include microbial lipopolysaccharides, flagellin, pyocyanin, pyochelin, pseudobactin, 2,4-diacetylphloroglucinol, and volatiles like 2,3-butanediol, N-alkylated benzylamine, salicylic acid (SA), etc. (Bakker et al. 2007; Hofte and Bakker 2007). In a few PGPR strains, production of SA may be essential for ISR, as in case of P. aeruginosa strain 7NSK2, SA-deficient mutant had lost ability to induce resistance to the leaf pathogen Botrytis cinerea in bean and TMV (De Meyer and Hofte 1997).
Most PGPR strains are producers of bioactive volatile organic compounds (VOCs), which along with other metabolites may perform several functions such as participating in competition, antagonizing bacterial and fungal growth, promoting growth in diverse microbial species, changing the environment and interactions with other microbes, and triggering ISR and growth promotion in plants (Beneduzi et al. 2012; Netzker et al. 2020; Ryu et al. 2003; Sharifi and Ryu 2018). Thus, microbes and their metabolites may prime the host plants for enhanced defence capacity that manifests in the expression of defence-related genes (Mauch-Mani et al. 2017).
Our ELISA results support reduced virus titre in the inoculated plants that received foliar treatment. The resistance induction governed by defence-related proteins may have interfered with virus replication or its movement, or both, as the viral RNA accumulated to low levels in plants that received foliar treatment with PM1. Although the plants that received treatment in the form of a soil drench showed a disease index and control rate of 86% and 8.5%, respectively, the serological assay and the semi-quantitative PCR results indicated a level of virus accumulation like that of the control set (Fig. 9). Furthermore, the symptomatic plants in the soil drench set carried a virus titre comparable to the control set, this in spite of having twice the number of plants compared to the control set with a disease severity index of 4. As is often the case, the symptom severity may not always correlate with virus concentration (Raupach et al. 1996). Hence a disease rating of either 4 or 5 translated into a similar level of virus titre.
Bacterial siderophores contribute to antibiosis and ISR. A pseudobactin siderophore from P. fluorescens strain WCS374r was found responsible for ISR elicitation in rice against the leaf blast fungus. The defence response involved a rapid accumulation of phenolic compounds and formation of hydrogen peroxide (De Vleesschauwer et al. 2008). It is also known that P. aeruginosa produces two major siderophores, pyochelin and pyocyanin, with reports suggesting the involvement of both in ISR (Bakker et al. 2007). The isolated strain PM1is also a producer of siderophore, as yet chemically undefined, with a possibility of its participation in ISR. But the probability of a major involvement in our case is low, as resistance induction was greater with foliar rather than root treatment. Since the cell-free supernatant was comparatively less efficacious than the broth culture, other bacterial components may have been involved in the induction of resistance.
PGPR strains can modify the root structures of the plants by producing phytohormones such as auxin and other signals, that lead to enhanced branching and development of root hair (Vacheron et al. 2013). Volatiles, including auxin, produced by PM1 isolate were also able to promote growth in C. tetragonoloba seedlings, by significantly promoting lateral root formation. Lateral root proliferation that was noted in plants treated with B. subtilis GB03, reportedly occurred via the auxin-dependent pathway (Zhang et al. 2007). PM1 also produces indole, which is known to modulate auxin production. Bacterial strains of Bacillus cereus and Klebsiella variicola isolated from the rhizosphere of tomato plants, produced IAA, gibberellic acid (GA), and kinetin, and an increase in shoot length and dry weight in tomato and mung bean upon application of these strains was noted (Sunera et al. 2020). GA is an important growth regulator and is involved in plant developmental processes. Inactivation of a transcription factor of the GRAS family, SIGRAS15, led to deactivation of GA biosynthetic genes and was associated with altered plant architecture (Naeem et al. 2020). Whether the observed growth promotion effects of isolate PM1 included mediation by GA production, was not determined.
Metabolites and airborne signals produced by several PGPR strains inhibit both fungal and bacterial species. Having determined the antiviral and growth promotion potential of PM1 isolate, we thought it worthwhile to check its antifungal and antibacterial potential as well, utilizing a few select organisms. PM1 volatiles could inhibit in vitro, the facultative parasite Fusarium oxysporum that is notorious for wilt diseases in various crops. With its greatest efficacy against Fusarium, amongst the organisms tested, development of wilt disease control protocol with the isolate PM1 could be developed. Formulations of PGPR have been employed for biological control. More recently, nanoparticles constructed with Penicillium oxalicum and cadmium acetate (CdONPs) demonstrated effective bacterial control in vitro (Asghar et al. 2020). Chitosan-coated iron oxide nanoparticles could inhibit post-harvest pathogens like Rhizopus oryzae causing fruit rot on strawberry (Saqib et al. 2019).
