Abstract
A novel actinomycete strain, designated as MG28T, was isolated from rhizosphere soil of Akebia trifoliate. The taxonomic position of the strain was investigated by using a polyphasic approach. BLAST search of the full-length 16S rRNA gene sequence of strain MG28T indicated it represented a member of the genus Streptomyces, and displayed 99.03%, 98.90%, 98.90%, 98.89%, 98.83% and less than 98.70% sequence similarities with S. phaeolivaceus GY16T, S. deccanensis KCTC 19241T, S. europaeiscabiei KACC 20186T, S. fructofermentans CGMCC 4.1593T, S. scabiei NRRL B-16523T and other species of the genus Streptomyces with validly published names, respectively. Phylogenomic analysis indicated that strain MG28T was closely related to Streptomyces deccanensis KCTC 19241T. However, the average nucleotide identity values and the digital DNA-DNA hybridization values between them indicated that strain MG28T represented a distinct species. Furthermore, strain MG28T was also distinctly differentiated from strain KCTC 19241T by morphological, physiological and biochemical characteristics. Therefore, strain MG28T (= MCCC 1K06895T = JCM 34922T) represents a novel species of the genus Streptomyces, for which the name Streptomyces akebiae sp. nov. is proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nowadays, plants and animals, as medicine source, play an important role (Alves 2012; Ho 2005), but it can't be ignored that microorganisms are also significant source of pharmaceutically drugs (Bérdy 2012; Debbab et al. 2010; Waditee-Sirisattha et al. 2016). In 2010, it was reported that more than 33,000 microbial secondary metabolites had been discovered. Among these bioactive compounds, approximately 13,600 were produced by actinomycetes, in which about 75% was produced by Streptomyces (Olano et al. 2009; Solecka et al. 2012). So, in recent decades, Streptomyces has gained more and more attention for their potential ability to produce new natural products (Bouizgarne et al. 2009; Goodfellow and Fiedler 2010). These reports encouraged us to search for novel Streptomyces with potentially useful applications in all kinds of samples. In previous studies, many novel Streptomyces such as S. rhizosphaericola (Vargas Hoyos et al. 2019), S. tritici (Zhao et al. 2018), S. triticagri (Han et al. 2020), S. triticirhizae (Han et al. 2020) and S. inhibens (Jin et al. 2019), S. fagopyri (Guo et al. 2020) and S. broussonetiae (Mo et al. 2020) have been isolated from the rhizosphere soil of plants. In recent years, during the process of investigating the diversity and function of microbes from the rhizosphere soil of Akebia trifoliate, one of strains, designated as MG28T, showed antifungal activity against plant fungi. The aim of the present work was to determine the taxonomic status of this strain. The results of polyphasic taxonomic studies indicated that strain MG28T represents a novel species of genus Streptomyces, for which the name Streptomyces akebia sp. nov. is proposed.
Materials and methods
Isolation and maintenance of the organism
The rhizosphere soil sample of Akebia trifoliate was collected from Changde city located in the northwest of Hunan Province, China (N29° 28′ 30.87″, E111° 27′ 57.70″). Strain MG28T was isolated using the methods as described by Mo et al. (2018). Strain MG28T was stored for short term deposition on ISP 4 agar medium (Shirling and Gottlieb 1966) slopes at 4 °C and suspended in sterile 30% (w/v) glycerol solution for long term deposition at − 80 °C. The type strain, Streptomyces deccanensis KCTC 19241T was purchased from the KCTC (Korean Collection for Type Cultures, Daejeon, Republic of Korea). Reference strain was grown under the same conditions for comparative testing.
