Abstract
During an investigation of microbial diversity in medicinal herbs, a novel actinomycete, strain NEAU-QHHV11T was isolated from the rhizosphere of Peucedanum praeruptorum Dunn collected from Xianglu Mountain in Heilongjiang Province, northeast China and characterized using a polyphasic approach. The organism was found to have typical characteristics of the genus Streptomyces. Phylogenetic analysis based on 16S rRNA gene sequence also indicated that strain NEAU-QHHV11T belongs to the genus Streptomyces and was most closely related to Streptomyces graminilatus NBRC 108882T (98.7 % sequence similarity) and Streptomyces turgidiscabies NBRC 16080T (98.7 % sequence similarity). The results of DNA–DNA hybridization and some phenotypic characteristics indicated that strain NEAU-QHHV11T could be distinguished from its close phylogenetic relatives. Thus, strain NEAU-QHHV11T represents a novel species of the genus Streptomyces, for which the name Streptomyces castaneus sp. nov. is proposed. The type strain is NEAU-QHHV11T (=CGMCC 4.7235T = DSM 100520T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Streptomyces was first described by Waksman and Henrici (1943) and now consists of more than 700 species with validly published names (http://www.bacterio.net/streptomycesa.html). Members of the genus are aerobic, Gram-positive bacteria which form an extensively branched substrate mycelium with aerial hyphae which differentiate into chains of spores and DNA with high G+C content (Lechevalier and Lechevalier 1970; Kämpfer 2012; Manfio et al. 1995). Indeed, Streptomyces is the largest genus of the phylum Actinobacteria (Kämpfer and Labeda 2006; Lodders and Kämpfer 2007) and constitutes the most predominant component of micro-organisms found in soils, sediments and decaying vegetation (Labeda et al. 2012). Peucedanum praeruptorum Dunn is one of the most popular traditional medicinal herbs which has been widely used in traditional Chinese medicine for over 1500 years and officially listed in the Chinese Pharmacopoeia (Liu et al. 2004). As part of a programme to discover actinomycetes with novel antibiotic production properties present in medicinal plants, strain NEAU-QHHV11T was isolated from the rhizosphere of P. praeruptorum Dunn. In this study, we performed a polyphasic taxonomy study on this strain and propose a novel species of the genus Streptomyces, Streptomyces castaneus sp. nov.
Materials and methods
Isolation and maintenance of the organism
Strain NEAU-QHHV11T was isolated from the rhizosphere soil of P. praeruptorum Dunn collected from Xianglu Mountain in Heilongjiang Province, northeast China (45°45′N127°29′E) in June 2014. The strain was isolated using the standard dilution plate method and grown on humic acid–vitamin agar (Hayakawa and Nonomura 1987) supplemented with cycloheximide (50 mg l−1) and nalidixic acid (20 mg l−1). After 21 days of aerobic incubation at 28 °C, the isolate was transferred and purified on the International Streptomyces Project (ISP) medium 3 (Shirling and Gottlieb 1966) and maintained as glycerol suspensions (20 %, v/v) at −80 °C. The type strains of Streptomyces graminilatus NBRC 108882T and Streptomyces turgidiscabies NBRC 16080T were purchased from the NITE Biological Resource Center (NBRC) and cultured under the same conditions for comparative analysis.
Morphological, cultural and physiological characteristics
Morphological characteristics were observed by light microscopy (Nikon ECLIPSE E200) and electron microscopy (Hitachi S-3400N) using cultures grown on ISP3 medium at 28 °C for 21 days. Samples for scanning electron microscopy were prepared using the method of Guan et al. (2015). Cultural characteristics were determined on ISP media 2–7 (Shirling and Gottlieb 1966) after 14 days at 28 °C. The ISCC–NBS colour charts were used to determine the designations of colony colours (Kelly 1964). Growth at different temperatures (4, 10, 15, 20, 28, 32, 37, 40 and 45 °C) was determined on ISP3 medium after incubation for 14 days. Growth tests for pH range (pH 3–12) and NaCl tolerance (0, 1, 2, 3, 4, 5, 6 and 7 % NaCl, w/v) were tested in ISP2 medium at 28 °C for 14 days on a rotary shaker. Production of catalase, esterase and urease was tested as described by Smibert and Krieg (1994). The utilization of sole carbon and nitrogen sources, decomposition of urea and cellulose, hydrolysis of starch and aesculin, reduction of nitrate, coagulation and peptonization of milk, liquefaction of gelatin and production of H2S were examined as described previously (Gordon et al. 1974; Yokota et al. 1993).