Nearly 96 metabolites were detected through GC–MS analysis of the cell-free extract. GC–MS metabolite profiling comes with its own set of problems. A single compound showed elution at different retention times, hence, could represent isomers or highly related compounds which could not be clearly distinguished (Fernie et al. 2011). Hence, only those metabolites with a similarity index greater than 90% were considered in this study. In addition to production of inorganic volatiles such as ammonia and HCN, isolate PM1 produced a range of VOCs (Table 3), contributing to antimicrobial activity, growth promotion, and ISR. For instance, 13-docosenamide, a fluorescein quencher that was produced maximally (18%), could be involved in interactions with other rhizosphere microbes (Tamilmanni et al. 2018). The aromatic benzaldehydes are known to be associated with enhanced plant growth and antifungal activity (De Vrieze et al. 2015). Pyrazine derivatives are associated with potent antimicrobial, antioxidant and anti-quorum sensing and cytotoxic activities (Mulner et al. 2020). Undecane, hexacosane, nonadecane, tetradecane, dodecane and 1-hexanol and pentadecane are volatiles that have previously been reported to promote growth (Cantore et al. 2015; Panichikkal et al. 2021), while hexadecane, 1-hexanol and pentadecane are involved in induction of systemic resistance (Park et al. 2013). Tridecane induces PR1 and VSP2 genes that trigger induced systemic resistance (Lee et al. 2012), while triacontanol works as a plant growth regulator. Bacterial volatiles can also regulate the jasmonic acid signalling pathway. They hence affect myriad functions in plants and influence the rhizosphere, however, considerable research is needed to establish the mechanism of volatile perception, the nature of the receptors that could be involved, and signalling pathways in plants that are downstream of initial perception (Pineda et al. 2013; Sharifi and Ryu 2016; Sharifi et al. 2018).
Opportunistic pathogens like P. aeruginosa and Klebsiella pneumoniae are widespread in the rhizosphere and are frequently isolated as potent PGPR strains, associated with a remarkable potential for producing antimicrobial metabolites and inducing resistance against bacteria and fungi. Given the PGPR potential and industrial importance of P. aeruginosa isolates, engineering non-pathogenic strains, through deletion of genes responsible for virulence, has also been successfully attempted (Valentine et al. 2020), and is needed to fully exploit this group of bacteria in biocontrol. The PM1 isolate of P. aeruginosa is one such candidate for further evaluation.
Data availability and materials
GenBank Accession ID OL321133.1.
References
Asghar M, Habib S, Zaman W, Hussain S, Ali H, Saqib S (2020) Synthesis and characterization of microbial mediated cadmium oxide nanoparticles. Microsc Res Techniq 83:1574–1584. https://doi.org/10.1002/jemt.23553
Backer R, Rokem JS, Ilangumaran G, Lamont J, Praslickova D, Ricci E, Subramanian S, Smith DL (2018) Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci 9:1–17
Bakker AW, Schippers B (1987) Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp-mediated plant growth-stimulation. Soil Biol Biochem 19:45–457
Bakker PA, Pieterse CM, van Loon LC (2007) Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology 97:239–243. https://doi.org/10.1094/PHYTO-97-2-0239
Barillot CDC, Sarde C-O, Bert V, Tarnaude E, Cochet L (2013) A standardized method for the sampling of rhizosphere and rhizoplane soil bacteria associated to a herbaceous root system. Ann Microbiol 63:471–476
Beckers GJ, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21:944–953
Beneduzi A, Ambrosini A, Passaglia LMP (2012) Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet Mol Biol 35:1044–1051
Beris D, Theologidis L, Skandalis N, Vassilakos N (2018) Bacillus amyloliquefaciens strain MB1600 induces salicylic acid dependent resistance in tomato plants against tomato spotted wilt virus and potato virus Y. Sci Rep 8:10320. https://doi.org/10.1038/s41598-018-28677-3
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bric JM, Bostock RM, Silversone SE (1991) Rapid in situ assay for indole acetic acid production by bacteria immobilization on a nitrocellulose membrane. App Environ Microbiol 57:535–538
