Abstract
In this article we describe the identification of endophytic bacteria belonging to three groups isolated from shoot tip cultures of banana cv. Grand Naine in a recent study (Thomas et al. 2008) based on partial 16S rRNA gene sequence homology analysis. The first group included banana stocks that displayed obvious colony growth on MS based tissue culture medium during the first in vitro passage. The second group constituted stocks that were tissue index-negative for cultivable bacteria initially but turned index-positive after a few to several (4–8) in vitro passages while the third group formed one sub-stock that turned index-positive after about 18 passages. The organisms belonged to about 20 different genera comprising of α, β, γ-proteobacteria, Gram-positive firmicutes and actinobacteria. Visibly expressing easily cultured organisms during the first in vitro passage included Enterobacter, Klebsiella, Ochrobactrum, Pantoea, Staphylococcus and Bacillus spp. Organisms of second group that were not detected or non-culturable originally constituted Brevundimonas, Methylobacterium, Alcaligenes, Ralstonia, Pseudomonas, Corynebacterium, Microbacterium, Staphylococcus, Oceanobacillus and Bacillus spp. while the third group that turned cultivable after extended in vitro culturing included mostly non-filamentous actinobacteria (Brachybacterium, Brevibacterium, Kocuria and Tetrasphaera spp.). The identification results suggested that the endophytes of second and third groups were not strictly obligate or fastidious microbes but those surviving in viable but-non-culturable (VBNC) state and displaying gradual activation to cultivable form during continuous tissue culturing. Several of the organisms isolated are known as beneficial ones in agriculture while some organisms have possible implications in human health. The use of tissue cultures for isolating uncommon endophytes is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microbial contamination is a major problem hindering tissue culture applications. There is a general tendency to attribute microbial contamination in tissue cultures to ineffective explant disinfection at culture initiation or inefficient sterile practices during culture handling. Our recent observations with banana and papaya shoot-tip cultures employing a combination of bacterial 16S rRNA gene-based molecular technique, light microscopy and cultivation-based approaches have shown the ubiquitous presence of endophytic bacteria in field shoots as well as apparently clean in vitro cultures across different genotypes either in culturable and/or non-culturable form (Thomas et al. 2007a, b, 2008). With bananas, only a small portion of field shoot tips showed visibly expressing or cultivable endophytes at culture initiation. The remaining apparently clean stocks harbored fastidious or viable but-non-culturable (VBNC) bacteria as brought out through microscopic/molecular observations and indexing of tissue segments or plating of tissue homogenate on different bacteriological media. These stocks displayed gradual activation of the originally non-culturable organisms to cultivation over subculture passages rendering almost all the apparently clean banana stocks as quiescent bacteria-harboring after a few to several (2–10) passages. A few stocks that stayed consistently index-negative however turned index-positive upon recurrent in vitro culturing over 2 years. Such covert bacteria harboring stocks showed substantial levels of colonization to the tune of 105–107 colony forming units g−1 tissue of cv. Grand Naine. This study has also brought out the quiescent survival of endophytic bacteria in unsuspecting suspension cultures and highlighted the implications of such association on micropropagation, in vitro gene banks and molecular profiling, and in tissue culture related issues like habituation and somaclonal variation (Thomas et al. 2008).
During the course of above study with banana ‘Grand Naine’, we have isolated bacteria belonging to three groups namely organisms that emerged as obvious contaminants during the first passage, those detected in apparently clean stocks after a few passages and those emerged after prolonged in vitro culturing (Thomas et al. 2008). This study was taken up with the objective of establishing the identity of organisms isolated in the above study.
Materials and methods
Bacteriological media and isolation of organisms
Bacteriological media used included nutrient agar (NA), Viss et al. (1991) 523 agar medium (VA) and trypticasein soy agar (TSA) in single-use 10-cm plates (Hi-Media Laboratories, Mumbai, India). Unless mentioned differently, NA or nutrient broth (NB) was used for all bacterial isolation and further culturing works. The nutrient plates used in culture indexing were pre-incubated at 37°C for 2–3 days to ensure their freedom from all accidental contaminants while that meant for dilution plating were prepared the same day. The plates were sealed hermetically in sterile polypropylene bags both before and after indexing/plating.
