Abstract
Key message
We developed T1AL·1PS and T1AS·1PL Robertsonian translocations by breakage-fusion mechanism based on wheat-A. cristatum 1P(1A) substitution line with smaller leaf area, shorter plant height, and other excellent agronomic traits
Abstract
Agropyron cristatum, a wild relative of wheat, is a valuable germplasm resource for improving wheat genetic diversity and yield. Our previous study confirmed that the A. cristatum chromosome 1P carries alien genes that reduce plant height and leaf size in wheat. Here, we developed T1AL·1PS and T1AS·1PL Robertsonian translocations (RobTs) by breakage-fusion mechanism based on wheat-A. cristatum 1P (1A) substitution line II-3-1c. Combining molecular markers and cytological analysis, we identified 16 spontaneous RobTs from 911 F2 individuals derived from the cross of Jimai22 and II-3-1c. Fluorescence in situ hybridization (FISH) was applied to detect the fusion structures of the centromeres in wheat and A. cristatum chromosomes. Resequencing results indicated that the chromosomal junction point was located at the physical position of Triticum aestivum chromosome 1A (212.5 Mb) and A. cristatum chromosome 1P (230 Mb). Genomic in situ hybridization (GISH) in pollen mother cells showed that the produced translocation lines could form stable ring bivalent. Introducing chromosome 1PS translocation fragment into wheat significantly increased the number of fertile tillers, grain number per spike, and grain weight and reduced the flag leaf area. However, introducing chromosome 1PL translocation fragment into wheat significantly reduced flag leaf area and plant height with a negative effect on yield components. The pre-breeding of two spontaneous RobTs T1AL·1PS and T1AS·1PL was important for wheat architecture improvement.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Common wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) is the most widely cultivated crop and is one of the most important foods around the world (IWGSC 2018; Li et al. 2019; Rahmatov et al. 2016). With the growth of population, wheat production must increase by more than 50% over current levels by 2050 to meet demand (Mottaleb et al. 2023). However, the long-term improvement of wheat varieties was mainly limited in the genetic recombination between regular varieties, which led to the exploitation and erosion of the genetic variation available and the decrease of species diversity (Qi et al. 2007; King et al. 2022). An effective means of increasing wheat yield is broadening its genetic diversity by transferring the alien genes of wild relatives into wheat (Bohra et al. 2022; Zhang et al. 2015). Agropyron cristatum (2n = 4x = 28, genomes PPPP), a tertiary gene pool of wheat improvement, provides a vast and untapped reservoir of genetic variation for agronomic traits of wheat. In addition to its resistance to biotic and abiotic stresses, A. cristatum carries many useful traits for improving wheat yield such as high floret numbers and tillers (Dong et al. 1992; Li et al. 1995). In our recently published study, we found that A. cristatum 1P addition line also reduced plant height and leaf area which were helpful for wheat plant architecture suitable for dense planting and increasing yield (Wang et al. 2022). But the addition lines may carry more deleterious effects and are not stably inherited by offspring. Hence, the development of translocation could be an effective method for transferring the alien genes from A. cristatum into wheat.
On account of translocation lines generally carrying smaller alien segments, they are genetically more stable and less likely to have deleterious effects (Cai et al. 1998; Faris et al. 2008; Jiang and Gill 1993). The use of substitution lines to induce the whole arm RobTs proved to be an effective way to produce translocation lines. The whole arm translocation lines created by spontaneous translocation have better compensation effects and genetic balance. Alien chromosomes in whole arm translocation lines can compensate for the loss of homoeologous wheat chromosomes. Breakage-fusion mechanism induced by the substitution line can produce spontaneous RobTs (Friebe et al. 2005). Centric misdivision followed by the fusion of the broken arms from different chromosomes results in whole-arm RobTs (Robertson 2005) which played a significant role in karyotype evolution (Friebe et al. 2005). It is reported that RobTs generally occur between some homoeologous chromosomes, which has the advantages of good compensation and is easier to be used in wheat breeding. A successful case is the widespread application of T1BL·1RS in wheat breeding due to its remarkable yield potential and disease resistance (Lukaszewski 1993; Friebe et al. 1996; Rabanus-Wallace et al. 2021; Villareal et al. 1991). In addition, Rahmatov et al. (2016) used breakage-fusion mechanism of univalent chromosomes to produce a new RobTs T2DS·2RL with stem rust resistance gene Sr59, and the kernel protein content of translocation includes segment 2DS, and 2RL has a significant improvement (Jagannath and Bhatia 1972; May and Appels 1978; Zeller and Koller 1981a). Ghazali et al. (2015) produced wheat–Thinopyrum bessarabicum whole arm RobTs T2EbS·2BL. Li et al. (2020) produced a spontaneous Robertsonian T4SlS·4BL translocation chromosome carrying Pm66 for powdery mildew resistance. Türkösi et al. (2018) developed a new T7BS·7HL RobTs conferring salt tolerance and (1, 3; 1, 4)-β-D-glucan content. Liu et al. (2016) developed a set of 13 compensating wheat-Ae. speltoides RobTs covering the S genome of Ae. speltoides except for the long arm of chromosome 4S. Qi et al. (2021) identified T6DS·6PL and T6PS·6DL translocation lines from wheat-A. cristatum 6P(6D) substitution line which could improve grain weight.
