Abstract
Key message
A complete set of six compensating Robertsonian translocation chromosomes involving barley chromosome 7H and three chromosomes of hexaploid wheat was produced. Grain β-glucan content increased in lines containing 7HL.
Abstract
Many valuable genes for agronomic performance, disease resistance and increased yield have been transferred from relative species to wheat (Triticum aestivum L.) through whole-arm Robertsonian translocations (RobT). Although of a great value, the sets of available translocations from barley (Hordeum vulgare L.) are limited. Here, we present the production of a complete set of six compensating RobT chromosomes involving barley chromosome 7H and three group-7 chromosomes of wheat. The barley group-7 long-arm RobTs had a higher grain β-glucan content compared to the wheat control. The β-glucan levels varied depending on the temperature and were higher under hot conditions. Implicated in this increase, the barley cellulose synthase-like F6 gene (CslF6) responsible for β-glucan synthesis was physically mapped near the centromere in the long arm of barley chromosome 7H. Likewise, wheat CslF6 homoeologs were mapped near the centromere in the long arms of all group-7 wheat chromosomes. With the set of novel wheat–barley translocations, we demonstrate a valuable increase of β-glucan, along with a resource of genetic stocks that are likely to carry many other important genes from barley into wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cell walls of cereal grains contain non-starch polysaccharides, which are essential sources of dietary fiber in the human diet. The soluble dietary fiber fraction (1-3)(1-4) mixed linked β-d-glucan (further β-glucan) is especially important due to its effects on human health (Cui et al. 2013; Sibakov et al. 2013). The United States Food and Drug Administration (2006) and European Food Safety Authority (2011) approved a health claim for barley β-glucan for lowering the risk of coronary heart disease by maintenance of normal blood LDL-cholesterol concentrations; prevention of obesity and type-2 diabetes by increase in satiety leading to a reduction in energy intake and reduction of post-prandial glycaemic responses. Though wheat products prevail in human consumption, wheat fiber has low levels of β-glucan and does not decrease blood LDL-cholesterol (Collins et al. 2010).
Cereal-mixed linkage (1-3)(1-4)-β-d-glucans are polymers of glucose residues joined by (1-3) and (1-4) linkages. Single (1-3) linkages are separated by two or three (1-4) linkages, resulting in trisaccharide (DP3) and tetrasaccharide (DP4) hydrolysis products after digestion with a specific lichenase enzyme. Grain of different cereals vary in β-glucans content, distribution among grain tissues and β-glucans structure, such as the ratios of DP3/DP4 units, amount of long cellulose-like fragments and molecular size. Barley and oat grain have the highest level of β-glucans, while wheat grain is very low in β-glucan and not a significant source of this polysaccharide. The content on a dry weight basis in a whole meal ranges between 3 and 20% in oats, and between 3 and 7% in barley, but in wheat it is less than 1% (Beresford and Stone 1983; Havrlentova and Kraic 2006; reviewed in Collins et al. 2010).
The solubility of β-glucans is essential for positive health effects and varies among cereals. It decreases with an increasing DP3/DP4 ratio. The β-glucan DP3/DP4 ratios from wheat, barley and oats are 4.5, 3.0 and 2.3, respectively. Also the ratio of DP3/DP4 depends on the grain tissue; β-glucans in the aleurone layer have higher ratios than those in the endosperm (reviewed by Izydorczyk and Dexter 2008; Collins et al. 2010; Cui et al. 2000, 2013; Shewry 2013). In barley, β-glucans are evenly distributed in the subaleurone layers and endosperm (Wirkijowska et al. 2012) and represent the major endosperm cell wall component, contributing about 75% of the total cell wall material (Fincher and Stone 1986). In wheat, β-glucans are concentrated in the subaleurone layer with little found in the endosperm (Jaaskelainen et al. 2013; Panato et al. 2017). Arabinoxylan is a major component of the wheat endosperm cell walls comprising 67%, whereas the β-glucan content is about 27% (Fincher and Stone 1986). Thus, a number of factors lead to wheat having lower β-glucan content with poorer solubility than oats or barley.