Thirty-seven bacterial isolates belonging to three groups that were retrieved from shoot-tip cultures of ‘Grand Naine’ described in the earlier experiment (Thomas et al. 2008) were taken up in this study. The first group included seven clones isolated after serial dilution and plating of bacterial growth that emanated from visibly contaminated stocks on tissue culture medium (TCM) at culture initiation (one from January 2004 batch and six from January 2005 set) (Table 1). The second group included 21 isolates retrieved from the bacteriological medium during tissue indexing of visibly clean stocks after 4–8 in vitro passages (Table 2). The third group included nine isolates that were detected during tissue indexing or homogenate-plating of one sub-stock from January 2004 batch (OC January 2004-11.4) (Table 3) that remained consistently index-negative but turned index-positive after about 20 passages spanning over 2 years, detected on VA and TSA but not on NA (Thomas et al. 2008).
Bacterial identification
Stringent laboratory sterile practices were followed during the course of in vitro culturing as well as bacterial isolation to rule out the accidental introduction of contaminants (Thomas et al. 2008). The identity of the organisms was determined based on partial (600–900 bp) 16S rRNA gene sequence analysis. Single colonies were selected after two to three rounds of dilution plating followed by one round of streaking on NA or TSA. DNA was extracted from overnight broth culture (2 ml) using a microbial DNA extraction kit (MOBIO Laboratories, Inc., Solano Beach, CA) or as per the CTAB protocol of Ausubel et al. (2005). 16S rDNA was amplified in 40 μl polymerase chain reaction (PCR) using 200 ng DNA (as determined using a ND1000 Spectrophotometer; Nanodrop Technologies, Inc., Wilmington, DE), 50 μM dNTPs and 1.0 unit of Taq DNA polymerase (Genie, Bangalore, India) and 20 pmol each of bacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R-Y (5′-GGYTACCTTGTTACGACTT-3′; Y = C/T) (Thomas et al. 2008). Thermocycling conditions consisted of initial one denaturation step of 94°C for 5 min followed by 32 amplification cycles of 94°C for 30 s, 55°C for 40 s, 72°C for 40 s followed by a final extension at 72°C for 5 min using an Eppendorf Mastercycler (Eppendorf AG, Hamburg, Germany).
PCR amplification was ascertained by running 6 μl samples in a 1% agarose gel. The remaining samples were purified using a PCR purification kit (Axygen Private Ltd., New Delhi) and the 16S rRNA gene was single-end sequenced using 27F at M/s Macrogen Inc., Seoul, Korea (http://www.macrogen.com) or at MWG Biotech, Bangalore, India (http://www.mwgdna.com). Similarity of partial 16S rRNA gene nucleotide sequences with known sequences in the NCBI Genbank database (http://www.ncbi.nlm.nih.gov/) was determined using BLAST version 2.2.15 (Altschul et al. 1997). The organism was assigned to a species if the sequence was ≥99% similar to a valid species sequence deposited with NCBI Genbank at the time of analysis (December 2006) as per Drancourt et al. (2000), or to genus if the species identity was not conclusive but the similarity was ≥97%. A dendrogram based on the neighbor joining method was created using ClustalW (1.75) package at the site http://www.bioinfo.cnio.es/treeapp/clustal-form.
Microscopy of microbial cultures
Colony observations and staining of pure cultures were carried out as per Cappuccino and Sherman (1996). Gram reaction was assessed as per Suslow et al. (1982) using 3% KOH. The slides were observed under oil immersion (1000×) using a Leica DM2000 microscope (Thomas et al. 2007b, 2008).
Accession number of nucleotide sequences
The partial 16S rRNA gene sequence data generated in this study have been deposited with the NCBI Genbank under the accession numbers DQ872436-DQ872463 in respect of the isolates retrieved during the first in vitro passage or detected after 4–10 in vitro passages, and DQ890503-DQ890513 for isolates that were detected after 2 years of continuous culturing.
