Abstract
Biological nitrogen fixation from the legume-rhizobia symbiosis is one of the main sources of fixed nitrogen on land environments. Diazotrophic bacteria taxonomy has been substantially modified by the joint use of phenotypic, physiological and molecular aspects. Among these molecular tools, sequencing and genotyping of genomic regions such as 16S rDNA and repetitive conserved DNA regions have boosted the accuracy of species identification. This research is a phylogenetic study of diazotrophic bacteria from sabiá (Mimosa caesalpiniifolia Benth.), inoculated with soils from five municipalities of the Brazilian Northeast. After bacterial isolation and morphophysiological characterization, genotyping was performed using REP, ERIC and BOX oligonucleotides and 16S rDNA sequencing for genetic diversity identification. A 1.5b Kb fragment of the 16S rDNA was amplified from each isolate. Morphophysiological characterization of the 47 isolates created a dendrogram, where isolate PE-GR02 formed a monophyletic branch. The fingerprinting conducted with BOX, ERIC and REP shows distinct patterns, and their compilation created a dendrogram with diverse groups and, after blasting in GenBank, resulted in genetic identities ranging from 77 to 99 % with Burkholderia strains. The 16S rDNA phylogenetic tree constructed with these isolates and GenBank deposits of strains recommended for inoculant production confirm these isolates are distinct from the previously deposited strains, whereas isolates PE-CR02, PE-CR4, PE-CR07, PE-CR09 and PE-GE06 were the most distinct within the group. Morphophysiological characterization and BOX, ERIC and REP compilation enhanced the discrimination of the isolates, and the 16S rDNA sequences compared with GenBank confirmed the preference of Mimosa for Burkholderia diazotrophic bacteria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mimosa caesalpiniifolia Benth. is a species of legume that is native to northeastern Brazil and is characterized by rapid growth, high regeneration capacity and drought resistance. It is considered a type of multi-use plant because it provides a food source for cattle, especially during the dry season in semiarid climates, and as a material for the construction of sturdy wooden posts and poles. This plant also shows great potential for reclamation due to its symbiotic diazotrophs (Babić et al. 2008). Nitrogen fixation by rhizobia-legume symbiosis is generally the main source of biologically fixed nitrogen for this species.
Many Burkholderia species occupy several ecological niches in various environments, including water, contaminated soil, plant rhizospheres, human respiratory tracts and hospital environments (Rufini et al. 2011). This genus encompasses approximately 30 distinct species originating from different locations. The interaction among Burkholderias and other organisms can be either pathogenic or symbiotic, or both, whereas several species, such as B. glathei, B. graminis, B. phenazinium, B. caribensis, B. caledonica, B. hospita, B. terrícola and B. sacchari, have unknown ecological roles. In addition, other Burkholderia species can be applied for plant growth promotion, biocontrol, bioremediation as well as nitrogen in soil environments, and this relationship dates back at least 58 MY (Sprent 2008). Elemental nitrogen is highly volatile and is one of the most expensive elements used for chemical fertilization. Therefore, rhizobia symbiosis is an alternative method for reducing fertilizer costs. Nodulation and nitrogen-fixing bacteria (NFB) are affected by biotic and abiotic factors (Gualter et al. 2008; Rufini et al. 2011). Among the biotic factors, genotypic characteristics of macro- and microsymbionts are important and influence the exchange of molecular signals, which reflect the different responses to host range, specificity and symbiotic efficiency. In this sense, the optimization of NFB is related to the joint selection of macro- and microsymbionts through knowledge of the genetic variability of their partners (Xavier et al. 2006). Whereas the traditional nomenclature of rhizobia should be properly used only for Rhizobium, Bradyrhizobium, Mesorhizobium, Sinorhizobium (Ensifer), Azorhizobium and Allorhizobium genera (Willems 2006), several other genera have been identified to participate in this symbiosis, such as Methylobacterium, Burkholderia, Ralstonia, Devosia, Phyllobacterium, Ochrobactrum, Cupriavidus and Pseudomonas (Moulin et al. 2001; Sy et al. 2001; Rivas et al. 2003; Trujillo et al. 2005; Valverde et al. 2005; Barrett and Parker 2006; Shiraishi et al. 2010). Although these genera do not fit the most traditional definition of rhizobia, a large body of literature continues to use this collective name for all bacteria that nodulate legumes and fix nitrogen for convenience and historical reasons (Liu et al. 2011). The use of classes of molecular markers and their variants has brought about a major evolution in the analysis of DNA profiles, conservation biology, population genetics and phylogenetic studies (Kochieva et al. 2006). In addition, molecular markers have also been used for the taxonomy and genetic diversity studies of microorganisms. In particular, amplification and sequencing of 16S rDNA genes have been shown to be reliable for separating genera and species of bacteria (Thomas et al. 2008). Fingerprinting using oligonucleotides that are specific for a genomic box element (BOX), enterobacterial repetitive intergenic consensus (ERIC) and repetitive extragenic palindromic (REP) techniques has been exemplified in studies on the genetic diversity of microorganisms in particular diazotrophs (Torres et al. 2008). However, the diverse search terms of diazotrophs are normally applied in a polyphasic approach by combining genetic information generated from the use of BOX elements, REP and ERIC sequences, genes such as 16S rDNA and phenotypic and physiological aspects for better genetic discrimination (Chagas Junior et al. 2010).