Buysens S, Heungens K, Poppe J, Hofte M (1996) Involvement of Pyochelin and Pyoverdin in suppression of Pythium-induced damping-off of Tomato by Pseudomonas aeruginosa 7NSK2. App Environ Microbiol 62:865–871
Cantore PL, Giorgio A, Iacobellis NS (2015) Bioactivity of volatile organic compounds produced by Pseudomonas tolaasii. Front Microbiol 6:1–14. https://doi.org/10.3389/fmicb.2015.01082
Cappuccino JC, Sherman N (1992) Microbiology: A laboratory manual. Benjamin Cummings Pub. Co., New York
Chaiharn M, Chunhaleuchanon S, Kozo A, Lumyong S (2008) Screening of rhizobacteria for their plant growth promoting activities. KMITL Sci Tech J 8:18–23
Clark MF, Adams AN (1977) Characteristics of the microplate method of enzyme linked immunosorbent assay for the detection of plant viruses. J Gen Virol 34:475–483. https://doi.org/10.1099/0022-1317-34-3-475
Collins CH, Lyne PM, Grange JM (1995) Collins and Lyne’s Microbiological methods. Butterworth-Heinemann Ltd., UK
Conrath U (2006) Systemic acquired resistance. Plant Signal Behav 1:179–184. https://doi.org/10.4161/psb.1.4.3221
Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR (2015) Priming for enhanced defense. Annu Rev Phytopathol 53:97–119
Cordovez V, Schop S, Hordijk K, Dupré de Boulois H, Coppens F, Hanssen I, Raaijmakers JM, Carrión VJ (2018) Priming of plant growth promotion byvolatiles of root-associated Microbacterium spp. Appl Environ Microbiol 84:e01865-18. https://doi.org/10.1128/AEM.01865-18
Dashti NH, Ali NY, Cherian VM, Montasser MS (2012) Application of plant growth-promoting rhizobacteria (PGPR) in combination with a mild strain of Cucumber mosaic virus (CMV) associated with viral satellite RNAs to enhance growth and protection against a virulent strain of CMV in tomato. Can J Plant Pathol 34:177–186
De Meyer G, Hofte M (1997) Salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 induces resistance to leaf infection by Botrytis cinerea on bean. Phytopathology 87:588–593
De Meyer G, Audenaert K, Hofte M (1999a) Pseudomonas aeruginosa 7NSK2-induced systemic resistance in tobacco depends on in planta salicylic acid accumulation but is not associated with PR1a expression. Eur J Plant Pathol 105:513–517
De Meyer G, Capieau K, Audenaert K, Buchala A, Metraux J-P, Hofte M (1999b) Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 activate the systemic acquired resistance pathway in bean. Mol Plant Microbe Interact 12:450–458
De Ronde D, Butterbach P, Kormelink R (2014) Dominant resistance against plant viruses. Front Plant Sci 5:307. https://doi.org/10.3389/fpls.2014.00307
De Vleesschauwer D, Djavaheri M, Bakker PAHM, Hofte M (2008) Pseudomonas fluorescens WCS374r-induced systemic resistance in rice against Magnaporthe oryzae is based on Pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response. Plant Physiol 148:1996–2012. https://doi.org/10.1104/pp.108.1
De Vrieze M, Pandey P, Bucheli TD, Varadarajan AR, Ahrens CH, Weisskopf L, Bailly A (2015) Volatile organic compounds from native potato-associated Pseudomonas as potential anti-oomycete agents. Front Microbiol 6:1–15. https://doi.org/10.3389/fmicb.2015.01295
Fernie AR, Aharon A, Willmitzer L, Stitt M, Tohge T, Kopka J, Carroll AJ, Saito K, Fraser PD, DeLuca V (2011) Recommendations for reporting metabolite data. Plant Cell 23:2477–2482
Gordon SA, Weber RP (1951) Colorimetric estimation of indoleacetic acid. Plant Physiol 26:192–195
Heatley NG (1944) A method for the assay of penicillin. Biochem J 38:61–65
Hibi T, Saito Y (1985) A dot immunobinding assay for the detection of tobacco mosaic virus in infected tissues. J Gen Virol 66:1191–1194. https://doi.org/10.1099/0022-1317-66-5-1191
Hofte M, Bakker P (2007) Competition for Iron and Induced Systemic Resistance by Siderophores of Plant Growth Promoting Rhizobacteria. In: Höfte Monica (ed) Microbial Siderophores. Springer, Berlin, Heidelberg, pp 121–133
Kloepper JW, Ryu CM, Zhang S (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Lee GH, Ryu C-M (2016) Spraying of Leaf-colonizing Bacillus amyloliquefaciens protects pepper from Cucumber mosaic virus. Plant Dis 100:2099–2105