Previously, we reported that II-3-1c was identified as wheat-A. cristatum 1P (1A) disomic substitution line with hair glume and reduced plant height phenotypes (Pan et al. 2017). Recently, we proved that the introgression of A. cristatum chromosome1P into wheat can improve plant architecture, including reducing plant height and leaf size of wheat (Wang et al. 2022). In this study, we developed two types of heritable whole arm translocation lines T1AL·1PS and T1AS·1PL from substitution line II-3-1c and evaluated their genetic effects of the alien translocation segment through genetic segregation population. These RobTs provide new germplasm resources for wheat architecture improvement.
Methods
Plant materials
Jimai22 is an elite commercial common wheat cultivar with a high yield and wide adaptability in the largest area in China. II-3-1c is a 1P(1A) disomic substitution line from the offspring of wheat-A. cristatum derivative II-3–1 (Pan et al. 2017). The RobTs were selected from an F2 population with 911 individuals, derived from the cross of Jimai22 and II-3-1c. The F2 and F3 progenies were genotyped by molecular markers for identification of putative RobTs. The secondary segregating population of F3 and F4 were originated from the heterozygous individuals of 1PS and 1PL RobTs. These wheat materials and RobTs were maintained at National Crop Genebank in Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS), Beijing, China.
Molecular markers analysis
Total genomic DNA was isolated from fresh leaf tissue using the CTAB protocol (Zhang et al. 2017). To identify the translocation events, we first use two chromosome 1PS-specific and two chromosome 1PL-specific EST-STS markers to genotype these F2 individuals (Table 1). The positive individuals were selected to generate F3 lines. According to the genotyping of EST-STS markers, we selected positive F3 lines for cytogenetics identification. To distinguish the homologous, heterologous and negative plants, we developed the co-dominant markers including EST-STS markers (Zhang et al. 2017) and Kompetitive Allele Specific PCR (KASP) markers (Table 1). These co-dominant markers were used for tracking the alien translocation fragment and genotyping in the genetic analysis.
SNP markers have been applied to high-throughput SNP genotyping arrays (Grewal et al. 2020) which are used to test F3:4 and F4:5 segregation population of 1PS translocation lines. Orthologous CDS sequences from reference genomes of diploid A. cristatum z1842 and wheat cv. Chinese Spring (CS) RefSeq v1.0 were aligned to identify putative interspecific SNPs. The primer design procedures were followed by Grewal et al. (2020) with some slight modifications. Each KASP PCR mixture is made by 50 ng of DNA template, 2.5 μl of HiGeno 2 × Probe Mix B (JasonGen Biotech Co., Ltd., Beijing, China), and 0.07 μl SNP-Specific primers. Thermal cycling conditions were 95 °C for 10 min; followed by 10 cycles of touchdown PCR: 95 °C for 20 s, 61 °C-55 °C for 40 s (drop 0.6 °C per cycle) and final 28–34 cycles of regular PCR: 95 °C for 20 s, 55 °C for 40 s. The fluorescence reading was performed on the BMG LABTECH microplate reader. EST-STS markers of A. cristatum chromosome1P were developed based on non-denaturing polyacrylamide gels according to the method of Zhang et al. (2017).
Cytogenetic analysis
To visualize P-genome chromatin in descendant of the 1P (1A) substitution line II-3-1c, non-denaturing fluorescence in situ hybridization (ND-FISH) was operated in root tip cells using oligonucleotide probes (oligo-pAc). ND-FISH use oligonucleotide probes (oligo-pSc119.2-1 and oligo-pTa535-1) to identify which chromosome fragment of wheat was translocated on chromosome 1P. The centromere was analyzed by FISH using two centromeric sequences oligo-pAcCR1 and oligo-CCS1 as probes (Sun et al. 2021; Han et al. 2017). The root tips of all the materials in this study were excised when these roots grew to 1.5–2 cm and were disposed with nitrous oxide gas and 90% acetic acid (King et al. 2017; Sun et al. 2021).
To observe the meiotic behavior of the translocation lines, we collect anthers containing pollen mother cells at the wheat booting stage. The anthers were disposed in Carnoy's Fluid (1:3 (v/v) acetic acid/ethanol) and saved in 70% ethanol for 48 h (Megyeri et al. 2013). Anthers with appropriate stages were squashed in 45% acetic acid and viewed under phase contrast and dispose it on the surface of liquid nitrogen for 1 min (Jauhar 2006; Li et al. 2000). All slides were observed by Zeiss Axio Imager Z2 upright epifluorescence microscope (Carl Zeiss Ltd, Germany) and captured with a CCD camera. The GISH and FISH were followed by previously described with some slight modifications (Fu et al. 2015; Han et al. 2006; Li et al. 2021; Sun et al. 2021).