Morphological, cultural and physiological characteristics
Cell morphology of strain MG28T were observed by scanning electron microscope (FEI-Quanta 450, America) after incubation on Gause's synthetic agar medium (Atlas 1993) for 21 days at 28 °C. The color of colonies and soluble pigments were observed on Gause's synthetic medium and ISP media (Shirling and Gottlieb 1966) after incubation at 28 °C for 21 days. Colors of substrate mycelia, aerial hyphae and soluble pigments were determined by the Color Standards and Color Nomenclature (Ridgway 1912). Growth at different temperatures (4, 10, 15, 20, 25, 30, 35, 40 and 45 °C) and the concentrations of NaCl (0–14%, w/v with an interval of 1% unit) was tested on ISP2 medium at 28 °C for 14 days. The pH range (pH 4.0–13.0 at intervals of 1.0 pH unit) for growth was tested by using ISP 2 as the basal medium using the buffer system as described by Xu et al. (2007). Carbon source utilization was tested by using ISP 9 medium (Shirling and Gottlieb 1966) supplemented with 0.5% carbon sources. The utilization of nitrogen sources was performed as described previously (Shirling and Gottlieb 1966). The other physiological characteristic including gelatin liquefaction, nitrate reduction, starch and aesculin hydrolysis, degradation tests for Tweens (20, 40, 60 and 80) were carried out according to the methods described by Xu et al. (2007) and Ruan and Huang (2011). The experiments were carried out in triplicate.
Chemotaxonomic characterisation
Biomass for chemotaxonomic analysis was collected by centrifugation from cultures grown in tryptic soy broth medium (TSB medium) in shake flasks for 7 days at 28 °C and then washed twice with distilled water. Cellular fatty acid composition analysis was carried out by China Center of Industrial Culture Collection (CICC) according to the protocol of the Sherlock Microbial Identification system (MIDI system; http://www.midi-inc.com/) and analyzed by GC (6968; Hewlett Packard) using the Microbial Identification software package (MIDI 2005). The isomers of diaminopimelic acid analysis and sugar analysis in whole-cell hydrolysates were performed according to the procedures described by Hasegawa et al. (1983) and Lechevalier and Lechevalier (1970). Menaquinones were extracted according to the method of Collins et al. (1977) and analysed by HPLC (Kroppenstedt 1985). The polar lipids were extracted, separated and analysed as described by Komagata and Suzuki (1987).
Phylogenetic analysis and genomic DNA-DNA correlation analysis
Genomic DNA extraction and PCR amplification of the 16S rRNA gene were performed as described by Weisburg et al. (1991) and Lane (1991). The PCR product was purified using a Mag Extractor PCR and Gel Clean up kit (toyobo) according to the manufacturers' instructions and sequenced directly using an automated DNA sequencing system (ABI 3730XL; Applied Biosystems) by Sangon Biotech (Shanghai). Whole-genome sequencing of strain MG28T and S. deccanensis KCTC 19241T was performed by Wuhan Benagen Technology Co., Ltd (Hubei, China) using a Nanopore PromethION sequencing system. The sequenced reads were assembled using SOAPdenovo (version 2.04) software (Xie et al. 2014). The 16S rRNA gene sequence of strain MG28T was compared with public databases and EzBioCloud database (http:// www.ezbiocloud.net/eztaxon; (Yoon et al. 2017). The genome sequences of strain MG28T and the closely related type strains with validly published names were used for reconstructing phylogenetic trees. Phylogenomic analysis was carried out using the Type (Strain) Genome Server (https://tygs.dsmz.de/) (Meier-Kolthoff and Göker 2019). The average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) values between the genomes of strain MG28T and its relatives were calculated using the JSpeciesWS online service (Richter et al. 2016) and the genome-to-genome distance calculator (Meier-Kolthoff et al. 2013), respectively. For calculating dDDH value, Formula 2 was used. The G + C content of the genomic DNA of strain MG28T was deduced from the genomic data.