Lee B, Farag MA, Park HB, Kloepper JW, Lee SH, Ryu C-M (2012) Induced resistance by a long-chain bacterial volatile: elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS ONE 7:1–11. https://doi.org/10.1371/journal.pone.0048744
Mauch-Mani B, Baccelli I, Luna E, Flors V (2017) Defence priming: an adaptive part of induced resistance. Annu Rev Plant Biol 68:485–512
Maurhofer M, Hase C, Meuwly P, Metraux J-P, Defago G (1994) Induction of systemic resistance of tobacco to tobacco necrosis virus by the root colonizing Pseudomonas fluorescence strain CHA0: Influence of the gacA gene and of pyoverdine production. Phytopathology 84:139–146
Merrick LF, Burke AB, Chen X, Carter AH (2021) Breeding with major and minor genes: genomic selection for quantitative disease resistance. Front Plant Sci 12:13667. https://doi.org/10.3389/fpls.2021.713667
Mulner P, Schwarz E, Dietel K, Junge H, Herfort S, Weydmann M, Lasch P, Cernava T, Berg G, Vater J (2020) Profiling for bioactive peptides and volatiles of plant growth promoting strains of the Bacillus subtilis complex of industrial relevance. Front Microbiol 11:1–22. https://doi.org/10.3389/fmicb.2020.01432
Naeem M, Waseem M, Zhu Z, Zhang L (2020) Downregulation of S1GRAS15 manipulates plant architecture in tomato (Solanum lycopersicum). Dev Genes Evol 230:1–12. https://doi.org/10.1007/s00427-019-00643-7
Netzker T, Shepherdson EMF, Zambri MP, Elliot MA (2020) Bacterial volatile compounds: Functions in communication, cooperation, and competition. Annu Rev Microbiol 74:409–430. https://doi.org/10.1146/annurev-micro-011320-015542
Oostendorp M, Kunz W, Dietrich B, Staub T (2001) Induced systemic resistance in plants by chemicals. Eur J Plant Pathol 107:19–28
Panichikkal J, Krishnankutty RE (2021) Root exudate components induced production of plant beneficial metabolites in rhizospheric Pseudomonas Spp. Rhizosphere 19:100366. https://doi.org/10.1016/j.rhisph.2021.100366
Park HB, Lee B, Kloepper JW, Ryu C-M (2013) One shot-two pathogens blocked: exposure of Arabidopsis to hexadecane, a long chain volatile organic compound, confers induced resistance against both Pectobacterium carotovorum and Pseudomonas syringae. Plant Signal Behav 8:7–9. https://doi.org/10.4161/psb.24619
Pieterse CMJ, van Wees SC, Hoffland E, van Pelt JA, van Loon LC (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8:1225–1237
Pieterse CMJ, van Wees SC, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC (1998) A novel signalling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10:1571–1580
Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Saskia CM, van Wees SC, Bakker PAHM (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52:347–375
Pikovskaya RI (1948) Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Microbiology 17:362–370
Pineda A, Soler R, Weldegergis BT, Shimwela MM, van Loon JJ, Dicke M (2013) Non-pathogenic rhizobacteria interfere with the attraction of parasitoids to aphid-induced plant volatiles via jasmonic acid signalling. Plant Cell Environ 36:393–404
Prasad V, Srivastava S (2017) Phytoproteins and induced antiviral defence in susceptible plants: The Indian context. In: Mandal B, Rao GP, Baranwal VK, Jain RK (eds) A Century of Plant Virology in India. Springer, pp 689–728
Preininger C, Sauer U, Bejarano A, Berninger T (2018) Concepts and applications of foliar spray for microbial inoculants. Appl Microbiol Biotechnol 102:7265–7282
Raupach GS, Liu L, Murphy JF, Tuzun S, Kloepper JW (1996) Induced systemic resistance of cucumber and tomato against cucumber mosaic cucumovirus using plant growth-promoting rhizobacteria (PGPR). Plant Dis 80:891–894
Ryu C-M, Farag MA, Hu C-H, Reddy MS, Wei HX, Pare PW, Kloepper JW (2003) Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci USA 100:4927–4932
Samad MYA, Razak CNA, Salleh AB, Zin Wan Yunus WM, Ampon K, Basri M (1989) A plate assay for primary screening of lipase activity. J Microbiol Methods 9:51–56. https://doi.org/10.1016/0167-7012(89)90030-4
Saqib S, Zaman W, Ullah F, Majeed I, Ayaz A, Munis MFH (2019) Organometallic assembling of chitosan-iron oxide nanoparticles with their antifungal evaluation against Rhizopus oryzae. Appl Organomet Chem 33:e5190. https://doi.org/10.1002/aoc.5190