Chemotaxonomic characterization
Biomass for chemical studies was prepared by growing strain NEAU-QHHV11T in tryptic soy broth (TSB; Lee and Whang 2014) in shake flasks at 28 °C for 7 days. Cells were harvested by centrifugation, then washed with distilled water and freeze-dried. The isomer of diaminopimelic acid (DAP) in the cell wall hydrolysates was derivatized and analysed by a HPLC method (McKerrow et al. 2000) using an Agilent TC-C18 Column (250 × 4.6 mm i.d. 5 µm) with a mobile phase consisting of acetonitrile: 0.05 mol l−1 phosphate buffer pH 7.2 (15:85, v/v) at a flow rate of 0.5 ml min−1. An Agilent G1321A fluorescence detector was used for peak detection with 365 nm excitation and 455 nm long pass emission filters. Whole-cell sugars were analysed according to the procedures developed by Lechevalier and Lechevalier (1980). Polar lipids were examined by two-dimensional TLC and identified using the method of Minnikin et al. (1984). Menaquinones were extracted from freeze-dried biomass and purified according to Collins (1985). Extracts were analysed by a HPLC–UV method (Wu et al. 1989) using an Agilent Extend-C18 Column (150 × 4.6 mm, i.d. 5 μm), typically at 270 nm. The mobile phase was acetonitrile–propyl alcohol (60:40, v/v). The presence of mycolic acids was checked by the acid methanolysis method as described previously (Minnikin et al. 1980). To determine cellular fatty acid compositions, strain NEAU-QHHV11T was cultivated in TSB medium in shake flasks at 28 °C for 7 days. Fatty acid methyl esters were extracted from the biomass as described by Gao et al. (2014) and were analysed by GC–MS using the method of Xiang et al. (2011).
DNA preparation, amplification and determination of 16S rRNA sequence
Biomass for molecular biological studies was prepared by growing the organism in TSB medium in shake flasks at 28 °C for 7 days. Extraction of chromosomal DNA and PCR amplification of the 16S rRNA gene sequence were carried out according to the procedure developed by Kim et al. (2000). The PCR product was purified and cloned into the vector pMD19-T (Takara) and sequenced using an Applied Biosystems DNA sequencer (model 3730XL). An almost full-length 16S rRNA gene sequence of strain NEAU-QHHV11T (1517 bp) was obtained and aligned with multiple sequences obtained from the GenBank/EMBL/DDBJ databases using Clustal X1.83 software (Thompson et al. 1997). Phylogenetic trees were constructed with the neighbour-joining (Saitou and Nei 1987) and maximum likelihood (Felsenstein 1981) algorithms using Molecular Evolutionary Genetics Analysis (MEGA) software version 6.06 (Tamura et al. 2013). The stability of the topology of the phylogenetic tree was assessed using the bootstrap method with 1000 repetitions (Felsenstein 1985). A distance matrix was generated using Kimura’s two-parameter model (Kimura 1980). All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). 16S rRNA gene sequence similarities between strains were calculated on the basis of pairwise alignments using the EzTaxon-e server (Kim et al. 2012).
DNA base composition and DNA–DNA hybridization
The G+C content of the genomic DNA was determined by the thermal denaturation (Tm) method (Mandel and Marmur 1968) with Escherichia coli JM109 DNA as the control. The DNA–DNA relatedness tests between strain NEAU-QHHV11T and S. graminilatus NBRC 108882T and S. turgidiscabies NBRC 16080T were carried out as described by De Ley et al. (1970) under consideration of the modifications described by Huss et al. (1983), using a model Cary 100 Bio UV/VIS spectrophotometer equipped with a Peltier-thermostatted 6 × 6 multicell changer and a temperature controller with in situ temperature probe (Varian). The concentration and purity of DNA samples were determined by measuring the optical density at 260, 280 and 230 nm. The DNA samples used for hybridization were diluted to OD260 around 1.0 using 0.1 × SSC (saline sodium citrate buffer) and then broken using a JY92-II ultrasonic cell disruptor (ultrasonic time 3 s, interval time 4 s, 90 times). The DNA renaturation rates were determined in 2 × SSC at 70 °C. The experiments were performed with three replications, and the DNA–DNA relatedness value was expressed as mean of the three values.