The β-glucan content in barley grain is a quantitative trait controlled by several loci (QTL) with different impacts. Using bi-parental mapping populations, Han et al. (1995) and Szucs et al. (2009) identified grain β-glucan QTLs on chromosomes 1H and 2H with a larger effect associated with the locus on 2H. Molina-Cano et al. (2007) located two QTLs on the distal end of chromosome arm 1HL, near the centromere on 5H and the largest QTL at the pericentromeric region of 7HL with up to 21.4% of variance explained. Li et al. (2008) found four regions associated with β-glucan content on chromosomes 2H, 3H, 6H and 7H with a major QTL explaining up to 39% of the phenotypic variance also located on chromosome 7H near the centromere and a minor QTL located at the end of 7HL with a 12.7% impact. Kim et al. (2011) located two major QTLs on chromosome 7H. Using association mapping, Shu and Rasmussen (2014) mapped QTLs on chromosomes 1, 4, 5, 6, and 7H; the major QTL was detected on chromosome 7H, 4H and 6H. Houston et al. (2014) found significant associations between grain β-glucan content and loci on chromosomes 1, 2, 3, 5, 6 and 7H, with the most significant loci on 2H and 3H. Five markers significantly associated with β-glucan content were found on chromosomes 2, 5 and 7H in genome-wide association studies by Pauli et al. (2015). Thus, in several studies the centromeric region of 7H has been consistently identified as a strong determinant of β-glucan content.
Though the genetic control of grain β-glucan content and metabolic pathways are not known completely, several genes involved in the synthesis of barley β-glucan have been identified. Grass-specific Cellulose synthase-like (Csl) F and H gene families are involved in the biosynthesis of β-glucans (Burton et al. 2006, 2008; Doblin et al. 2009). Nine CslF gene sequences, one CslF pseudogene and one HvCslH1 gene were found in the sequence assembly of the barley genome (Schreiber et al. 2014). Some of these genes were co-localized with the grain β-glucan QTLs. A cluster of five CslF genes (HvCslF3, 4, 8, 10 and 12) and HvCslH1 were mapped near the centromere on chromosome arm 2HS. Two genes HvCslF9 and HvCslF6 were mapped near the centromere on 1HS and 7HL, respectively. HvCslF13, HvCslF7 and HvCslF11 were located at the distal ends of 2HL, 5HL and 7HL, respectively (Burton et al. 2008; Doblin et al. 2009; Schreiber et al. 2014). Analysis of the CslF and H gene expression during grain development showed the transcripts of CslF9 and CslF6 predominated in the endosperm with transient increase of CslF9 transcripts at the beginning of endosperm development and a high expression of CslF6 throughout endosperm development with increase at later stages. The transcript levels of the CslF8, CslF10 and CslH1 were low throughout endosperm development (Burton et al. 2008; Doblin et al. 2009; Schreiber et al. 2014). The importance of HvCslF6 for β-glucan accumulation in grain was confirmed by producing plants with complete lack of β-glucan in grain caused by a mutation in the HvCslF6 locus (Tonooka et al. 2009) and by nearly doubling the content of β-glucan in grain by overexpression of CslF6 in barley transformed with CslF6 cDNA (Burton et al. 2011). Overexpression of CslF4 in the same experiment increased grain β-glucan content by 50% (Burton et al. 2011). Additional candidate genes influencing synthesis of grain β-glucan are HvGlb1 and HvGlb2 coding (1,3;1,4)-β-d-glucan endohydrolase isoenzyme EI and EII and co-localized with significant QTLs at the pericentromeric region of 1HL (Han et al. 1995) and the distal end of 7HL (Li et al. 2008; Kim et al. 2011; Houston et al. 2014). A transient increase of HvGlb1 transcripts at the beginning of endosperm development was observed in some barley cultivars (Burton et al. 2008).