Results and discussion
The endophytes that emerged as obvious contaminants during the first in vitro cycle of cultured shoot tips belonged to the genera of Enterobacter, Klebsiella, Ochrobactrum, Bacillus, Pantoea or Staphylococcus, falling under three bacterial subdivisions as per Bergy’s Manual of Systematic Bacteriology (2005) namely α- and γ-proteobacteria and firmicutes. Generally one and occasionally two organisms (e.g. January2005-OC03) were isolated from a single stock soon after its detection as a contaminated culture (Table 1).
The organisms isolated from stocks that showed bacterial activation after a few to several passages belonged to α-proteobacteria (Brevundimonas, Methylobacterium), β-proteobacteria (Alcaligenes, Ralstonia), γ-proteobacteria (Pseudomonas), actinobacteria (Corynebacterium, Microbacterium), spore-forming firmicutes (Oceanobacillus picturae, Bacillus fusiformis, Bacillus neonatiensis, Bacillus pumilus) and non-spore forming firmicutes (Staphylococcus epidermidis and Staphylococcus arlettae) (Table 2). The organisms derived from the sub-stock January 2004-OC11.4 during 19th passage that had remained index-negative for almost 2 years included Brachybacterium and Brevibacterium spp., both non-filamentous actinobacteria. Dilution plating of the tissue homogenate from this sub-stock during the 20th passage yielded five diverse colony types on TSA from the shoot part which included Brevibacterium, Kocuria, Tetrasphaera spp. and Brachybacterium sp. (2×) and two from corm (Brachybacterium and Staphylococcus spp.) (Table 3). Although the tissue homogenate did not show any colony development on NA upon plating, once activated on TSA, the above isolates showed the ability to grow on NA. However, TSA continued to be more suitable based on the extent of colony growth.
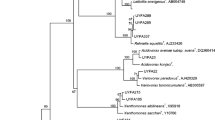
The endophytic organisms isolated in this study displayed considerable diversity including Gram-negative α, β, γ-proteobacteria, Gram-positive low G + C spore-forming firmicutes, non-spore forming firmicutes and Gram-positive and Gram-negative actinobacteria, distributed under 20 genera and 24 species. The organisms isolated in the three groups tended to cluster together under the respective group (Fig. 1). No common organism was detected with different stocks taken up in this study. The isolates retrieved during the first in vitro passage as easily cultivable contaminants included more of Gram-negative organisms (75%) endorsing the earlier reports with endophytes (Bell et al. 1995; Reiter et al. 2002; Thomas et al. 2007a) while those present initially as fastidious or VBNC organisms but turned cultivable subsequently displayed a Gram-positive predominance (53 and 67% in group two and three, respectively). It was noteworthy that 90% of the belatedly activated organisms (after 2 years) belonged to the sub-division of actinobacteria, to which spore-forming Gram-positive filamentous actinomycetes belong. The isolates in this study did not show filamentous growth on nutrient media, and one organism (Brachybacterium sp.) appeared clearly Gram-negative (Table 3).
Dendrogram using Clustal W (1.75) program based on the partial 16S rDNA sequence data of different endophytic bacteria isolated from in vitro cultured corm tips of banana ‘Grand Naine’. The name of the organism is followed by the NCBI accession no. and a, b, c represent the organisms retrieved as easily cultivable ones during the first in vitro passage, those activated after 4–10 in vitro passages and that emerged in a part of the stocks after 2 years of continuous in vitro culturing, respectively. Scale bar represents 0.01 substitutions per nucleotide position
Earlier studies at INIBAP Musa Germplasm Centre cited covert bacterial association in only a small proportion of stocks (4.5–6%) which was detected during culture screening through explant base streaking on nutrient medium (Van den Houwe et al. 1998; Van den Houwe and Swennen 2000; Strosse et al. 2004). Majority of the organisms in the above studies (60–65%) belonged to Gram-positive spore forming Bacillus spp. (B. circulans, B. pumilus, B. sphaericus, B. subtilis and B. stearothermophilus) which have supposedly accumulated in the cultures as heat resistant spores over time. The rest included Gram-positive Mycobacterium sp. (35%) or Gram-negative Acinetobacter sp. (Van den Houwe et al. 1998; Van den Houwe and Swennen 2000). The deviant observations at INIBAP could be contributed by the absence of detailed tissue screening, the low culture incubation temperatures followed (16°C) or that the organisms were yet to be activated to cultivation.