Junction region identification by resequencing translocation lines
To explore junction regions of segments in fine detail, we resequence the homozygous T1AL·1PS and T1AS·1PL translocation lines. Resequence data were used for characterizing A. cristatum introgressions into wheat based on comparative mapping depths. By mapping raw reads of T1AL·1PS and T1AS·1PL translocation lines to a combined reference genome made up of the pseudomolecules of A. cristatum Accession No. z1842 (unpublished) and CS Ref seq v1.0, we can get accurate translocation breakpoints according to the sequencing depth and the distribution of the reads. The genomic DNA sample was fragmented by sonication to a size of ~ 350 bp, then DNA fragments were end-polished, A-tailed, and ligated with the full-length adapters for Illumina sequencing with further PCR amplification. Subsequently, we used the Illumina Novaseq platform to generate 10 × sequencing coverage raw sequences with a 150 bp read length. After sequence quality checking and filtering, we retained 62.6 Gb and 69.8 Gb of high-quality genomic data for sequence alignment. The remaining high-quality reads were mapped to the mixed reference genomes using Burrows-Wheeler Aligner software (Li et al. 2009) with the command ‘mem -t 4 -k 32 -M’. To reduce mismatches generated by PCR amplification before sequencing, duplicated reads were removed using SAMtools (v0.1.1) (Li et al. 2009). Reads depth of each genome site were calculated by SAMtools (v0.1.1) (Li et al. 2009) with the command ‘samtools depth’. Then, average values of reads depth were estimated for 1-Mb sliding windows with a 1-Mb step size along each chromosome. Based on the average values, we create fit curve using the R package ggplot2 (Wickham 2016).
Evaluation of agronomic traits
F3:4 and F4:5 family segregation populations of heterozygous T1AL·1PS and T1AS·1PL translocation plants were planted in the experimental station of the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Xinxiang, Henan Province during the 2020–2021 and 2021–2022 sowing seasons. The plant density was 30 cm by 10 cm with a 2 m length. Leaf length and width of flag leaf, top second leaf, top third leaf, plant height, fertile tillering number, spike length, total spikelet number, kernel number per spikelet, and grain number per spike were measured for each plant.
Results
The development of the translocation lines
In previous study, we have proved that the common wheat-A. cristatum 1P addition line carries genes that reduce the size of flag leaves. To produce chromosome 1P translocation lines, we developed an F2 population by crossing substitution line 1P(1A) II-3-1c with commercial wheat cv. Jimai22. Six EST-STS markers and two KASP markers specific to chromosome arms 1PS and 1PL, were used to genotype 911 F2 individuals for identification of putative translocation lines (Table 2). As a result, a total of 32 plants were identified lacking chromosome1PS, and 28 plants lacking chromosome1PL. These 60 F2 plants were selected to generate F3 lines, and the progenies of F3 lines were genotyped by EST-STS markers. These F3 lines, which were showing > 50% transmission rate of chromosome arm, were selected for further GISH analysis. Finally, 9 T1AL·1PS and 7 T1AS·1PL spontaneous compensating wheat-A. cristatum translocations were obtained (Fig. 1). The frequency of translocations was 1.77% in the F2 population. The efficiency of identification was up to 26.7% in terms of the positive plants after P-chromosome-specific molecular marker detection. Therefore, the study provides a highly efficient method for identifying the translocations by molecular marker detection.
Chromosome composition of RobTs
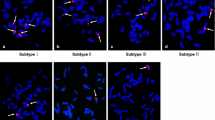
Based on the marker results, 60 F3 families were identified as potential wheat-A. cristatum RobTs. To identify the chromosome composition, GISH analysis was employed for the cytogenetics detection for these selected F3 lines. The result showed that 9 F3 families were short-arm RobTs, 7 F3 families were long-arm RobTs, and the remaining plants were telosomic addition lines (Table 2, Table S1). GISH/FISH analysis revealed that there are 42 chromosomes in the root tip somatic cells of these RobTs. The GISH/FISH pattern showed that 3 and 6 short-arm translocation lines were homozygous and heterozygous for the compensating RobT T1AL·1PS, respectively, (Fig. 2a, c) and 4 and 3 long-arm translocation lines were homozygous and heterozygous for the compensating RobT T1AS·1PL, respectively (Fig. 2b, d).
GISH/FISH pattern of mitotic metaphase chromosomes homozygous for wheat-A. cristatum Robertsonian translocation. GISH pattern for the T1AL·1PS translocation line (a) and T1AS·1PL translocation line (b), FISH pattern for the T1AL·1PS translocation line (c) and T1AS·1PL translocation line (d). GISH pattern for the pollen mother cell in T1AL·1PS translocation line (e) and T1AS·1PL translocation line (f). A. cristatum genomic DNA was used as GISH probe (red), and chromosomes were counterstained with DAPI (blue). Tam535 Cy5 (red) and Psc119.2 (green) were used as FISH probes; chromosomes were counterstained with DAPI (blue). A. cristatum chromosome segments are labeled by white arrows in the images. Scale bar = 10 nm (color figure online)
To identify homologous chromosome pairing behavior in metaphase I of meiosis of RobT lines T1AL·1PS and T1AS·1PL, GISH analysis was conducted in pollen mother cells. We found that 42 chromosomes paired as 21 bivalents at meiotic metaphase I and the translocation chromosomes formed a stable ring bivalent (Fig. 2e, f). The cytogenetics results suggested that the translocation chromosome arms of A. cristatum 1PS and 1PL could be stably inherited.