Recent studies have found that Burkholderia species were the main bacterial symbiont for Mimosa legumes (Chen et al. 2008). For example, whereas Reis Junior et al. (2010) observed more than one species of Burkholderia in the same Mimosa species, Bontemps et al. (2010) found Mimosa species of Central Brazil, which were located over 1,800 km apart, nodulated with species of Burkholderia. Both studies covered a large number of Mimosa species but included few samples of each species. Therefore, this work aims to study the diversity of M. caesalpiniifolia (Benth.) rhizobial isolates from five sources that are over 300 km apart from each other and from different ecological conditions of the Brazilian Northeast in order to evaluate whether rhizobial diversity is dependent on its origin.
Materials and methods
Sample collection
Soil samples were collected in March 2010 from “sabiá” (M. caesalpiniifolia Benth.) woodlots from five municipalities of the Brazilian Northeast: Crato—Ceará State (7°12′S 28.80′W; light, semi-arid, hot tropical to sub-humid, tropical climates), Gravatá (8°16′S 35°32′W) and Itambé (7°24′S 35°11′W) from Pernambuco State; (rainy tropical climate); Serra Talhada—Pernambuco State (7°55′S 48.36′W; semi-arid, tropical climate) and Mossoró—Rio Grande do Norte State (5°12′S 31.68′W; hot, semi-arid tropical climate). PE-CR, PE-GR, PE-IT, PE-MO and PE-ST isolate codes were used to denote these regions, respectively. From each woodlot, a compound sample was collected from the soil near the representative plants. Soil samples were then used for rhizobial isolation and fertility characterization according to the Brazilian standard methods (Embrapa 1999).
All soil sampling was conducted after proper permission was given, by the land owner for the Gravatá, Pernambuco farm, the Pernambuco Agronomical Institute (IPA) Research Station Managers at Itambé and Serra Talhada, both in Pernambuco), the Crop Science Department Director at the Federal Rural University of the Semi-Arid, at Mossoró—Rio Grande do Norte, and the Campus Director of Ceará Education, Science and Technology Institute at Crato—Ceará.
All of the respective managers were duly informed of the nature of the soil and plant sampling, and its ultimate objectives, and agreed with the intended use and methods.
Inoculation
“Sabiá” seeds were disinfected with 2 % sodium hypochloride for 2 min, washed twice in autoclaved distilled water, and immersed for 10 min in 70 °C water to break seed dormancy. Subsequently, the seeds were germinated in autoclaved, washed sand and transplanted into Leonard jars 20 days after germination (GAD). Sand/vermiculite (1:2) and 10 % Hoagland nutrient solution without nitrogen, as described by Hoagland and Arnon (1950), were placed in the upper portion of the Leonard jars. When transplanted, each seedling was inoculated with 5 g of soil, and 10 replicates per origin and five non-inoculated controls were included. Nodule harvest took place 60 days after seed germination, and the nodules were stored in polypropylene tubes with silica gel.
The dried nodules were rehydrated with sterile water for 12 h and isolated according to standard methods in petri dishes containing YMA media with Congo red as described by (Vincent 1970). After purification, the nodules were cultivated in YMA media with bromothymol blue and morphophysiologically characterized following Table 1.
Molecular characterization
The bacterial isolates were cultivated in 5 mL of TY media at 28 °C and agitated for 24 h at 200 rpm for DNA extraction. A 1-mL sample of the bacterial broth was transferred to 1.5 mL polypropylene tubes and centrifuged for 3 min at 13,000 rpm. Next, the precipitate was used for DNA extraction using the Wizard® Genomic DNA Purification kit (Promega Biotecnologia do Brasil), following the manufacturer’s recommendation. After extraction, the quantity and integrity of the DNA was evaluated by 0.8 % agarose gel electrophoresis. The DNA was diluted in ultrapure, autoclaved water at 1:100 (30–50 ng) and stored at −20 °C. PCR was conducted using the REP-1 (5′-IIII CGI CGI CAT CIG GC-3′), REP-2 (5′-ICG ICT ATC IGG CCT AC-3′), ERIC-1(5′-ATG TAA GCT CCT GGG GAT TCA C-3′), ERIC-2 (5′-AAG TAA GTG ACT GGG GTG AGC G-3′) and BOX-A1R (5′-CTA CGG CAA GGC GAC CGC TGA CG-3′) primers. Amplification reactions had a final volume of 10 μL including 5.0 µL of GoTaq Colorless Master Mix (Promega), 0.4 mM of each primer and 100 ng of sample DNA. The reaction mixtures were placed in a Biometra® thermocycler using the following conditions: a denaturation cycle for 7 min at 95 °C, followed by a 35- (BOX and REP primers) or 30-cycle (ERIC primer) denaturation for 8 min at 94 °C, annealing for 1 min at 40 °C (REP), 53 °C (ERIC) or 55 °C (BOX) and a final extension cycle for 16 min at 72 °C. The final reaction products were analyzed by electrophoresis in a 1.5 % agarose gel in 0.5X TBE buffer, running at 90 volts with 1.25 µL of SYBR Green and 2.5 µL of bromophenol blue. A 1 Kb ladder (Promega molecular) was used to determine the size of the products. Visualization and documentation of the amplification results were achieved using a LPIX-HE photodocumenter and the Labimagem 1D software (Loccus Biotecnologia).