A wide range of Gram-positive and Gram-negative bacteria including Bacillus, Enterobacter, Pseudomonas, Staphylococcus, Xanthomonas, Corynebacterium, Micrococcus, Brevibacillus, Microbacterium, Pantoea, Brevundimonas, Sphingomonas, Agrobacterium, Methylobacterium spp. have been reported in tissue cultures of various plant species (Leifert and Cassells 2001; Herman 2004; Thomas 2004a, b, 2006; Kulkarni et al. 2007; Thomas et al. 2007a). Habiba et al. (2002) isolated four Gram-positive (Cellulomonas uda, C. flavigena, Corynebacterium paurometabolum and Bacillus megaterium) and three Gram-negative isolates (Klebsiella, Erwinia and Pseudomonas spp.) from table banana cultures. The present study with banana has identified some uncommon endophytic organisms such as Ochrobactrum intermedium, Alcaligenes faecalis, Ralstonia mannitolilytica, Oceanobacillus picturae, Bacillus neonatiensis, Brachybacterium, Brevibacterium, Kocuria rosea, Tetrasphaera spp. etc.
The identification results have indicated that the group two and group three organisms in this study (Brevundimonas, Methylobacterium, Alcaligenes, Ralstonia, Pseudomonas, Corynebacterium, Microbacterium, Staphylococcus, Oceanobacillus, Bacillus, Brachybacterium, Brevibacterium, Kocuria, Tetrasphaera spp.) did not really belong to the obligate or fastidious type that has complex or undeciphered growth requirements. On the other hand, it unraveled the capability of these organisms to enter the VBNC state as endophytes and their gradual activation to cultivable form during continuous in vitro culturing. Many bacteria are known to enter the VBNC state in response to changes in environmental conditions or stress, and cells can remain so for long periods (Xu et al. 1982; Colwell et al. 1985; Alexander et al. 1999) and the same has been reported with endophytic bacteria (Reiter et al. 2002; Reiter and Sessitsch 2006). Several factors are known to induce the organisms to VBNC state and this phenomenon in Gram-negative bacteria is akin to sporulation in Gram-positive bacteria. The factor(s) that contributed to the activation of VBNCs to cultivation have not been understood (Thomas et al. 2008). Change in medium pH from acidic to alkaline, release of tissue breakdown products, gradual inoculum build up with time, exposure of internal tissue during recurrent subculturing etc. appeared some possibilities (Thomas 2004a, b; Thomas et al. 2006). Basaglia et al. (2007) attained the resuscitation of VBNC cells of Sinorhizobium meliloti through exposure to a mix of antibiotics. Some of the antibiotic-supplied watermelon stocks in our earlier studies showed obvious growth on the TCM suggesting that controlling of certain bacteria by one antibiotic led to the emergence of other bacteria (Thomas 2004b; Thomas et al. 2006). In papaya, supply of tissue constituents resulted in significant growth enhancement in the endophytic isolates, Microbacterium, Pantoea, Enterobacter, Brevundimonas, Sphingomonas, Methylobacterium, Agrobacterium and Bacillus spp. (Thomas et al. 2007a).
The delayed detection of bacteria in the cultures prima facie suggested the possibility of lateral introduction. However, hermetic sealing of culture vessels together with stringent sterility checks, microscopic detection of bacterial cells including spores in the tissue sap at culture initiation (Thomas et al. 2008), similar observations with different stocks and batches and the fact that most of the organisms isolated in this study were not common laboratory contaminants discounted such a possibility. When chanced upon organisms like S. epidermidis, which could be associated with human skin, we undertook mock inoculations using several nutrient plates, ruling out the possibility of lateral introduction.