Whole genome sequencing uncovers the junction point of translocation
To reveal the junction point of translocation between chromosome1P and chromosome1A, we conducted whole genome sequencing (WGS) of RobT lines T1AL·1PS and T1AS·1PL. The CS RefSeq v1.0 and the diploid A. cristatum assembly were combined as an integrated mixed genome and the Illumina paired-end reads of translocation lines were mapped to this mixed genome. With an average read depth of 10 × Illumina paired-end reads, the macro-level visualization of RobT lines T1AL·1PS and T1AS·1PL showed that the junction region occurred at the position of Triticum aestivum chromosome1A 212.5 Mb and A. cristatum chromosome1P 230 Mb (Fig. 3 and Fig.S1), respectively.
Resequencing-based whole genome uncovered the breakpoint of translocation. Combining the Chinese Spring wheat genome and diploid A. cristatum genome as mixed references, raw reads of homozygous RobTs were mapped on group 1 chromosomes. Gain of a large region of chromosome 1P from A. cristatum (light blue line) and loss of a large region on chromosome 1A of wheat (light blue line) in translocation lines. Resequencing depth is about 10 × depth per million versus ‘Chinese Spring’. The resequencing result of T1AL·1PS (a) and T1AS·1PL (b) (color figure online)
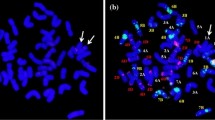
Since the centromere position was located on 210.2–215.8 M in cChromosome1A of the Chinese Spring genome based on CENH3 ChIP-seq (IWGSC 2018; Coombes et al.2023), the junction of RobT lines T1AL·1PS and T1AS·1PL was the same region of centromere. Meanwhile, the centromere of A. cristatum chromosome1P is also predicted at the position of 230–238 Mb (unpublished). Hence, the WGS data supported that the junction positions are located in the region of centromere on wheat chromosome1A and A. cristatum chromosome 1P. To investigate the detailed structures of the centromeres in T1AS·1PL and T1AL·1PS translocation lines, we carried out FISH analysis using two centromeric sequences oligo-pAcCR1 from A. cristatum and oligo-CCS1 from wheat as probes (Wang et al. 2022). The results showed that the translocated chromosomes of T1AS·1PL and T1AL·1PS translocation lines contained fusion centromeres and approximately half of the centromere was derived from A. cristatum while the other half was derived from wheat (Fig. 4). These results further proved that T1AL·1PS and T1AS·1PL translocation lines in this study are compensating RobTs.
Phenotypic characterization of T1AL·1PS and T1AS·1PL translocation lines
To evaluate the genetic effect of the chromosome arms of 1PS and 1PL, the KASP and co-dominant EST-STS markers were developed to genotype the genetic segregation populations (Fig. 5) and the plant architecture traits of the homozygous positive plants and negative plants in segregation population were investigated. The identification results are shown in Table S3 and Fig. 5. Compared with the negative plants, the flag leaf length of the T1AL·1PS plants reduced significantly, but the flag leaf width increased significantly. The similar effects were also observed in the top second leaves of T1AL·1PS plants (Fig. 6). For the T1AS·1PL plants, the length and width of flag leaf had a significant decrease in two growing seasons, and the plant height also decreased significantly. These results suggest that A. cristatum 1PS and 1PL translocated fragments in RobT lines can bring the same plant architecture changes as that of the addition line of A. cristatum 1PS and 1PL reported previously (Wang et al. 2022).
Genotype calling of the KASP marker of T1AL·1PS (a) and amplification patterns of EST-STS marker in T1AS·1PL (b). A red dots correspond to negative plants (W:W), green dots correspond to heterozygous plants (W:P), blue dots correspond to homozygous plants (P:P), and black dots correspond to non-template controls (NTC). b Marker represents DNA marker, Z559 is A. cristatum, JM22 is common wheat cultivar, II-3-1c is 1P(1A) substitution line, 66501–6 is homozygous positive plant, 66706–1 is heterozygous plant, and 66701–3 and 66691–3 are homozygous negative plant (color figure online)
To evaluate the agronomic traits of the RobT lines, we completed two years of replicate phenotype investigation by using F3:4 and F4:5 family segregation populations of heterozygous T1AL·1PS and T1AS·1PL translocation plants (Table S3). Among three yield component factors, the thousand-grain weight, the number of fertile tillers, and the grain number per spike increased significantly after the introduction of the translocated chromosome1PS, which suggested that the introduction of 1PS may increase the yield component factors. Compared with the negative plants, the thousand-grain weight, the number of fertile tillers, and the grain number per spike of positive plants with T1AL·1PS increased significantly by 3.5, 3.1, and 7.4 g, respectively. However, the introduction of the chromosome 1PL showed an obvious negative effect on grain weight. Our results indicated that the 1PS translocation segment can be directly used in wheat breeding for elite plant architecture improvement, but the 1PL segment needs to overcome its linkage drag with the reduction in the grain weight (Fig.S2).