For the 16S rDNA amplification, we utilized primers fD1 (5′-AGA GTT TGA TCC TGG CTC AG-3′) and rD1 (5′-AAG GAG GTG ATC CAG CC-3′) as described by Weisburg et al. (1991). Amplification reactions took place in a final volume of 25 μL with the same proportions as the reactions described above. A Biometra® thermocycler set at the following conditions was used: initial denaturation at 94 °C for 2 min, followed by 30 cycles of 94 °C for 1 min, 60 °C for 2 min, 72 °C for 1 min and a final extension at 72 °C for 10 min. The amplified products were evaluated for amplification in a 1.5 % agarose gel. The PCR products of the 16S rDNA amplification were purified as follows: For every 25 μL of amplified product, 2 μL ammonium acetate and 52 μL of 100 % absolute ethanol were added. To precipitate the DNA, it was centrifuged at 13,000 rpm for 20 min. The supernatants were discarded, and the pellet was washed with 70 % ethanol. To eliminate the ethanol remains, the samples were evaporated at room temperature for 1 h and the precipitate was re-suspended in 20 μL of sterile water. A 10 μL of the amplified product was used for the cut reaction by adding 2.5 U of each enzyme and 10 % of 10X reaction buffer until reaching a final volume of 30 μL (Lyra et al. 2013). The solution was stored at −20 °C and sent to the commercial facilities of Macrogen, South Korea, for sequencing.
Data analysis
Morphophysiological characteristics were transformed into a binary matrix according to Table 2, and REP, ERIC and BOX profile bands were scored individually for the absence/presence of each band that appeared in any of the respective gels using the Jaccard coefficient. Cluster analysis used the NTSYS-pc 2.1 (Rohlf 2008) implementation of the UPGMA algorithm. A 60 % similarity cutoff was adopted for morphophysiological characteristic clustering according to Melloni et al. (2006), whereas a 63 % cutoff was adopted for rep-PCR according to Antunes (2010).
16S rDNA sequence quality was evaluated using Chromas 2.33 (Technelysium Pty Ltd, Tewantin, Australia), and the sequences were aligned to the NCBI reference strains for Burkholderia sabiae, B. xenovarans, B. andropogonis, B. japonicum, B. galegae, Sinorhizobium meliloti and Rhizobium etli bv. mimosae using the BioEdit 7.0.9.0 software through ClustalW (Larkin et al. 2007). The matrix originated in the ClustalW was converted using the MEGA v.4.1 (Beta) software (Tamura et al. 2007) using the standard genetic code. The phylogenetic tree was constructed using the Kimura 2 algorithm by neighbor-joining (Saitou and Nei 1987), with 1,000 bootstrap replicates and pairwise deletion.
Results and discussion
Forty-seven isolates were obtained: 10 each from Gravatá and Serra Talhada, and nine each from the remaining origins. All isolates exhibited fast growth, and 51 % increased the pH of media. Fast-growing bacteria also dominated for cowpea (Vigna unguiculata); however, in these cases, the culture media were usually acidified (Silva et al. 2007; Leite et al. 2009; Medeiros et al. 2009). This faster bacterial growth is common in semi-arid regions and may be a survival strategy to enhance growth during the short rainy season (Santos et al. 2007).
A single morphophysiological group at 60 % similarity, as described by Melloni et al. (2006), for caupi and common bean (Phaseolus vulgaris) rhizobia was found. This group was split into subgroups IA and IB with 70 % similarity, whereas isolate PE-GR02 was an external, monophyletic group (Fig. 1). Subgroup IA had 29 isolates in 10 subgroups of 100 % internal similarity and 76–87 % similarity between subgroups, with the PE-ST10 isolate as a monophyletic branch. Subgroup IB had four subgroups with isolates from all five origins. Isolates PE-ST04 and PE-GR07 had 100 % similarity, although their origins were 300 km apart and had different climatic and soil fertility regimens. The same pattern was seen for isolates PE-MO04 and PE-ST08, which were also 300 km apart; however, these regions were climatically more similar.