Results and discussion
Morphological and physiological characterization of the strain MG28T
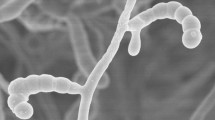
The morphological characteristics showed that strain MG28T had the typical characteristics of the genus Streptomyces. Cells of strain MG28T was Gram-stain-positive and aerobic. Strain MG28T formed an extensively branched substrate hyphae and aerial mycelium on Gause's synthetic agar medium, and produced straight to flexuous spore chains consisting of rod spores with smooth-surfaced (Fig. 1). Strain MG28T was observed to grow well on all tested media. Melanin and diffusible pigment were produced on ISP 6 and ISP 7 agar media, respectively. Other cultural characteristics of strain MG28T were shown in Table 1. Strain MG28T was found to grow at 4–45 °C (optimum, 28 °C), pH 6.0–11.0 (optimum, pH 7.0) and tolerate up to 5.0% (w/v) NaCl. Detailed physiological characteristics about sole carbon source utilization about strains are presented in the species description.
Chemotaxonomic characteristics
The chemotaxonomic studies revealed that strain MG28T exhibited characteristics typical of members of the genus Streptomyces. For example, the cell wall contained ll-diaminopimelic acid as the diagnostic amino acid, and the whole-cell hydrolysates were galactose and glucose. The predominant cellular fatty acids (> 5%) of strain MG28T were Sum in Feature 3 (C16:1ω7C/C16:1ω6C) (19.7%), iso-C16:0 (18.9%), anteiso-C15:0 (17.8%), C16:0 (7.0%), iso-C15:0 (5.3%) and anteiso-C17:0 (5.2%). Other fatty acids present in smaller amounts (> 1%) were iso-C14:0 (4.8%), anteiso-C17:1ω5C (2.9%), C14:0 (2.9%), iso H-C16:1 (2.3%), Sum in Feature 9 (10-methyl C16:0) (2.0%), cyclo-C17:0 (1.4%) and anteiso-C13:0 (1.0%). The menaquinones were MK-9 (4.6%), MK-9(H2) (30.1%), MK-9(H4) (32.8%), MK-9(H6) (18.3%) and MK-9(H8) (12.9%), which is also typical of members of the genus Streptomyces. The DNA G + C content of strain MG28T was 70.8%, within the range 69–78% of the member of the genus Streptomyces (Wright and Bibb 1992). The polar lipids consisted of diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylinositol mannosides (PIM), phosphatidyl glycerol (PG) and unidentified spots (L1) (Fig. S1, available in the online version of this paper).
Phylogenic analysis
Based on the full-length 16S rRNA gene sequence (1527 bp) analysis, strain MG28T represented a member of the genus Streptomyces, and shared 99.03% similarity with S. phaeolivaceus GY16T, 98.90% similarities with S. deccanensis KCTC 19241T and S. europaeiscabiei KACC 20186T, 98.89% similarity with S. fructofermentans CGMCC 4.1593T, 98.83% similarity with S. scabiei NRRL B-16523T and less than 98.7% similarity with other type species of the genus Streptomyces. Considering that the phylogenomic analysis exhibited better resolution than the phylogenetic analysis based on 16S rRNA gene sequence (Rodriguez et al. 2018), so, in the present work, the phylogenomic analysis was carried out in order to clarify taxonomic status of strain MG28T. It was shown in Fig. 2, strain MG28T was clustered together with S. deccanensis KCTC 19241T, suggesting that it was closely related to S. deccanensis KCTC 19241T. However, the ANIm/ANIb and dDDH values between strain MG28T and S. deccanensis KCTC 19241T were 92.9/91.4% and 47.0%, much less than 95–96% and 70% cut-off points recommended for delineating species (Richter and Rosselló-Móra 2009; Chun et al. 2018). In addition, the cultural, physiological and biochemical characteristics were dissimilar enough to distinguish strain MG28T from S. deccanensis KCTC 19241T (Tables 1 and 2). For example, strain MG28T forms a gray aerial mycelium and yellow substrate mycelium with no diffusible pigment on Gause's synthetic medium, while S. deccanensis KCTC 19241T develops white aerial mycelium and kaiser brown substrate mycelium with apricot buff diffusible pigment. In addition, the appearance of the straight to flexuous spore's chain with smooth surfaces of strain MG28T is clearly distinguishable from the straight spore's chain with hairy surfaces of S. deccanensis KCTC 19241T. Meanwhile, according to the description of Stackebrandt and Ebers (2006), if 16S sequence similarity between two strains ≥ 98.7%, ANI or dDDH values need to be calculated to evaluate their DNA-DNA relatedness in delineating new species. In the present work, ANIm/ANIb and dDDH values between strain MG28T and S. phaeolivaceus GY16T, between strain MG28T and S. europaeiscabiei KACC 20186T, between strain MG28T and S. fructofermentans CGMCC 4.1593T, between strain MG28T and S. scabiei NRRL B-16523T were 85.8–90.7%/(80.2–91.4%) and 23.2–38.8% (Table 3) [37, 38], respectively, well below the 95–96% and 70% cut-off point recommended for delineating prokaryote species. In conclusion, strain MG28T represents a novel Streptomyces species, for which the name Streptomyces akebiae sp. nov. is proposed.