Results and discussion
EzTaxon-e analysis of the 16S rRNA gene sequence (1517 bp) revealed that strain NEAU-QHHV11T belongs to the genus Streptomyces, with the highest 16S rRNA gene sequence similarity to S. graminilatus NBRC 108882T (98.7 %) and S. turgidiscabies NBRC 16080T (98.7 %). Phylogenetic analysis based on 16S rRNA gene sequences showed that strain NEAU-QHHV11T formed a separate branch with S. graminilatus NBRC 108882T and S. turgidiscabies NBRC 16080T in the neighbour-joining tree (Fig. 1) and a similar topology was also recovered with the maximum likelihood algorithm (Fig. S4). DNA–DNA hybridization was employed to further clarify the relatedness between strain NEAU-QHHV11T and S. graminilatus NBRC 108882T and S. turgidiscabies NBRC 16080T. The levels of DNA–DNA relatedness between them were 49.7 ± 0.5 and 39.1 ± 0.9 %, respectively. These values are below the threshold value of 70 % recommended by Wayne et al. (1987) for assigning strains to the same genomic species. The G+C content of the genomic DNA of strain NEAU-QHHV11T was 71.5 ± 0.3 mol %.
Neighbour-joining tree based on 16S rRNA gene sequences (1421 bp, by omitting unaligned regions) showing the relationship between strain NEAU-QHHV11T and related taxa. Asterisks indicate branches of the tree that were also recovered using the maximum likelihood method. Bootstrap values >50 % (based on 1000 replications) are shown at branch points. Bar, 0.002 substitutions per nucleotide position
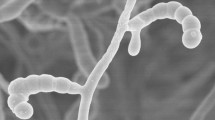
The morphological characteristics observation of a 3-week-old culture of strain NEAU-QHHV11T grown on ISP3 medium revealed that it had the typical characteristics of the genus Streptomyces. Aerial mycelium and substrate mycelium were well developed without fragmentation. Long spore chains were observed, and the spores were found to be cylindrical, smooth and non-motile (Fig. S1). Cultural characteristics of strain NEAU-QHHV11T are shown in Table S1. Good growth was observed on ISP2, ISP3, ISP4 and ISP7 media; moderate growth was observed on ISP5 and ISP6 media. The colour of the colonies varied from yellowish white to deep purplish red. Purplish red soluble pigments were observed on ISP3 medium (Fig. S2), which could apparently distinguish the isolate from S. graminilatus NBRC 108882T and S. turgidiscabies NBRC 16080T. Melanoid pigments were not observed on ISP6 or ISP7 media. Growth of strain NEAU-QHHV11T was observed at 10–32 °C (optimum 28 °C), pH range 5–11 (optimum pH 7) and in the presence of 0–4 % NaCl (w/v). Hydrolysis of starch and Tween 20 were positive, which could also differentiate strain NEAU-QHHV11T from its closely related strains. Other physiological and biochemical characteristics of strain NEAU-QHHV11T compared with S. graminilatus NBRC 108882T and S. turgidiscabies NBRC 16080T are listed in Table 1.
Strain NEAU-QHHV11T contained ll-diaminopimelic acid as the cell wall diamino acid, ribose and glucose as whole-cell sugars and MK-9(H8) (51.6 %) and MK-9(H10) (38.6 %) as the predominant menaquinones, with smaller amounts of MK-9(H6) (6.7 %), MK-9(H0) (1.8 %) and MK-9(H2) (1.3 %). The polar lipids were found to be diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, phosphatidylinositol mannoside and an unidentified phospholipid (Supplementary Fig. S3). The major cellular fatty acids were determined to be composed of iso-C16:0 (27.8 %), C17:0 cyclo (14.0 %), anteiso-C15:0 (9.2 %), iso-C15:0 (8.9 %), anteiso-C17:0 (7.7 %), C16:0 (5.9 %) and C16:1ω5c (5.1 %), and minor amounts of C16:1ω7c (4.7 %), iso-C17:0 (4.2 %), C18:1ω2c (3.0 %), iso-C14:0 (2.4 %), C15:0 (1.9 %), C18:0 (1.9 %) and C17:1ω9c (1.5 %) were also present. Strain NEAU-QHHV11T contained major amounts of iso- and anteiso-fatty acids, which are in accord with the fatty acid composition of Streptomyces spp. However, the presence of C17:0 cyclo as major fatty acids is rare in the genus Streptomyces (Kämpfer 2012).