REP primers formed up to 30 bands that ranged from 100 to 5,000 bp and formed four groups and a monophyletic branch consisting of isolate PE-MO08 (Fig. 2), although this isolate was morphophysiologically similar to isolates PE-MO10, PE-GR09 and PE-IT02 (Fig. 1). Group I had internal similarity that ranged from 63 to 93 % and nine isolates, including seven from PE-MO and one each from the PE-GR and PE-CR isolates, with the highest similarity between two of the PE-MO isolates. Group II was the largest with 22 isolates: one from PE-MO, all PE-ST isolates, four each from the PE-CR and PE-GR isolates and three each from the PE-IT isolate. The internal similarities of Group II ranged from 66 to 100 % among the PE-ST08, PE-ST09 and PE-ST10 isolates. Three additional PE-ST isolates had 97 % similarity to this subgroup, which indicated that this origin had the lowest diversity according to this method. Group III, which was the smallest group, had only the PE-CR04, PE-CR07 and PE-GR10 isolates, and whereas the PE-CR isolates exhibited 90 % similarity, PE-GR was only 68 % similar to these isolates. Group IV consisted of the remaining 12 isolates, which included two PE-CR, four PE-GR and six PE-IT isolates. Whereas the internal similarities of PE-IT ranged from 74 to 93 % and the internal similarities of PE-GR ranged from 89 to 96 %, PE-CR10 exhibited 82 % similarity to the PE-GR group and only 70 % similarity to PE-CR05, which were from the same origin. Although the PE-ST isolates were found in a single group, this grouping was not observed for any of the other origins, which was similar to what was observed by Stocco et al. (2008) for the rhizobial diversity of common beans from 23 areas of the southern Brazil state of Santa Catarina.
ERIC primers created up to 41 bands with molecular weights that ranged from 150 to 4,000 bp, which confirmed its high discrimination power as suggested by Rademaker and de Bruijn (1996). These primers created five groups (Fig. 3) and two monophyletic branches for PE-GR07 (V) and PE-GR02 (VII), which was also isolated on the morphophysiological tree (Fig. 1). Group I included all PE-ST isolates, as was found when using the REP primers (Fig. 2), and the internal similarities of this group ranged from 83 to 100 %. Group I also contained seven of the nine PE-MO isolates, which had 100 % similarity between the PE-MO03 and MO05 isolates, and this pair had 83 % similarity when using the REP primers (Fig. 2). This finding again demonstrates that each primer generated different groupings. Group II had 11 isolates, including one PE-MO, two PE-IT, three PE-CR and five PE-GR isolates, and was the most geographically dispersed. The highest internal similarity was found between two of the PE-GR isolates (93 %). Group III had 13 isolates, with three each from the PE-CR and PE-GR isolates and seven PE-IT isolates. This group had the highest internal similarity of 96 % between the PE-IT03 and PE-IT04 isolates. Group IV had only two isolates, which were from PE-CR and showed 76 % similarity, whereas Group VI also had only two isolates, but these isolates were from PE-MO and PE-CR and had 68 % similarity. The origins of Group IV and VI are located more than 320 km and are ecologically different.
Whereas Menna et al. (2009) indicated BOX as the most differentiating of the primers, in this instance, it generated the lowest number of bands (Melloni et al. 2006) with molecular weights ranging from 200 to 5,000 bp. BOX primers also generated the largest single grouping of all three primers (Fig. 4). The dendrogram formed three groups and a single monophyletic branch that contained PE-GR10 (Group II), which was grouped with two PE-CR isolates (70 % similar when using the REP primers (Fig. 2) and with the PE-GR05 isolate (76 % similar when using the BOX primers (Fig. 3). Group I had 24 isolates that included all PE-ST isolates, eight of nine of the PE-MO and six of the PE-GR isolates. Grouping all of the PE-ST isolates into a single group replicated the observation that was seen when using the other primers (Figs. 2, 3), but with this primer, five similarity groupings of 100 % were found: three for a total of seven of the PE-ST isolates and two for another five of the PE-MO isolates. This total was again the highest number of 100 % similarities that was found for any of the primers. Group III had 14 isolates, two from PE-GR, five from PE-CR, of which two had 100 % similarity, and seven from PE-IT, with another two that were 100 % similar. Group IV had the largest geographical distribution, with a PE-MO, a PE-GR, two PE-IT and four PE-CR isolates, and had the highest similarity (97 % for a PE-CR and PE-IT pair), although their origins were 450 km apart.
The compiled dendrogram REP-, ERIC- and BOX-PCR (Fig. 5) presented six groups and two monophyletic branches, which agreed with the suggestions of Albino et al. (2006) and Torres et al. (2008) that the joint use of different primers enhanced the discrimination power of this technique. Group I included all PE-ST and eight of the PE-IT isolates, which again confirmed that the PE-ST origin had the lowest diversity and a 67–100 % internal similarity range. Group II had only two isolates: one each from PE-CR and PE-GR (68 % similarity), whereas Group III had five PE-GR isolates with similarities that ranged from 70 to 92 %. Group IV had a PE-GR, two PE-IT and three PE-CR isolates, the highest geographical range and internal similarities that ranged from 70 to 88 %. Group VI again had only two PE-GR isolates with 64 % similarity, and Group VII had five PE-CR and seven PE-IT isolates with similarities ranging from 67 to 94 %. Two monophyletic branches consisting of PE-GR02 and PE-MO10 were also created.