Not much is known about β-glucan synthesis and its genetic control in wheat. A genome-wide association study of β-glucan content in grain of tetraploid wheat (Triticum turgidum L. 2n = 4x = 28, genome AABB) revealed seven genomic regions associated with the trait, located on chromosomes 1A, 2A (two), 2B, 5B and 7A (two). In five of the seven QTL sites, candidate genes encoding glycosyl transferases (GT) or glycosyl hydrolases (GH) were identified (Marcotuli et al. 2016). Schreiber et al. (2014) found 34 TaCslF sequences in the hexaploid wheat (Triticum aestivum L. 2n = 6x = 42, genome AABBDD) sequence database (International Wheat Genome Sequencing Consortium). Ten sets of TaCslF gene sequences similar to 10 barley HvCslF genes were found in all three wheat genomes, located on the same homoeologous chromosomes as in barley. Four additional genes include one B-genome duplicate of HvCslF8 paralogue and two B-genome and one D-genome duplicates of HvCslF13 paralogue. Three sequences similar to HvCslF6 were found on all three wheat group-7 chromosomes. Nemeth et al. (2010) showed the influence of TaCslF6 genes on β-glucan content in grain of hexaploid wheat by RNA interference suppression. In transgenic plants with suppressed TaCslF6, the grain β-glucan content was decreased by 30–52%.
Adding barley to wheat-based products to elevate the β-glucan content can increase their potential health benefits (reviewed in Collins et al. 2010). However, barley flour performance, appearance and flavor are limiting factors when making baked products (Limberger-Bayer et al. 2014).
Transferring barley traits to wheat can also be accomplished through the production of transgenic plants. The transformation of barley with HvCslF4 and HvCslF6 genes increased the β-glucan content in grain or leaf tissues (Burton et al. 2011). The transformation with rice OsCslF and barley HvCslH1 cDNAs resulted in the deposition of β-glucan into leaf cell walls of transgenic Arabidopsis (Burton et al. 2006; Doblin et al. 2009). However, this approach has some limitations. Foods from transgenic crops are not generally accepted by consumers. Besides, many countries have restrictions or prohibition to the use of transgenic organisms in food production (Law Library of Congress (U.S.) 2014).
Another possible approach to transfer barley traits to wheat is interspecific hybridization and chromosome engineering. A set of six barley-wheat disomic addition lines and 13 ditelosomic addition lines were developed by Islam et al. (1978, 1981) and Islam (1983) using hexaploid spring wheat cultivar Chinese Spring and two-rowed spring barley Betzes. Later, several other wheat and barley cultivars were successfully used for the interspecific hybridization (McGuire and Qualset 1990; Molnar-Lang et al. 2012; Turkosi et al. 2016). The possibility to increase the β-glucan content in wheat grain by introgressing barley genes was investigated by Cseh et al. (2011, 2013). Almost two-fold increase of β-glucan content in grain in comparison with wheat parent was detected in the wheat–barley non-compensating Robertsonian translocation line (RobT) T4BS·7HL produced from wheat Asakaze komugi and six-rowed winter barley Manas hybrid, but it was still two times less than in the barley parent (Cseh et al. 2011). These results indicated that the HvCslF6 gene is present on the long arm of chromosome 7H and is functional in a wheat background. The expression of two barley genes HvCslF9 and HvCslF6 in winter wheat Mv9kr1—winter two-rowed barley Igri 1HS ditelosomic line and 7H disomic addition line was detected in grain (Cseh et al. 2013) and resulted in β-glucan increase by 22 and 60%, respectively, compared to the wheat parent, but was still about five times less than in the barley parent.
In our work, we produced a complete set of hexaploid wheat lines with compensating RobT chromosomes involving barley chromosome 7H and three homoeologous group-7 wheat chromosomes and analyzed their effect on β-glucan content in wheat grain.
Materials and methods
Plant material
The material used in this study included barley (Hordeum vulgare L.) cv. Betzes (WGRC accession TA9001), wheat (Triticum aestivum L.) cv. Chinese Spring (CS, TA3008); a CS-Betzes 7H chromosome addition line (TA3697) and CS-Betzes 7HS and 7HL ditelosomic addition lines (TA3588 and TA3587) from a set of lines produced by Islam et al. (1978, 1981) and the CS monosomic stocks CSM7A, CSM7B and CSM7D (TA3047, TA3054 and TA3061, respectively). All lines are maintained at the Wheat Genetics Resources Center at Kansas State University.