Discussion
During the domestication and artificial selection of crops, plant architectures suitable for density planting were continuously considered. Growing evidence has shown that high-density planting is an effective measure for increasing crop yield per unit of land area (Cao et al. 2022). Small leaves allow better light to capture in the canopy under high-density planting, thus enhancing photosynthesis efficiency decreasing disease and increasing the grain yield. Throughout the past half century, maize yields have increased in part because plants have been grown at increasing densities (Duvick 2005). In wheat, the ideal plant architecture of varieties with high-yield potential is expected to be small leaves, strong stems, and large spikes for improving the light transmission and absorption efficiency under population competition in China (Ru et al. 2015). Wild ancestors are reservoirs of valuable traits, including diverse forms of high-yield potential and resistance to both biotic and abiotic stresses, which remain crucial for the adaptation of modern cultivars to future climates. Crop wild relatives have been used for decades in crop improvement to enhance plant performance (Bohra et al. 2022). Tian et al. (2019) reported that introgressing the wild UPA2 allele from teosinte into modern hybrids enhances high-density maize yields. Plant architecture improvement is a new direction in crop breeding (Wang et al. 2022), but the pre-breeding of plant architecture in wheat has rarely been reported. In the wheat pre-breeding using wild relatives, the transfer of rye chromosome1RS into wheat can provide substantive yield and stress tolerance benefits, and the famous T1BL·1RS translocations have been widely used in bread wheat breeding programs worldwide (Lukaszewski 1990; Rabanus-Wallace et al. 2021).
Here, we developed T1AL·1PS and T1AS·1PL RobTs by transferring alien chromosome1P of A. cristatum into wheat. We found that introducing 1PS fragment into wheat can increase the number of tillers, the grain number per spike and grain weight, and reduce the area of flag leaf of wheat without yield penalty. However, introducing 1PL fragment into wheat reduce significantly the area of flag leaf, top second leaf, top third leaf and plant height, but the yield component factors TKW was reduced. These novel germplasms with good plant architecture from A. cristatum under natural selection pave the way for plant architecture improvement adapting to high-density planting.
Many wheat-alien translocations have been produced for targeted chromosomes through the centromere breakage-fusion mechanism (Ardalani et al. 2016; Li et al. 2000; Sears 1952). The hybrid centromere in translocation lines has a more stable structure and could transmit to the next generation (Friebe et al. 2005; Wang et al. 2017). According to statistics, most existing wheat-related species translocation lines were obtained by spontaneous translocation (Li et al. 2000). However, the frequency of generating spontaneous translocation lines is low and the high-throughput identification methods are lacking. Many studies have been reported on the frequency of generating spontaneous translocation lines between wild relatives and wheat. Friebe et al. (2005) reported that the frequency of RobTs observed in progenies derived from double-monosomic 1Ht/1A plants is 1%. A higher frequency of about 5% RobTs involving chromosome 2B of wheat and 2Eb of Th. bessarabicum was reported (Ghazali et al. 2015). The frequency of compensating RobTs was 0.83% among progenies of double-monosomic wheat- Ae. Searsii introgression plants (Liu et al. 2011a, b). Two short-arm wheat-Th. Intermedium RobT plants have been identified from 1152 F2 plants, and two long-arm wheat-Th. Intermedium RobT plants were generated from 840 F2 plants (Liu et al. 2011a, b). Liu et al. (2016) reported that the average frequency of recovered RobTs between wheat and Aegilops speltoides is 3.2%, ranging from 0.7% for chromosome 6S to 6.8% for 5S. The frequency of group-7 compensating translocation chromosomes was 0.3–0.9% by crossing the monosomic stocks CSM7A, CSM7B, and CSM7D and 7H addition line (Danilova et al. 2018). A compensating wheat-Thinopyrum elongatum RobTs with a positive effect on flour quality T1AS·1EL derived flour is better suited to bread-making than Chinese Spring- and Norin 61-derived flour and that this is because of its greater gluten diversity (Tanaka et al. 2017). The mean frequency of recovery cRobTs for the D.villosum was 2.3% among 2323 individuals from seven cross progenies of double-monosomic plants (Liu et al. 2011a, b).
In the present study, the frequency of the compensating RobTs recovery between A. cristatum 1P and wheat 1A is 1.76% and is up to 26.7% with the aid of specific molecular marker detection. The frequency is close to that of wheat‑Aegilops speltoides RobTs 1A/1S (Liu et al. 2016). The frequency observed in this study means that about 300 progenies would be sufficient to isolate five compensating RoBTs by the breakage-fusion mechanism of univalent chromosomes. If molecular markers specific for wild relatives were used for assistance selection, the frequency of recovered RobTs is up to 26.7%. Hence, our study provides an effective universal strategy for the recovery of compensating RobTs. The two RobTs T1AL·1PS and T1AS·1PL were important pre-breeding lines for wheat architecture improvement.