Phylogenetic tree based on whole genome sequences of MG28T and related reference strains. Tree inferred with FastME 2.1.6.1 (Vincent et al. 2015) from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of GBDP distance formula d5. The numbers above branches are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 96.0%. The tree was rooted at the midpoint (Farris 1972)
Description of Streptomyces akebiae sp. nov.
Streptomyces akebiae (a.ke'bi.ae. N.L. gen. n. akebiae referring to come from Akebia trifoliata)
Good growth on Gause's synthetic medium and ISP 2-7. The aerial mycelia are gray and the substrate mycelia are yellow on Gause's synthetic agar medium. The aerial mycelia differentiate into straight to flexuous spore chains (which are) consisting of rod spores with smooth-surfaced. Growth occurs at pH 6.0–11.0 (optimum, 7.0), 4–45 °C (optimum, 28 °C) and 0–5.0% NaCl (w/v). Positive in tests for aesculin hydrolysis, nitrate reduction, gelatin liquefaction, urease and degradation of starch, negative for Tweens (20, 40, 60 and 80) decomposition. Cellobiose, d-mannitol, d-ribose, d-xylose, glycerol, l-rhamnose, and sucrose can be used as sole carbon source for growth, but not l-arabinose, maltose or myo-inositol. l-arginine, l-cystine and l-phenylalanine can be utilized as sole nitrogen source, but not for creatine, l-methionine, l-serine or l-valine. The cell wall contains alanine, glutamic acid, glycine and ll-diaminopimelic acid, and the whole-cell hydrolysates are galactose and glucose. The polar lipids are diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylinositol mannosides (PIM), phosphatidyl glycerol (PG) and unidentified spots (L1). The menaquinones are MK-9, MK-9(H2), MK-9(H4), MK-9(H6) and MK-9(H8). The major fatty acids (> 5%) are C16:1ω7C/C16:1ω6C, iso-C16:0, anteiso-C15:0, C16:0, iso-C15:0 and anteiso-C17:0. The genomic DNA G+C content of the type strain is 70.8%.