In conclusion, it is evident from the genotypic and phenotypic data that strain NEAU-QHHV11T represents a novel species of the genus Streptomyces, for which the name Streptomyces castaneus sp. nov. is proposed.
Description of Streptomyces castaneus sp. nov
Streptomyces castaneus (cas.ta’ne.us. L. masc. adj. castaneus, chestnut coloured).
Aerobic, Gram-staining positive, non-motile actinobacteria that forms well-developed substrate mycelium and aerial hyphae. Long spore chains appear as straight, and spores are smooth, cylindrical and non-motile. Grows well on ISP 2, ISP 3, ISP 4 and ISP 7 media, moderately on ISP 5 and ISP 6 media. Purplish red soluble pigment is observed on ISP3 medium and grows at 15–32 °C, pH range 5–11 and in the presence of 0–4 % NaCl (w/v) with optimum at 28 °C and pH 7. Positive for liquefaction of gelatin, coagulation and peptonization of milk, production of esterase, decomposition of urea, hydrolysis of starch and aesculin and negative for decomposition of cellulose, reduction of nitrate and production of H2S. d-Galactose, d-glucose, meso-inositol, lactose, d-maltose, d-mannitol, d-mannose, d-raffinose, l-rhamnose, d-sorbitol and d-sucrose are utilized as sole carbon sources, while l-arabinose, d-fructose, d-ribose and d-xylose are not metabolized. l-Alanine, l-arginine, l-asparagine, l-aspartic acid, l-glutamic acid, l-glutamine, glycine, l-proline, l-serine and l-threonine are utilized as sole nitrogen sources, while creatine and l-tyrosine are not metabolized. The cell wall contains ll-diaminopimelic acid as diagnostic diamino acid, and the whole-cell hydrolysates contain ribose and glucose. The major menaquinones are MK-9(H8) and MK-9(H10). The polar lipids profile contains diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, phosphatidylinositol mannoside and an unidentified phospholipid. The major fatty acids are C16:0, C17:0 cyclo, anteiso-C15:0, iso-C15:0, anteiso-C17:0 and C16:1ω5c (>5 %). The DNA G+C content of the type strain is 71.5 mol %.
The type strain is NEAU-QHHV11T (=CGMCC 4.7235T = DSM 100520T), which was isolated from the rhizosphere soil of Peucedanum praeruptorum Dunn collected from Xianglu Mountain in Heilongjiang Province, northeast China. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain NEAU-QHHV11T is KP230425.
References
Collins MD (1985) Isoprenoid quinone analyses in bacterial classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–284
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gao RX, Liu CX, Zhao JW, Jia FY, Yu C, Yang LY, Wang XJ, Xiang WS (2014) Micromonospora jinlongensis sp. nov., isolated from muddy soil in China and emended description of the genus Micromonospora. Antonie Van Leeuwenhoek 105:307–315
Gordon RE, Barnett DA, Handerhan JE, Pang C (1974) Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol 24:54–63
Guan XJ, Liu CX, Zhao JW, Fang BZ, Zhang YJ, Li LJ, Jin PJ, Wang XJ, Xiang WS (2015) Streptomyces maoxianensis sp. nov., a novel actinomycete isolated from soil in Maoxian, China. Antonie van Leeuwenhoek 107:1119–1126
Hayakawa M, Nonomura H (1987) Humic acid-vitamin agar, a new medium for the selective isolation of soil actinobacteria. J Ferment Technol 65:501–509
Huss VAR, Festl H, Schleifer KH (1983) Studies on the spectrometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Kämpfer P (2012) Genus I Streptomyces. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology, vol 5, 2nd edn. The Actinobacteria Springer, New York, pp 1455–1767
Kämpfer P, Labeda DP (2006) International committee on systematics of prokaryotes; subcommittee on the taxonomy of the Streptomycetaceae: minutes of the meeting, 25 July 2005, San Francisco, CA, USA. Int J Syst Evol Microbiol 56:495
Kelly KL (1964) Inter-society color council-national bureau of standards color-name charts illustrated with centroid colors. US Government Printing Office, Washington