Cytogenetic analyses
Genomic in situ hybridization (GISH) was performed according to Zhang et al. (2001) with modifications described by Danilova et al. (2017). The GISH probe mixture (20 μl/slide) contained 40 ng of barley gDNA probe labeled with fluorescein-12-dUTP (PerkinElmer, Waltham, MA, USA), 1 ng of Cy5-(GAA)9 oligonucleotide probe (synthesized by Integrated DNA Technologies, Inc., Coralville, IA, USA) and no wheat-blocking DNA.
Fluorescent in situ hybridization (FISH) with cDNA probe 7S-4 (tplb0015e09), physically mapped to the short arm of group-7 wheat chromosomes and mixture of oligonucleotide probes FAM-(GAA)9 and FAM-pAs1 (Danilova et al. 2012, 2014), labeling microsatellite (GAA) n and D-genome specific Afa family tandem repeat pAs1 (Nagaki et al. 1995), was used to identify chromosomes and determine the structure of the translocations. Physical position of the CslF6 gene and cDNA sequences, used for KASP marker development, were verified by FISH. Chromosome preparations, probe labeling and the FISH procedure were as described previously (Kato et al. 2004, 2006) with minor modifications (Danilova et al. 2012). The cDNA probes were labeled with Texas red-5-dCTP, (PerkinElmer, Waltham, MA, USA).Wheat full-length cDNA clones were obtained from the National BioResource Project-Wheat, Yokohama, Japan. Barley full-length cDNA clones AK353650 and AK365097 were obtained from the Genebank of the National Institute of Agrobiological Sciences, Tsukuba, Japan. The sequence of cDNA AK365097 was verified by BLAST (Altschul et al. 1990) and was found to be 99% similar to EU267181, barley cellulose synthase-like CslF6 mRNA. The average relative distance of a cDNA FISH site from the centromere was measured and calculated on five chromosomes using the MicroMeasure 3.3 software (Reeves and Tear 2000). To paint barley chromosomes using FISH, a probe to barley-specific copia-like retroelement BARE-1 (Manninen and Schulman 1993; Waugh et al. 1997; Wicker et al. 2009) was developed using sequence Z17327 and BARE LTR primers (Table 1). Chromosome preparations were mounted and counterstained with propidium iodide (PI) or 4′,6- diamidino-2-phenylindole (DAPI) in Vectashield (Vector Laboratories, Burlingame, CA, USA).
Images were captured using a Zeiss Axioplan 2 microscope with a cooled charge-coupled device camera CoolSNAP HQ2 (Photometrics, Tucson, AZ, USA) and AxioVision 4.8 software (Carl Zeiss, Thornwood, NY, USA). Images were processed using the Adobe Photoshop software (Adobe Systems Incorporated, San Jose, CA, USA).
Molecular markers analysis
Genomic DNA was isolated from leaves of 2-week-old seedlings using a BioSprint 96 workstation according to the Qiagen BioSprint DNA Plant Handbook protocol (QIAGEN Inc., Valencia, CA, USA).
The 8-µl Kompetitive Allele Specific PCR (KASP) reaction mixture contained 4 µl of DNA sample with a concentration of 10–20 ng/μl, 0.11 μl of 72 × primer mix and 4 μl of a 2 × KASP master mix (LGC Genomics, Beverly, MA, USA). The KASP PCR used a PTC-221 MJ Research thermal cycler under following conditions: primer pair 5—initial denaturation 94 °C, 15 min and 45 cycles of 94 °C—20 s, 56 °C—1 min; primer pair 10—initial denaturation 94 °C—15 min, then 10 cycles 94 °C—20 s, 63 °C (drop 0.6º C every cycle) for 1 min followed by 43 cycles of 94 °C—20 s, 56 °C—1 min. The PCR results were read using a BioRad CFX96 machine (BioRad Laboratories, Inc., Hercules, CA, USA), after reading cycle: 37 °C for 1 min, with Bio-Rad CFX Manager 3.1 software. KASP PCR primer sequences are listed in Table 1. Primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA, USA). For developing KASP markers, sequences with determined physical positions were selected on wheat cytogenetic map (Danilova et al. 2014) or barley physical map (Mayer et al. 2012; Kersey et al. 2016).