The higher diversity that was observed with any of the primers confirms that this technique was useful for strain fingerprinting, as was suggested in the literature (Grange and Hungria 2004; Hungria et al. 2006; Kaschuk et al. 2006; Grange et al. 2007; Menna et al. 2009). This observation also agreed with Rademaker et al. (2000), who stated that the best results were found when more than one primer was used. However, whereas the joint analysis formed eight groups, ERIC, when used alone, formed seven groups, which indicated a relatively small gain when using all primers and suggested that a cost x benefit analysis would be advisable if a large number of strains were to be fingerprinted. Polyphasic approaches that combine genetic information by using the BOX element, REP and ERIC sequences and genes such as 16S, when combined with phenotypic and physiological aspects, should be used to improve the accuracy of gene discrimination and have proven useful in a variety of studies on rhizobia (Reis Junior et al. 2010).
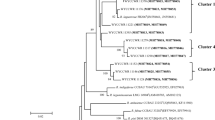
Nineteen of the isolates did not sequence satisfactorily and were not included in the dendrogram. The 16S rDNA phylogenetic tree (Fig. 6) agreed with the compiled dendrogram from the BOX, ERIC and REP primers but not with the morphophysiological dendrogram (Fig. 1). This discrepancy was due to large differences between all origins with the exception of Mossoró, which was the only origin with similar grouping patterns for all of the employed techniques. In general, the isolates had 77–99 % identities with Burkholderia. Only 23 isolates had similarities of at least 97 % with eight of the accesses of the Burkholderia strains that were found in the National Center for Biotechnology Information (NCBI), which indicated that they might be identified at the species level (Table 3) (Kemp and Aller 2004; Lozupone and Knight 2005; Vos 2011). However, other authors (Menna et al. 2006; Binde et al. 2009) suggested that 15 nucleotides with differences or similarities lower than 99 % should be considered to be indicative of a new bacterial species.
16S rDNA phylogenetic dendrogram of “sabiá” (Mimosa caesalpiniifolia Benth.) rhizobial isolates from five origins in the Brazilian Northeast. Brackets indicate NCBI access numbers for the isolates. Branch values are the percentages of 1,000 bootstrap replicates; ID values indicate homology percentage with the indicated NCBI entry
When a 99 % similarity level is considered the cutting point, nine isolates could be identified as belonging to Burkholderia sp. tpig4.4 (PE-MO06), Burkholderia sabiae Br3407 (PE-MO10), Burkholderia sp. JPY 584 (PE-GR07), Burkholderia diazotrophica (PE-GR09), Burkholderia sp. SEMIA 6385 (PE-IT01), Burkholderia sp. SEMIA 6398 (PE-MO07, PE-ST04) and Burkholderia sp. NKMU-JPY359 (PE-ST01, PE-CR01) (Table 3). Although the distance between the origins of the isolates is 400 km between PE-MO07–PE-ST04 and 200 km between PE-ST01–PE-CR01, the results agreed with the conclusions of Bontemps et al. (2010) that Burkholderia was the main bacterial symbiont of Mimosa species in Brazil.
A group with isolates from five municipalities with similarities from 97 to 99 % was formed with representative strains Burkholderia sp. SEMIA 6398, Burkholderia diazotrophica, Burkholderia sp. DPU-3, Burkholderia tpig4.4 and Burkholderia sp. NKMU-JPY359 (Fig. 6) and a nearby group with six PE-IT strains, of which five had 97–99 % similarities to Burkholderia sp. SEMIA 6395, one had 98 % similarity to Burkholderia sp. DPU-3 and one had 97 % similarity to Burkholderia sabiae (Table 3).
Four PE-CR and one PE-GR isolates had identities between 77 and 89 % with Burkholderia entries in NCBI (Table 3) and might be new species of the genus; however, their low similarity indicated that further studies must be conducted before new species were proposed with certainty (Djedidi et al. 2011). Furthermore, most PE-ST isolates had high similarities, within the 97–99 % range, with the Burkholderia sp. strains SEMIA 6385, DPU-3 and Burkholderia sabiae.
In all cases for which over 97 % similarities where found between Burkholderia sp. entries in NCBI, none were confined to a single origin, even when considering the large geographical and ecological differences between them. Strains SEMIA6398 and DPU-3 were found in Serra Talhada and Mossoró (320 km apart, both semi-arid); NKMU isolates were found in Serra Talhada and Crato (150 km apart, different climates); BR3407 isolates were found in Itambé and Mossoró (340 km apart, different climates); SEMIA6485 isolates were found in Crato, Itambé and Gravatá (distances ranging from 100 to 470 km, all climates were relatively humid); and DPU-3 isolates were found in Itambé, Mossoró and Serra Talhada (distances ranging from 100 to 390 km, different climates). This observation agreed with previous findings that a single bacterial species might be found across very different environmental or geographical conditions (Martínez-Romero and Caballero-Mellado1996; Moreira et al. 1998; Vos 2011).