Kim SB, Brown R, Oldfield C, Gilbert SC, Iliarionov S, Goodfellow M (2000) Gordonia amicalis sp. nov., a novel dibenzothiophene-desulphurizing actinobacterial. Int J Syst Evol Microbiol 50:2031–2036
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vanncanneyt M, Swings J, Kim SB, Liu Z, Chun J, Tamura T, Oguchi A, Kikuchi T, Kikuchi H, Nishii T, Tsuji K, Yamaguchi Y, Tase A, Takahashi M, Sakane T, Suzuki KI, Hatano K (2012) Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek 101:73–104
Lechevalier MP, Lechevalier H (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol 20:435–443
Lechevalier MP, Lechevalier HA (1980) The chemotaxonomy of actinomycetes. In: Dietz A, Thayer DW (eds) Actinobacterial taxonomy special publication. Society of Industrial Microbiology, Arlington 6:227–291
Lee HJ, Whang KS (2014) Streptomyces graminilatus sp. nov., isolated from bamboo litter. Int J Syst Evol Microbiol 64:528–532
Liu R, Feng L, Sun A, Kong L (2004) Preparative isolation and purification of coumarins from Peucedanum praeruptorum Dunn by high-speed counter-current chromatography. J Chromatogr A 1057:89–94
Lodders N, Kämpfer P (2007) Streptomycetaceae: phylogeny, ecology and pathogenicity. In: Encyclopedia of life science. Wiley, pp 1–9. doi:10.1002/9780470015902.a0020392
Mandel M, Marmur J (1968) Use of ultraviolet absorbance temperature profile for determining the guanine plus cytosine content of DNA. Methods Enzymol 12B:195–206
Manfio GP, Zakrzewska-Czerwinska J, Atalan E, Goodfellow M (1995) Towards minimal standards for description of Streptomyces species. Biotechnologia 7:242–283
McKerrow J, Vagg S, McKinney T, Seviour EM, Maszenan AM, Brooks P, Seviour RJ (2000) A simple HPLC method for analysing diaminopimelic acid diastereomers in cell walls of Gram-positive bacteria. Lett Appl Microbiol 30:178–182
Minnikin DE, Hutchinson IG, Caldicott, AB, Goodfellow M (1980) Thin-layer chromatography of methanolysates of mycolic acid-containing bacteria. J Chromatogr 188:221–233
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal K, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Miyajima K, Tanaka F, Takeuchi T, Kuninaga S (1998) Streptomyces turgidiscabies sp. nov. Int J Syst Evol Microbiol 48:495–502
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 607–654
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Gibson TJ, Plewniak F (1997) The clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 5:4876–4882
Waksman SA, Henrici AT (1943) The nomenclature and classification of the actinomycetes. J Bacteriol 46:337–341
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG (1987) International committee on systematic bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wu C, Lu X, Qin M, Wang Y, Ruan J (1989) Analysis of menaquinone compound in microbial cells by HPLC. Microbiology [English translation of Microbiology (Beijing)] 16:176–178
Xiang WS, Liu CX, Wang XJ, Du J, Xi LJ, Huang Y (2011) Actinoalloteichus nanshanensis sp. nov., isolated from the rhizosphere of a fig tree (Ficus religiosa). Int J Syst Evol Microbiol 61:1165–1169
Yokota A, Tamura T, Hasegawa T, Huang LH (1993) Catenuloplanes japonicus gen. nov., sp. nov., nom. rev., a new genus of the order Actinomycetales. Int J Syst Bacteriol 43:805–812
Acknowledgments
This work was supported in part by grants from the National Outstanding Youth Foundation (No. 31225024), the National Key Technology R&D Program (No. 2012BAD19B06), the National Natural Science Foundation of China (No. 31471832, 31171913, 31500010, 31572070 and 31372006) and Chang Jiang Scholar Candidates Program for Provincial Universities in Heilongjiang (CSCP).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Erko Stackebrandt.
Shuyu Zhou and Zhilei Li have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, S., Li, Z., Bai, L. et al. Streptomyces castaneus sp. nov., a novel actinomycete isolated from the rhizosphere of Peucedanum praeruptorum Dunn. Arch Microbiol 199, 45–50 (2017). https://doi.org/10.1007/s00203-016-1274-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-016-1274-9