There is an emerging interest in endophytes as agents in plant growth promotion and stress alleviation (Azevedo et al. 2000; Hallmann 2001; Thomas et al. 2007a) and several of the organisms isolated in this study have been reported as potential beneficial organisms (Hallmann 2001). Streptomycete antagonists of fusarium wilt in Musa have been isolated from banana roots (Cao et al. 2005). Growth promotion in rice by Enterobacter cloacae (Mehnaz et al. 2001), bioconrol effect by Pantoea agglomerans (Nunes et al. 2002) are some other documented examples. Actinomycetes are known to promote plant growth and are involved in biocontrol (Conn and Franco 2004). On the other hand, some of the organisms isolated in this study have possible implications in human health as they showed high sequence identity (99–100%) to organisms associated with diseases or infections, such as Ralstonia mannitolilytica isolated from respiratory secretions of cystic fibrosis patients (Coenye et al. 2002), Staphylococcus epidermidis from chronic wounds (Frank et al. 2005), Corynebacterium amycolatum from a urinary tract infected patient (Goldenberger et al. 2002) or Bacillus neonatiensis from a patient with neonatal sepsis (Ko et al. 2005). Human pathogens have been isolated as endophytes in other studies as well, which included dreaded Salmonella and Nocaridia spp. (Rosenblueth and Martínez-Romero 2006). Several bacteria isolated here are often reported from environmental samples, which is a reflection of the poor knowledge about the endophytes rather than the possibility of their lateral entry as contaminants. It is quite likely that the endophytes reach the soil, sewage, sludge, untreated water etc. at the end of the life cycle of the host or through plant debris and get isolated frequently from environmental samples. One species may be present in diverse environments but the strains could be different. For instance, three morphotypes of Curtobacterium citreum that we isolated from chrysanthemum showed the highest similarity to an isolate from deep sea (Panicker et al. 2007). 16S rRNA gene based identification cannot discriminate between different strains.
This study also demonstrates the utility of tissue culture system in harnessing some less common organisms or novel endophytes by bringing the VBNC cells to cultivation and making them amenable for future exploitation. The fact that a part of these organisms would perpetuate as endophytes with the new suckers while the rest returns to the soil at the culmination of normal life span of banana plants (12–15 months) and become a part of soil microbial community adds credence for these observations to soil and environmental microbiology and offers a new vista to study microbe–plant cyclic association.
In summary, endophytic bacteria isolated from in vitro cultures of banana belonged about 24 species falling under 20 genera. The study has identified several endophytic organisms with potential ability to survive in VBNC state. Such organisms turned cultivable after recurrent in vitro culturing of stock plants displaying a Gram-positive predominance, including actinobacteria and spore-forming Bacillus spp. The organisms that were easily cultivable and expressed as visible contaminants during the first in vitro passage included common endophytes with a Gram-negative predominance. The isolated organisms included those reportedly useful in plant growth promotion or biocontrol but also the ones with implications in human health. The study thus demonstrates the utility of tissue culture system in isolating uncommon endophytes and to explore into endophytic microbiology.