Conclusions
The ERIC marker was more polymorphic than the REP and BOX; however, the dendrogram that compiled data from the three markers generated the highest number of groups. All studied isolates sequenced showed homology with genus Burkholderia, with identities for these isolates ranging from 77 to 99 %. Whereas the rhizobial diversity differed between the origins, the lowest was detected in Serra Talhada, although similarities were found between isolates from all of the origins. Although the low similarity of some of the isolates with NCBI accesses indicated the probable existence of new species, all those with higher than 97 % 16S rDNA similarity were similar to Burkholderia species, which confirmed this genus position as the main bacterial symbiont of M. caesalpiniifolia in this study.
References
Albino U, Saridakis DP, Ferreira MC, Hungria M, Vinuesa P, Andrade G (2006) High diversity of diazotrophic bacteria associated with the carnivorous plant Drosera villosa var. villosa growing in oligotrophic habitats in Brazil. Plant Soil 287:199–207. doi:10.1007/s11104-006-9066-7
Antunes JEL (2010) Diversidade genética e eficiência simbiótica de isolados de rizóbios nativos em feijão-fava (Phaseolus lunatus L.). Dissertation, Universidade Federal do Piauí
Babić KH, Schauss K, Hai B, Sikora S, Redžepović S, RadI V, Schloter M (2008) Influence of different Sinorhizobium meliloti inocula on abundance of genes involved in nitrogen transformations in the rhizosphere of alfalfa (Medicago sativa L.). Environ Microbiol 10:2922–2930. doi:10.1111/j.1462-2920.2008.01762.x
Barrett CF, Parker MA (2006) Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. nodule bacteria on two Mimosa spp. in Costa Rica. Appl Environ Microbiol 72:1198–1206. doi:10.1128/AEM.72.2.1198-1206.2006
Binde DR, Menna P, Bangel EV, Barcellos FG, Hungria M (2009) Rep-PCR fingerprinting and taxonomy based on the sequencing of the 16S rRNA gene of 54 elite commercial rhizobial strains. Appl Microbiol Biotechnol 83:897–908. doi:10.1007/s00253-009-1927-6
Bontemps C, Elliott GN, Simon MF, Reis Júnior FB, Gross E, Lawton RC, Neto NE, Loureiro MF, Faria SM, Sprent JI, James EK, Young PW (2010) Burkholderia species are ancient symbionts of legumes. Mol Ecol 19:44–52. doi:10.1111/j.1365-294X.2009.04458.x
Chagas Junior A, Oliveira L, Oliveira A (2010) Caracterização fenotípica de rizóbios nativos isolados de solos da Amazônia e eficiência simbiótica em feijão caupi. Acta Sci Agron 32:161–169. doi:10.4025/actasciagron.v32i1.900
Chen WM, Faria SM, Chou JH, James EK, Elliott GN, Sprent JI, Bontemps C, Young JP, Vandamme P (2008) Burkholderia sabiae sp. nov., isolated from root nodules of Mimosa caesalpiniifolia. Int J Syst Evol Microbiol 58:2174–2179. doi:10.1099/ijs.0.65816-0
Djedidi S, Yokoyama T, Tomooka N, Ohkama-Ohtsu N, Risal CP, Abdelly C, Sekimoto H (2011) Phenotypic and genetic characterization of rhizobia associated with alfalfa in the Hokkaido and Ishigaki regions of Japan. Syst Appl Microbiol 34:453–461. doi:10.1016/j.syapm.2011.04.006
Embrapa (1999) Manual de análises químicas de solos. EMBRAPA, Brasília
Grange L, Hungria M (2004) Genetic diversity of indigenous common bean (Phaseolus vulgaris) rhizobia in two Brazilian ecosystems. Soil Biol Biochem 36:1389–1398. doi:10.1016/j.soilbio.2004.03.005
Grange L, Hungria M, Graham PH, Martínez-Romero E (2007) New insights into the origins and evolution of rhizobia that nodulate common bean (Phaseolus vulgaris) in Brazil. Soil Biol Biochem 39:867–876. doi:10.1016/j.soilbio.2006.10.008
Gualter RMR, Leite LFC, Araújo ASFd, Alcântara RMCM, Costa DB (2008) Inoculação e adubação mineral em feijão-caupi: efeitos na nodulação, crescimento e produtividade. Sci Agrar 9:469–474
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp stn, Berkeley
Hungria M, Chueire LMO, Megías M, Lamrabet Y, Probanza A, Guttierrez-Mañero FJ, Campo RJ (2006) Genetic diversity of indigenous tropical fast-growing rhizobia isolated from soybean nodules. Plant Soil 288:343–356. doi:10.1007/s11104-006-9125-0