Schaad NW, Jones JB, Chun W (2001) Laboratory guide for identification of plant pathogenic bacteria, 3rd edn. American Phytopathological Society Press St, Paul, USA, p 372
Schwyn B, Neilands JB (1987) Universal chemical assay for detection and determination of siderophore. Anal Biochem 160:47–56
Sharifi R, Ryu C-M (2016) Are bacterial volatile compounds poisonous odors to a fungal pathogen Botrytis cinerea, alarm signals to Arabidopsis seedlings for eliciting induced resistance, or both? Front Microbiol. https://doi.org/10.3389/fmicb.2016.00196
Sharifi R, Ryu C-M (2018) Revisiting bacterial volatile-mediated plant growth promotion: lessons from the past and objectives for the future. Ann Bot 122:349–358
Srinivasan K, Mathivanan N (2009) Biological control of sunflower necrosis virus disease with powder and liquid formulations of plant growth promoting microbial consortia under field conditions. Biol Control 51:395–402
Sunera Amna, Saqib S, Uddin S, Zaman W, Rehman SU, Munis MFH, Chaudhary HJ (2020) Characterization and phytostimulatory activity of bacteria isolated from tomato (Lycopersicon esculentum Mill) rhizosphere. Microb Pathog 140:10396. https://doi.org/10.1016/j.micpath.2020.103966
Tamilmani E, Radhakrishnan R, Sankaran K (2018) 13-Docosenamide release by bacteria in response to glucose during growth-fluorescein quenching and clinical application. Appl Microbiol Biotechnol 102:6673–6685. https://doi.org/10.1007/s00253-018-9127-x
Teather RM, Wood PJ (1982) Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol 43:777–780. https://doi.org/10.1128/aem.43.4.777-780.1982
Vacheron J, Desbrosses G, Bouffaud M-L, Touraine B, Moenne-Loccoz Y, Muller D, Legendre L, Wisniewski-Dye F, Prigent-Combaret C (2013) Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci 4:1–19. https://doi.org/10.3389/fpls.2013.00356
Valentine ME, Kirby BD, Withers TR, Johnson SL, Long TE, Hao Y, Lam JS, Niles RM, Yu HD (2020) Generation of a highly attenuated strain of Pseudomonas aeruginosa for commercial production of alginate. Microb Biotechnol 13:162–175. https://doi.org/10.1111/1751-7915.13411
Van Loon LC, Bakker PAHM, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483
Zehnder GW, Yao C, Murphy JF, Sikora EJ, Kloepper JW (2000) Induction of resistance in tomato against cucumber mosaic cucumovirus by plant growth-promoting rhizobacteria. BioControl 45:127–137. https://doi.org/10.1023/A:1009923702103
Zhang H, Kim MS, Krishnamachari V, Payton P, Sun Y, Grimson M, Farag MA, Ryu CM, Allen R, Melo IS, Pare PW (2007) Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226:839–51. https://doi.org/10.1007/s00425-007-0530-2
Acknowledgements
Ashish Kumar Gupta gratefully acknowledges University Grants Commission-Council of Scientific and Industrial Research National Eligibility Test Junior Research Fellowship (19/06/2016(i)EU-V). Jyoti Verma gratefully acknowledges University Grants Commission Rajiv Gandhi National Fellowship (F1-17.1/2017–18/RGNF-2017–18-SC-4TT-47935/(SA-III/Website).
Funding
This study was funded by University Grants Commission-Council of Scientific and Industrial Research National Eligibility Test Grant No. 19/06/2016(i)EU-V and Rajiv Gandhi National Fellowship Grant No. F1-17.1/2017–18/RGNF-2017–18-SC-4TT-47935/(SA-III/Website).
Author information
Authors and Affiliations
Contributions
SS and VP conceived the work and designed the experiments. AKG carried out bacterial isolation, identification including rRNA sequencing, and RT-PCR-related work. AKG, JV, and AS performed the plant growth promotion experiments. AKG, SS, and VP analysed the results. AKG wrote the first draft of the manuscript that was edited and finalized in its current form by SS and VP. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Ethical approval
The results reported herein are purely based on plants, and no humans/animals were used for this research.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gupta, A.K., Verma, J., Srivastava, A. et al. Pseudomonas aeruginosa isolate PM1 effectively controls virus infection and promotes growth in plants. Arch Microbiol 204, 494 (2022). https://doi.org/10.1007/s00203-022-03105-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03105-3