Abbreviations
- BIM:
-
Bacteriological indexing medium
- NA:
-
Nutrient agar
- OC:
-
Original culture
- TCM:
-
Tissue culture medium
- TSA:
-
Trypticasein soy agar
- VA:
-
Viss et al. (1991) agar
- VBNC:
-
Viable but non-culturable
References
Alexander E, Pham D, Steck TR (1999) The viable-but-nonculturable condition is induced by copper in Agrobacterium tumefaciens and Rhizobium leguminosarum. Appl Environ Microbiol 65:3754–3756
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (2005) Current protocols in molecular biology. vol 1. Wiley, New York, pp 2.4.1–2.4.2
Azevedo JL, Maccheroni W Jr, Pereira JO, Araújo WL (2000) Endophytic microorganisms: a review on insect control and recent advances on tropical plants. Electronic J Biotech 3:40–65
Basaglia M, Povolo S, Casella S (2007) Resuscitation of viable but not culturable Sinorhizobium meliloti 41 pRP4-luc: effects of oxygen and host plant. Curr Microbiol 54:167–174
Bell CR, Dickie GA, Harvey WLG, Chan JWYF (1995) Endophytic bacteria in grapevine. Can J Microbiol 41:46–63
Cao L, Qiu Z, You J, Tan H, Zhou (2005) Isolation and characterization of endophytic streptomycete antagonists of fusarium wilt pathogen from surface sterilized banana roots. FEMS Microbiol Lett 247:147–152
Cappuccino JG, Sherman N (1996) Microbiology—a laboratory manual, 4th edn. Addison Wesley Longman Inc., Harlow
Coenye T, Goris J, Spilker T, Vandamme P, LiPuma JJ (2002) Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J Clin Microbiol 40:2062–2069
Colwell RR, Brayton PR, Grimes DJ, Roszak DR, Huq SA, Palmer LM (1985) Viable, but non-culturable, Vibrio cholerae and related pathogens in the environment: implications for release of genetically engineered microorganisms. Biotechnol 3:817–820
Conn VM, Franco CMM (2004) Effect of microbial inoculants on the indigenous actinobacterial endophyte population in the roots of wheat as determined by terminal restriction fragment length polymorphism. Appl Environ Microbiol 70:6407–6413
Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Rauult D (2000) 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol 38:3623–3630
Frank DN, Wysocki A, Specht-Glick DD, Rooney A, Feldman RA, St Amand AL, Pace NR, Trent J (2005) Comparison of culture and molecular-based identification of microbial constituents of chronic human wounds (NCBI acc no. DQ170425)
Goldenberger D, Grubenmann M, Dutly F (2002) Corynebacterium amycolatum isolated from urine of a 27-year-old female with recurrent urinary tract infection (NCBI acc no. AY831726)
Habiba U, Reza S, Saha ML, Khan MR, Hadiuzzaman S (2002) Endogenous bacterial contamination during in vitro culture of banana: identification and prevention. Plant Tiss Cult 12:117–124
Hallmann J (2001) Plant interactions with endophytic bacteria. In: Jeger MJ, Spence NJ (eds) Biotic interactions in plant-pathogen associations, CABI Publishing, Oxon, Wallingford, pp 87–119
Herman EB (2004) Recent advances in plant tissue culture viii. Microbial contaminants in plant tissue cultures: solutions and opportunities 1996–2003. Agritech Consultants, Inc., Shrub Oak, USA
Ko KS, Oh WS, Song J-H (2005) Two novel Bacillus species isolated from a patient with neonatal sepsis: Bacillus neonatiensis sp. nov. and Bacillus koguryoae sp. nov. (NCBI acc no. AY904032)
Kulkarni AA, Kelkar SM, Watwe MG, Krishnamurthy MV (2007) Characterization and control of endophytic bacterial contaminants in in vitro cultures of Piper spp., Taxus baccata subsp. wallichiana, and Withania somnifera. Can J Microbiol. 53:63–74
Leifert C, Cassells AC (2001) Microbial hazards in plant tissue and cell cultures. In vitro Cell Dev Biol Plant 37:133–138
Mehnaz S, Mirza MS, Haurat J, Bally R, Normand P, Bano A, Malik KA (2001) Isolation and 16S rRNA sequence analysis of the beneficial bacteria from the rhizosphere of rice. Can J Microbiol 47:110–1177
Nunes C, Usall J, Texido N, Fons E, Vinas I (2002) Post-harvest biological control by Pantoea agglomerans (CPA-2) on Golden Delicious apples. J Appl Microbiol 92:247–255
Panicker B, Thomas P, Janakiram T, Venugopalan R (2007) Influence of cytokinin levels on in vitro propagation of shy suckering chrysanthemum ‘Arka Swarna’ and activation of endophytic bacteria. In vitro Cell Dev Biol Plant 43:614–622