Kaschuk G, Hungria M, Andrade DS, Campo RJ (2006) Genetic diversity of rhizobia associated with common bean (Phaseolus vulgaris L.) grown under no-tillage and conventional systems in Southern Brazil. Appl Soil Ecol 32:210–220. doi:10.1016/j.apsoil.2005.06.008
Kemp PF, Aller JY (2004) Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol Ecol 47:161–177. doi:10.1016/S0168-6496(03)00257-5
Kochieva EZ, Ryzhova NN, Legkobit MP, Khadeeva NV (2006) RAPD and ISSR analyses of species and populations of the genus Stachys. Russ J Genet 42:723–727. doi:10.1134/S1022795406070039
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Leite J, Seido SL, Passos SR, Xavier GR, Rumjanek NG, Martins LMV (2009) Biodiversity of rhizobia associated with cowpea cultivars in soils of the lower half of the São Francisco River Valley. Rev Bras Ciênc Solo 33:1215–1226
Liu XY, Wu W, Wang ET, Zhang B, Macdermott J, Chen WX (2011) Phylogenetic relationships and diversity of β-rhizobia associated with Mimosa species grown in Sishuangbanna, China. Int J Syst Evol Microbiol 61:334–342. doi:10.1099/ijs.0.020560-0
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi:10.1186/1471-2105-7-371
Lyra MCCP, Santos DC, Mondragon-Jacobo C, Silva MLRB, Mergulhão ACES, Martínez-Romero E (2013) Isolation and molecular characterization of endophytic bacteria associated with the culture of forage cactus (Opuntia spp.). J Appl Biol Biotechnol 1:11–16. doi:10.7324/JABB.2013.1103
Martínez-Romero E, Caballero-Mellado J (1996) Rhizobium phylogenies and bacterial genetic diversity. Crit Rev Plant Sci 15:113–140. doi:10.1080/07352689.1996.10393183
Medeiros EV, Martins CM, Lima JAM, Fernandes YTD, Oliveira VR, Borges WL (2009) Diversidade morfológica de rizóbios isolados de caupi cultivado em solos do estado do Rio Grande do Norte. Acta Sci Agron 31:529–535. doi:10.4025/actasciagron.v31i3.793
Melloni R, Moreira FMS, Nóbrega RSA, Siqueira JO (2006) Eficiência e diversidade fenotípica de bactérias diazotróficas que nodulam caupi [Vigna unguiculata (L.) Walp] e feijoeiro (Phaseolus vulgaris L.) em solos de mineração de bauxita em reabilitação. Rev Bras Ciênc Solo 30:235–246
Menna P, Hungria M, Barcellos FG, Bangel EV, Hess PN, Martínez-Romero E (2006) Molecular phylogeny based on the 16S rRNA gene of elite rhizobial strains used in Brazilian commercial inoculants. Syst Appl Microbiol 29:315–332. doi:10.1016/j.syapm.2005.12.002
Menna P, Pereira AA, Bangel EV, Hungria M (2009) Rep-PCR of tropical rhizobia for strain fingerprinting, biodiversity appraisal and as a taxonomic and phylogenetic tool. Symbiosis 48:120–130. doi:10.1007/BF03179991
Moreira FMS, Haukka K, Young JPW (1998) Biodiversity of rhizobia isolated from a wide range of forest legumes in Brazil. Mol Ecol 7:889–895
Moulin L, Munive A, Dreyfus B, Boivin-Masson C (2001) Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 411:948–950
Rademaker JLW, de Bruijn FJ (1996) Characterization and classification of microbes by REP-PCR genomic fingerprinting and computer assisted pattern analysis. In: Caetano-Anollés G, Gresshoff PM (eds) DNA Marker: protocols, applications and overviews. Wiley, New York, pp 1–26
Rademaker JLW, Hoste B, Louws FJ, Kersters K, Swings J, Vauterin L, Vauterin P, Bruijn FJ (2000) Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int J Syst Evol Microbiol 50:665–677. doi:10.1099/00207713-50-2-665
Reis Junior FB, Simon MF, Gross E, Boddey RM, Elliott GN, Neto NE, Loureiro MF, Queiroz LP, Scotti MR, Chen WM, Norén A, Rubio MC, Faria SM, Bontemps C, Goi SR, Young JPW, Sprent JI, James EK (2010) Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol 186:934–946. doi:10.1111/j.1469-8137.2010.03267.x
Rivas R, Willems A, Subba-Rao NS, Mateos PF, Dazzo FB, Kroppenstedt RM, Martínez-Molina E, Gillis M, Velázquez E (2003) Description of Devosia neptuniae sp. nov. that nodulates and fixes nitrogen in symbiosis with Neptunia natans, an aquatic legume from India. Syst Appl Microbiol 26:47–53. doi:10.1078/072320203322337308
Rohlf FJ (2008) NTSYSpc: Numerical Taxonomy System, ver. 2.20. Exeter Publishing, Setauket