Data availability
Data and materials supporting the current study can be obtained by contacting the corresponding authors.
Code availability
Not applicable.
References
Ardalani S, Mirzaghaderi G, Badakhshan H (2016) A Robertsonian translocation from Thinopyrum bessarabicum into bread wheat confers high iron and zinc contents. Plant Breed 135:286–290. https://doi.org/10.1111/pbr.12359
Bohra A, Kilian B, Sivasankar S, Caccamo M, Mba C, McCouch SR, Varshney RK (2022) Reap the crop wild relatives for breeding future crops. Trends Biotechnol 40(4):412–431. https://doi.org/10.1016/j.tibtech.2021.08.009
Cai X, Jones SS, Murray TD (1998) Molecular cytogenetic characterization of thinopyrum and wheat-Thinopyrum translocated chromosomes in a wheat-Thinopyrum amphiploid. Chromosome Res 6:183–189. https://doi.org/10.1023/A:1009255516850
Cao Y, Zhong Z, Wang H, Shen R (2022) Leaf angle: a target of genetic improvement in cereal crops tailored for high-density planting. Plant Biotechnol J 20(3):426–436. https://doi.org/10.1111/pbi.13780
Coombes B, Fellers JP, Grewal S, Rusholme-Pilcher R, Hubbart-Edwards S, Yang CY, Joynson R, King IP, King J, Hall A (2023) Whole-genome sequencing uncovers the structural and transcriptomic landscape of hexaploid wheat/Ambylopyrum muticum introgression lines. Plant Biotechnol 21(3):482–496. https://doi.org/10.1111/pbi.13859
Danilova TV, Friebe B, Gill BS, Poland J, Jackson E (2018) Development of a complete set of wheat-barley group-7 Robertsonian translocation chromosomes conferring an increased content of β-glucan. Theor Appl Genet 131:377–388. https://doi.org/10.1007/s00122-017-3008-z
Dong YS, Zhou RH, Xu SJ, Li LH, Caudero Y, Wang RC (1992) Desirable characteristics in perennial triticeae collected in china for wheat improvement. Hereditas 116:176–178. https://doi.org/10.1111/j.1601-5223.1992.tb00224.x
Duvick DN (2005) The contribution of breeding to yield advances in maize (Zea mays l.). Adv Agron 86:83–145. https://doi.org/10.1016/S0065-2113(05)86002-X
Faris JD, Xu SS, Cai X, Friesen TL, Jin Y (2008) Molecular and cytogenetic characterization of a durum wheat-Aegilops speltoides chromosome translocation conferring resistance to stem rust. Chromosome Res 16:1097–1105. https://doi.org/10.1007/s10577-008-1261-3
Friebe B, Jiang J, Raupp WJ, Mcintosh RA, Gill BS (1996) Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91:59–87. https://doi.org/10.1007/BF00035277
Friebe B, Zhang P, Linc G, Gill BS (2005) Robertsonian translocations in wheat arise by centric misdivision of univalents at anaphase i and rejoining of broken centromeres during interkinesis of meiosis ii. Cytogenet Genome Res 109:293–297. https://doi.org/10.1159/000082412
Fu S, Chen L, Wang Y, Li M, Tang Z (2015) Oligonucleotide probes for nd-fish analysis to identify rye and wheat chromosomes. Sci Rep 5:10552. https://doi.org/10.1038/srep10552
Ghazali S, Mirzaghaderi G, Majdi M (2015) Production of a novel Robertsonian translocation from Thinopyrum bessarabicum into bread wheat. Tsitol 49:38–42. https://doi.org/10.3103/S0095452715060031
Grewal S, Edwards SH, Yang CY, King, (2020) Rapid identification of homozygosity and site of wild relative introgressions in wheat through chromosome-specific kasp genotyping assays. Plant Biotechnol J. https://doi.org/10.1111/pbi.13241
Han F, Lamb JC, Birchler JA (2006) High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci USA 103:3238–3243. https://doi.org/10.1073/pnas.0509650103
Han H, Liu W, Lu Y, Zhang J, Yang X, Li X, Hu Z, Li L (2017) Isolation and application of P genome-specific DNA sequences of Agropyron Gaertn. in Triticeae. Planta 245:425–437. https://doi.org/10.1007/s00425-016-2616-1
Iwgsc IWGSC, Bellec A, Berges H, Vautrin S, Alaux M, Alfama F et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. https://doi.org/10.1126/science.aar7191
Jagannath DR, Bhatia CR (1972) Effect of rye chromosome 2 substitution on kernel protein content of wheat. Theor Appl Genet 42:89–92. https://doi.org/10.1007/bf00277949
Jauhar P (2006) Cytological analyses of hybrids and derivatives of hybrids between durum wheat and Thinopyrum bessarabicum, using multicolour fluorescent GISH. Plant Breed 125:19–26. https://doi.org/10.1111/j.1439-0523.2006.01176.x