Reiter B, Sessitsch A (2006) Bacterial endophytes of the wildflower Crocus albiflorus analyzed by characterization of isolates and by a cultivation-independent approach. Can J Microbiol 52:140–149
Reiter B, Pfeifer U, Schwab H, Sessitsch A (2002) Response of endophytic bacterial communities in potato plants to infection with Erwinia carotovora subsp. atroseptica. Appl Environ Microbiol 68:2261–2268
Rosenblueth M, Martínez-Romero E (2006) Bacterial endophytes and their interaction with hosts. Mol Plant Microbe Interact 8:827–837
Strosse H, Van den Houwe I, Panis B (2004) Banana cell and tissue culture—a review. In: Jain SM, Swennen R (eds) Banana improvement: Cellular, molecular biology, and induced mutations, (http://www.fao.org/docrep/007/ae216e/ae216e00.html)
Suslow TV, Schroth MN, Isaka M (1982) Application of a rapid method for Gram differentiation of plant pathogenic and saprophytic bacteria without staining. Phytopathol 72:917–918
Thomas P (2004a) A three-step screening procedure for detection of covert and endophytic bacteria in plant tissue cultures. Curr Sci 87:67–72
Thomas P (2004b) In vitro decline in plant cultures: detection of a legion of covert bacteria as the cause for degeneration of long-term micropropagated triploid watermelon cultures. Plant Cell Tiss Org Cult 77:173–179
Thomas P (2006) Isolation of an ethanol-tolerant endospore-forming Gram-negative Brevibacillus sp. as a covert contaminant in grape tissue cultures. J Appl Microbiol 101:764–774
Thomas P, Prabhakara BS, Pitchaimuthu M (2006) Cleansing the long-term micropropagated triploid watermelon cultures from covert bacteria and field testing the plants for clonal fidelity and fertility during the 7–10 year period in vitro. Plant Cell Tiss Org Cult 85:317–329
Thomas P, Kumari S, Swarna GK, Gowda TKS (2007a) Papaya shoot tip associated endophytic bacteria isolated from in vitro cultures and host-endophyte interaction in vitro and in vivo. Can J Microbiol 53:380–390
Thomas P, Kumari S, Swarna GK, Prakash DP, Dinesh MR (2007b) Ubiquitous presence of fastidious endophytic bacteria in field shoots and index-negative apparently clean shoot-tip cultures of papaya. Plant Cell Rep 26:1491–1499
Thomas P, Swarna GK, Patil P (2008) Ubiquitous presence of normally non-cultivable endophytic bacteria in field shoot tips of banana and their gradual activation to quiescent cultivable form in tissue cultures. Plant Cell Tiss Org Cult, doi:10.1007/s11240-008-9340-x
Van den Houwe I, Swennen R (2000) Characterization and control of bacterial contaminants in in vitro cultures of banana (Musa spp.). Acta Hort 530:69–79
Van den Houwe I, Gun J, Swennen R (1998) Bacterial contamination in Musa shoot tip cultures. Acta Hort 490:485–492
Viss PR, Brooks EM, Driver JA (1991) A simplified method for the control of bacterial contamination in woody plant tissue culture. In vitro Cell Dev Biol Plant 27:42
Xu HS, Roberts N, Singleton FL, Attwell RW, Grimes DJ, Colwell RR (1982) Survival and viability of nonculturable E. coli and V. cholerae in the estuarine and marine environment. Microbial Ecol 8:313–323
Acknowledgments
This study was supported by a grant from the Department of Biotechnology, Govt. of India under the project ‘Identification of covert endophytic microbes in plant tissue cultures and their management and control’. Contributions from D. P. Prakash and V. Bidari during their brief association with the project and laboratory assistance by N. Shivrudriah and B. Hanumanthraju are acknowledged. This publication bears IIHR Contribution No. 10/2007.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supply of live bacterial cultures or genetic material for research purpose is subject to their revival from glycerol stocks (as some of the organisms showed poor tolerance) and the requestor obtaining written permission from the Director General, Indian Council of Agricultural Research, New Delhi-110001.
Rights and permissions
About this article
Cite this article
Thomas, P., Swarna, G.K., Roy, P.K. et al. Identification of culturable and originally non-culturable endophytic bacteria isolated from shoot tip cultures of banana cv. Grand Naine. Plant Cell Tiss Organ Cult 93, 55–63 (2008). https://doi.org/10.1007/s11240-008-9341-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-008-9341-9