Rufini M, Ferreira PAA, Soares BL, Oliveira DP, de Andrade MJB, Moreira FMS (2011) Simbiose de bactérias fixadoras de nitrogênio com feijoeiro-comum em diferentes valores de pH. Pesq Agropec Bras 46:81–88
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Santos CERS, Stamford NP, Borges WL, Neves MCP, Rumjanek NG, Nascimento LR, Freitas ADS, Vieira IMMB, Bezerra RV (2007) Faixa hospedeira de rizóbios isolados das espécies Arachis hypogaea, Stylosanthes guyanensis e Aeschynomene americana. Rev Bras Ciênc Agrárias 2:20–27
Shiraishi A, Matsushita N, Hougetsu T (2010) Nodulation in black locust by the Gammaproteobacteria Pseudomonas sp. and the Betaproteobacteria Burkholderia sp. Syst Appl Microbiol 33:269–274. doi:10.1016/j.syapm.2010.04.005
Silva VN, Silva LESF, Figueiredo MVB, Carvalho FG, Silva MLRB, Silva AJN (2007) Caracterização e seleção de populações nativas de rizóbios de solo da região semi-árida de Pernambuco. Pesq Agropec Trop 37:16–21
Sprent JI (2008) 60 Ma of legume nodulation. What’s new? What’s changing? J Exp Bot 59:1081–1084. doi:10.1093/jxb/erm286
Stocco P, Santos JCP, Vargas VP, Hungria M (2008) Assessment of biodiversity in rhizobia symbionts of common bean (Phaseolus vulgaris L.) in Santa Catarina, Brazil. Braz Rev Bras Ciênc Solo 32:1107–1120
Sy A, Giraud E, Samba R, de Lajudie P, Gillis M, Dreyfus B (2001) Nodulation of certain legumes on the genus Crotalaria by the new species Methylobacterium. Can J Microbiol 47:503–508. doi:10.1139/cjm-47-6-503
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi:10.1093/molbev/msm092
Thomas P, Swarna GK, Roy PK, Patil P (2008) Identification of culturable and originally non-culturable endophytic bacteria isolated from shoot tip cultures of banana cv. Grand Naine. Plant Cell Tiss Organ Cult 93:55–63. doi:10.1007/s11240-008-9341-9
Torres AR, Araújo WL, Cursino L, Hungria M, Plotegher F, Mostasso FL, Azevedo JL (2008) Diversity of endophytic enterobacteria associated with different host plants. J Microbiol 46:373–379. doi:10.1007/s12275-007-0165-9
Trujillo ME, Willems A, Abril A, Planchuelo AM, Rivas R, Ludeña D, Mateos PF, Martínez-Molina E, Velázquez E (2005) Nodulation of Lupinus albus by strains of Ochrobactrum lupini sp. nov. Appl Environ Microbiol 71:1318–1327. doi:10.1128/AEM.71.3.1318-1327.2005
Valverde A, Velázquez E, Fernández-Santos F, Vizcaíno N, Rivas R, Mateos PF, Martínez-Molina E, Igual JM, Willems A (2005) Phyllobacterium trifolii sp. nov., nodulating Trifolium and Lupinus in Spanish soils. Int J Syst Evol Microbiol 55:1985–1989. doi:10.1099/ijs.0.63551-0
Vincent JM (1970) A manual for the practical study of root-nodule bacteria. Blackwell, Oxford
Vos M (2011) A species concept for bacteria based on adaptive divergence. Trends Microbiol 19:1–7. doi:10.1016/j.tim.2010.10.003
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Willems A (2006) The taxonomy of rhizobia: an overview. Plant Soil 287:3–14. doi:10.1007/s11104-006-9058-7
Xavier GR, Martins LMV, Ribeiro JRA, Rumjanek NG (2006) Especificidade simbiótica entre rizóbios e acessos de feijão-caupi de diferentes nacionalidades. Rev Caatinga 19:25–33
Acknowledgments
We are grateful to Walter Fabricio Silva Martins (Liverpool University) and Alice Cassetari (São Paulo University) for help in this manuscript. The authors thank Higher Education Personnel Improvement Coordination (CAPES), National Research Council (CNPq) and Pernambuco State Science and Technology Funding Foundation (FACEPE) for research grants conceded to the second author which funded the research. The first author thanks FACEPE for his masters’ grant, and the second and third authors thank CNPq for his research fellowship, while the third and fourth authors thank CAPES for their post-doctoral and masters grants.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Rights and permissions
About this article
Cite this article
Martins, P.G.S., Junior, M.A.L., Fracetto, G.G.M. et al. Mimosa caesalpiniifolia rhizobial isolates from different origins of the Brazilian Northeast. Arch Microbiol 197, 459–469 (2015). https://doi.org/10.1007/s00203-014-1078-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-014-1078-8