Jiang J, Gill BS (1993) Sequential chromosome banding and in situ hybridization analysis. Genome 36(4):792–795. https://doi.org/10.1139/g93-104
King J, Grewal S, Yang CY, Hubbart S, Scholefield D, Ashling S et al (2017) A step change in the transfer of interspecific variation into wheat from Amblyopyrum muticum. Plant Biotechnol J. https://doi.org/10.1111/pbi.12606
King J, Grewal S, Fellers JP, King IP (2022) Exploring untapped wheat genetic resources to boost food security. In: Reynolds MP, Braun HJ (eds) Wheat improvement: food security in a changing climate. Springer, Cham, pp 3–15
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al (2009) The sequence alignment/ map format and samtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Li G, Zhang T, Yu Z, Wang H, Yang E, Yang Z (2021) An efficient Oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. Plant J 105:978–993. https://doi.org/10.1111/tpj.15081
Li H, Dong Z, Ma C, Xia Q, Tian X, Sehgal S, Koo DH, Friebe B, Ma P, Liu W (2020) A spontaneous wheat-Aegilops longissima translocation carrying Pm66 confers resistance to powdery mildew. Theor Appl Genet 133:1149–1159. https://doi.org/10.1007/s00122-020-03538-8
Li H, Zhou Y, Xin W, Wei Y, Guo ZJ, L, (2019) Wheat breeding in northern China: achievements and technical advances. Crop J 7:718–729. https://doi.org/10.1016/j.cj.2019.09.003
Li L, Dong Y, Zhou R, Li X, Pei L (1995) Cytogenetics and self -fertility of hybrids between Triticum aestivum L. and Agropyron cristatum (L.) Gaertn. Acta Genet Sin 22:109–114
Li YW, Li HJ, Liang H, Tang SX, Jia X (2000) Fluorescence in situ hybridization applied to the meiotic analysis and spontaneous chromosome translocation in the pollen mother cells of hybrids of Triticum-Haynaldia. Acta Genet Sin 27:317–324
Liu C, Qi L, Liu W, Zhao W, Wilson J, Friebe B, Gill BS (2011a) Development of a set of compensating Triticum aestivum-Dasypyrum villosum Robertsonian translocation lines. Genome 54(10):836–844. https://doi.org/10.1139/g11-05
Liu W, Jin Y, Rouse M, Friebe B, Gill B, Pumphrey MO (2011b) Development and characterization of wheat-Ae. searsii Robertsonian translocations and a recombinant chromosome conferring resistance to stem rust. Theor Appl Genet 122:1537–1545. https://doi.org/10.1007/s00122-011-1553-4
Liu W, Koo DH, Friebe B, Gill BS (2016) A set of Triticum aestivum-Aegilops speltoides Robertsonian translocation lines. Theor Appl Genet 129:2359–2368. https://doi.org/10.1007/s00122-016-2774-3
Lukaszewski AJ (1990) Frequency of 1RS/1AL and 1RS/1BL translocations in the United States wheats. Crop Sci 30:1151–1153. https://doi.org/10.2135/cropsci1990.0011183X003000050041x
Lukaszewski AJ (1993) Reconstruction in wheat of complete chromosomes 1B and 1R from the 1RS.1BL translocation of ‘kavkaz’ origin. Genome 36:821–824. https://doi.org/10.1139/g93-109
May CE, Appels R (1978) Rye chromosome 2R substitution and translocation lines in hexaploid wheat. Cereal Res. Commun. 6(3):231–234
Megyeri M, Molnár-Láng M, Molnár I (2013) Cytomolecular identification of individual wheat-wheat chromosome arm associations in wheat-rye hybrids. Cytogenet Genome Res 139:128–136. https://doi.org/10.1159/000346047
Mottaleb KA, Kruseman G, Frija A, Sonder K, Lopez-Ridaura S (2023) Projecting wheat demand in China and India for 2030 and 2050: implications for food security. Front Nutr 9:1077443. https://doi.org/10.3389/fnut.2022.1077443
Pan C, Li Q, Lu Y, Zhang J, Liu W (2017) Chromosomal localization of genes conferring desirable agronomic traits from Agropyron cristatum chromosome 1P. PLoS ONE 12:0175265. https://doi.org/10.1371/journal.pone.0175265
Qi L, Friebe B, Zhang P, Gill BS (2007) Homoeologous recombination, chromosome engineering and crop improvement. Chromosome Res 15(1):3–19. https://doi.org/10.1007/s10577-006-1108-8
Qi K, Han H, Zhang J, Zhou S, Li X, Yang X, Liu W, Lu Y, Li L (2021) Development and characterization of novel Triticum aestivum-Agropyron cristatum 6P Robertsonian translocation lines. Mol Breed 41(10):59. https://doi.org/10.1007/s11032-021-01251-y
Rabanus-Wallace MT, Hackauf B, Mascher M, Lux T, Stein N (2021) Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat Genet 5:564–573. https://doi.org/10.1038/s41588-021-00807-0