The type strain, MG28T (= MCCC 1K06895T = JCM 34922T) was isolated from rhizosphere soil of Akebia trifoliate, in Changde of Hunan Province, China. The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequence and genome sequence of strain MG28T are OM368590 and CP080647, respectively.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- MCCC:
-
Marine Culture Collection of China
- ISP:
-
International Streptomyces Project
References
Alves R (2012) Relationship between fauna and people and the role of ethnozoology in animal conservation. Ethnobiol Conserv 1:1–69. https://doi.org/10.15451/ec2012-8-1.2-1-69
Atlas RM (1993) Handbook of microbiological media. Edited by Parks LC. CRC Press, Boca Raton
Bouizgarne B, Lanoot B, Loqman S, Sproer C, Klenk HP, Swings J, Ouhdouch Y (2009) Streptomyces marokkonensis sp. nov., isolated from rhizosphere soil of Argania spinosa L. Int J Syst Evol Microbiol 59:2857–2863. https://doi.org/10.1099/ijs.0.011387-0
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo MEF (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. https://doi.org/10.1099/ijsem.0.002516
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230. https://doi.org/10.1099/00221287-100-2-221
Debbab A, Aly AH, Lin WH, Proksch P (2010) Bioactive compounds from marine bacteria and fungi. Microb Biotechnol 3(5):544–563. https://doi.org/10.1111/j.1751-7915.2010.00179.x
Farris JS (1972) Estimating phylogenetictrees from distance matrices. Am Nat 106:645–668
Goodfellow M, Fiedler HP (2010) A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie Van Leeuwenhoek 98:119–142. https://doi.org/10.1007/s10482-010-9460-2
Guo YH, Tang XK, Hu SR, Li KQ, Zhou ML, Gao J (2020) Steptomyces fagopyri sp. nov. a novel actinomycete isolated from rhizospheric soil of Fagopyrum dibotrys. Int J Syst Evol Microbiol 70:6437–6443. https://doi.org/10.1099/ijsem.0.004555
Han C, Yu Z, Zhao J, Shi H, Hu J, Wang X (2020) Streptomyces triticagri sp. nov., and Streptomyces triticirhizae sp. nov., two novel Actinobacteria isolated from the rhizosphere soil of wheat (Triticum aestivum L.). Int J Syst Evol Microbiol 70:126–138. https://doi.org/10.1099/ijsem.0.003727
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322. https://doi.org/10.2323/jgam.29.319
Ho J (2005) Information resources on human-animal relationships past and present. AWIC Resour Ser 30:1–94
János B (2012) Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot (tokyo) 65:385–395. https://doi.org/10.1038/ja.2012.27
Jin L, Zhao Y, Song W, Duan L, Jiang S, Wang X (2019) Streptomyces inhibens sp. nov., a novel actinomycete isolated from rhizosphere soil of wheat (Triticum aestivum L.). Int J Syst Evol Microbiol 69:688–695. https://doi.org/10.1099/ijsem.0.003204
Komagata K, Suzuki KI (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207. https://doi.org/10.1016/S0580-9517(08)70410-0
Kroppenstedt RM (1985) Fatty acid and menaquinone analysis of actinomycetes and related organisms. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 173–199
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Lechevalier MP, Lechevalier H (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol 20:435–443. https://doi.org/10.1099/00207713-20-4-435
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:14. https://doi.org/10.1186/1471-2105-14-60
MIDI (2005) Sherlock microbial identification system operating manual, Version 6.0. Newark DE: MIDI Inc.
Mo P, Zhao JR, Li KQ, Tang XK, Gao J (2018) Streptomyces manganisoli sp. nov., a novel actinomycete isolated from manganese-contaminated soil. Int J Syst Evol Microbiol 68:1890–1895. https://doi.org/10.1099/ijsem.0.002762
Mo P, Liu J, Zhao YL, Xu ZG (2020) Streptomyces phaeolivaceus sp. nov. and Streptomyces broussonetiae sp. nov. isolated from the leaves and rhizosphere soil of Broussonetia papyrifera. Int J Syst Evol Microbiol 70:6458–6467. https://doi.org/10.1099/ijsem.0.004556
Olano C, Méndez C, Salas JA (2009) Antitumor compounds from marine actinomycetes. Mar Drugs 7:210–248. https://doi.org/10.3390/md7020210
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Richter M, Rosselló-Móra R, Glöckner FO, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32(6):929–931. https://doi.org/10.1093/bioinformatics/btv681