Rahmatov M, Rouse MN, Nirmala J, Danilova T, Friebe B, Steffenson BJ et al (2016) A new 2DS·2RL Robertsonian translocation transfers stem rust resistance gene Sr59 into wheat. Theor Appl Genet 129:1383–1392. https://doi.org/10.1007/s00122-016-2710-6
Robertson W (2005) Chromosome studies. I. Taxonomic relationships shown in the chromosomes of Tettegidae and Acrididiae : V-shaped chromsomes and their signivicance in Acrididae, Locustidae, and Grillidae: chromosomes and variations. J Morphol 27:179–331. https://doi.org/10.1002/jmor.1050270202
Ru Z, Feng S, Li G (2015) High-yield potential and effective ways of wheat in yellow & huai river valley facultative winter wheat region. Sci Agr 48:3388–3393. https://doi.org/10.3864/j.issn.0578-1752.2015.17.006
Sears ER (1952) Misdivision of univalents in common wheat. Chromosoma 4:535–550. https://doi.org/10.1007/BF00325789
Sun Y, Lyu M, Han H, Zhou S, Lu Y, Liu W, Yang X, Li X, Zhang J, Liu X, Li L (2021) Identification and fine mapping of alien fragments associated with enhanced grain weight from Agropyron cristatum chromosome 7P in common wheat backgrounds. Theor Appl Genet 134:3759–3772. https://doi.org/10.1007/s00122-021-03927-7
Tanaka H, Nabeuchi C, Kurogaki M, Garg M, Saito M, Ishikawa G, Nakamura T, Tsujimoto H (2017) A novel compensating wheat-Thinopyrum elongatum Robertsonian translocation line with a positive effect on flour quality. Breed Sci 67(5):509–517. https://doi.org/10.1270/jsbbs.17058
Tian J, Wang C, Xia J, Wu L, Xu G, Wu W, Li D, Qin W, Han X, Chen Q, Jin W, Tian F (2019) Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365(6454):658–664. https://doi.org/10.1126/science.aax5482
Türkösi E, Darko E, Rakszegi M, Molnár I, Molnár-Láng M, Cseh A (2018) Development of a new 7BS.7HL winter wheat-winter barley Robertsonian translocation line conferring increased salt tolerance and (1,3;1,4)-β-D-glucan content. PLoS ONE 13:0206248. https://doi.org/10.1371/journal.pone.0206248
Villareal RL, RajaramMujeebkgazi SA, Toro E (1991) The effect of chromosome 1b/1r translocation on the yield potential of certain spring wheats (Triticum aestivum L.). Plant Breed 106:77–81. https://doi.org/10.1111/j.1439-0523.1991.tb00482.x
Wang X, Han B, Sun Y, Kang X, Zhang M, Han H, Zhou S, Liu W, Lu Y, Yang X, Li X, Zhang J, Liu X, Li L (2022) Introgression of chromosome 1P from Agropyron cristatum reduces leaf size and plant height to improve the plant architecture of common wheat. Theor Appl Genet 135:1951–1963. https://doi.org/10.1007/s00122-022-04086-z
Wickham H (2016) ggplot2-Elegant Graphics for Data Analysis, 2nd edn. Springer, New York
Zeller FJ, Koller OL (1981) Identification of a 4A/7R and a 7B/4R wheat-rye chromosome translocation. Theor Appl Genet 59:33–37. https://doi.org/10.1007/BF00275773
Zhang J, Liu W, Lu Y, Liu Q, Yang X, Li X, Li L (2017) A resource of large-scale molecular markers for monitoring Agropyron cristatum chromatin introgression in wheat background based on transcriptome sequences. Sci Rep 7:11942. https://doi.org/10.1038/s41598-017-12219-4
Zhang R, Hou F, Feng Y, Zhang W, Zhang M, Chen P (2015) Characterization of a Triticum aestivum-Dasypyrum villosum T2VS·2DL translocation line expressing a longer spike and more kernels traits. Theor Appl Genet 128:2415–2425. https://doi.org/10.1007/s00122-015-2596-8
Funding
This research was funded by the National Natural Science Foundation of China, grant number NSFC No. 31971879, and the Chinese Agriculture Research System, grant number CARS-03.
Author information
Authors and Affiliations
Contributions
Bohui Han and Xiao Wang have contributed equally to this work. LHL and JPZ conceived the research. BHH performed the research and wrote the paper. XW analyzed the data. YYS, XLK, MZ, and JWL collected the data, and HMH, SHZ, YQL, WHL, XMY, and XQL participated in the preparation of the reagents and materials in this study. All authors contributed to and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Communicated by Ian D Godwin.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, B., Wang, X., Sun, Y. et al. Pre-breeding of spontaneous Robertsonian translocations for density planting architecture by transferring Agropyron cristatum chromosome 1P into wheat. Theor Appl Genet 137, 110 (2024). https://doi.org/10.1007/s00122-024-04614-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-024-04614-z