Ridgway R (1912) Color standards and color nomenclature. Ridgway, Washington, DC, pp 1–43 (plate I–LII)
Rodriguez RL, Gunturu S, Harvey WT, Rossello-Mora R, Tiedje JM, Cole HR, Konstantinidis KT (2018) The microbial genomes atlas (MiGA) webserver: taxonomic and gene diversity analysis of archaea and bacteria at the whole genome level. Nucleic Acids Res 46:W282–W288. https://doi.org/10.1093/nar/gky467
Ruan J, Huang Y (2011) Rapid Identification and systematics of actinobacteria. Science Press, Beijing
Shirling EB, Gottlieb D (1966) Methods for characterisation of Streptomyces species. Int J Syst Bacteriol 16:313–340. https://doi.org/10.1099/00207713-16-3-313
Solecka J, Zajko J, Postek M, Rajnisz A (2012) Biologically active secondary metabolites from actinomycetes. Cent Eur J Biol 7:373–390. https://doi.org/10.2478/s11535-012-0036-1
Stackebrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155
Vargas Hoyos HA, Nobre Santos S, Da Silva LJ, Paulino Silva FS, Bonaldo Genuario D, Domingues Zucchi T, Soares Melo I (2019) Streptomyces rhizosphaericola sp. nov., an actinobacterium isolated from the wheat rhizosphere. Int J Syst Evol Microbiol 69:2431–2439. https://doi.org/10.1099/ijsem.0.003498
Vincent L, Richard D, Olivier G (2015) FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol 32:2798–2800. https://doi.org/10.1093/molbev/msv150
Waditee-Sirisattha R, Kageyama H, Takabe T (2016) Halophilic microorganism resources and their applications in industrial and environmental biotechnology. AIMS Microbiol 2:42–54. https://doi.org/10.3934/microbiol.2016.1.42
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Wright F, Bibb MJ (1992) Codon usage in the G+C-rich Streptomyces genome. Gene 113:55–65. https://doi.org/10.1016/0378-1119(92)90669-G
Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S, Zhou X, Lam T, Li Y, Xu X, Wong G, Wang J (2014) SOAPdenovo-trans: denovo transcriptome assembly with short RNA-seq reads. Bioinformatics 30:1660–1666. https://doi.org/10.1093/bioinformatics/btu077
Xu LH, Li WJ, Liu ZH, Jiang CL (2007) Actinomycetes systematics: principles, methods and practices. Science Press, Beijing
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Zhao J, Shi L, Li W, Wang J, Wang H, Han T, Tian Y, Xiang W, Wang X (2018) Streptomyces tritici sp. nov., a novel actinomycete isolated from rhizosphere soil of wheat (Triticum aestivum L.). Int J Syst Evol Microbiol 68:492–497. https://doi.org/10.1099/ijsem.0.002493
Acknowledgements
The authors thank Shuang Wan and Wangjie Xu (Wuhan Benagen Technology Co., Ltd, Hubei, China), Yongpeng Yang (Marine Culture Collection of China, MCCC, Xiamen, China) and CICC (China Centre of Industrial Culture Collection, Beijing, China) for providing excellent technical assistance.
Funding
This work was supported by Education Department of Hunan Province (21C0515), Natural Science Foundation of Hunan Province (2021JJ50024), Key projects of Hunan Provincial Department of Education (19A340), the Doctoral Start-up project of Hunan University of Arts and Science (21BSQD09 and 21BSQD10) and Innovation Team of Microbial Technology in Hunan University of Arts and Science.
Author information
Authors and Affiliations
Contributions
WZ and JG conceived the idea of the study; PM, KL, JZ, FZ, YC, XL, XL, KH, WZ and JG analysed the data; WZ and JG interpreted the results. PM, KL, WZ and JG wrote the paper. All authors discussed the results and revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animal experiments by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession numbers for the full-length 16S rRNA gene sequence and genome sequence of strain MG28T and Streptomyces deccanensis KCTC 19241T are OM368590 and OM807210, and CP080647 and CP092431, respectively.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mo, P., Li, K., Zhou, J. et al. Streptomyces akebiae sp. nov., a novel actinomycete isolated from rhizosphere soil of Akebia trifoliate. Antonie van Leeuwenhoek 115, 1297–1305 (2022). https://doi.org/10.1007/s10482-022-01772-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-022-01772-2