Compensation test and β-glucan content
The ability of barley chromosome arms 7HS and 7HL to compensate for a homoeologous part of wheat genome was tested on plants homozygous for the RobTs. To estimate fertility and test compensation, plants were grown in a greenhouse with a 16 h light: 8 h dark photoperiod at 21 °C day and 18 °C night until spike emergence; then temperatures were increased to 28 °C day and 23 °C night until maturity. Fertility was estimated by the average number of seed per spikelet calculated from three heads harvested from five plants. Confidence intervals were calculated for a level equal to 95%.
Seed used for β-glucan measurements were collected from plants grown in two growth chambers CMP6050, (Conviron, Canada) with photoperiod 14 h light: 10 h dark and temperature settings 20 °C day, 17 °C night till plants start heading (45 days); after that temperatures were changed in one chamber to 30 °C day and 20 °C night till maturity, while in the other chamber stayed unchanged. The β-glucan content on a dry weight basis was measured in whole grain flour in four replicates using a β-Glucan Assay Kit for mixed linkage 1,3:1,4-β-d-glucan in cereal grains (Megazyme, Ireland) according to the manufacturer’s instructions (AACC Method 32-23.01). The grain samples were milled using a coffee grinder. Because the uniform, fine milling is essential for the test, we verified the size of the flour particles using 0.5-mm sieve and found that samples passed the 0.5-mm mesh. For the test, grain from five plants of each genotype, grown under the same conditions, was combined in equal ratios to 20-g samples which were used for the moisture measurement, calculation of 1000 kernel weight (TKW) and grinding. Moisture was measured in the grain using near infrared light spectroscopy with DA 7250 NIR Analyzer (Perten, Sweden) in three replicates; the average moisture value was used for all calculations. Statistical data analysis, calculation of average values, confidence at level equal 95% and correlation coefficients were performed using Microsoft Excel functions and ANOVA analysis tools.
Results
Physical mapping of CslF6 gene and barley cDNA
The HvCslf6 cDNA AK365097 FISH probe was hybridized to the wheat–barley chromosome 7H addition line and signals were detected near the centromere (relative distance 0.1) on the long arms of all group-7 wheat and barley chromosomes (Fig. 1). This result is in agreement with BLASTN search (Altschul et al. 1990) against the CS wheat flow-sorted chromosome sequence database (Mayer et al. 2014); sequences with approximately 95% nucleotide identity to cDNA AK365097 were found in the 7L arms of all three wheat genomes.
FISH mapping. CslF6 loci (cDNA AK365097) were detected near the centromere on the long arms of all group-7 wheat and barley chromosomes. The chromosomes were identified based on pattern produced by simultaneous labeling of (GAA) n - and pAs1-repeats (green signals). CslF6 is labeled in red, chromosomes counterstained with DAPI are blue. The average distance of CslF6 sites from the centromere was measured for 7HL at 0.13 ± 0.03; for 7AL at 0.11 ± 0.03; for 7BL at 0.04 ± 0.02 and for 7DL at 0.11 ± 0.02
FISH was used to verify the physical position of cDNA sequences selected for molecular marker development. The barley cDNA AK353650 used for the development of 7HS-specific marker was detected on the distal ends of chromosome arms 7HS, 7AS, 7DS and 4AL at relative distance 0.9 (supplemental Fig. S1). The presence of this probe on the long arm of chromosome 4A is explained by a species-specific cyclic translocation involving chromosomes 4A, 5A and 7B, which is present in all accessions of T. turgidum L. and T. aestivum (Naranjo et al. 1987; Devos et al. 1995; Mickelson-Young et al. 1995; Miftahudin et al. 2004).
Development of 7H-specific KASP markers
KASP markers with determined physical positions were developed for barley and wheat chromosome arms 7S and 7L using sequences of wheat cDNA tplb0013b07 physically mapped on 7L near the centromere (fraction length (FL) 0.2) (Danilova et al. 2014) and barley cDNA AK353650, physically mapped on 7HS close to the telomere (FL 0.9), respectively. The position of the barley sequence AK353650 was detected on the barley physical map of chromosome 7H and verified by FISH as described above. Barley sequences of cvs. Morex, Barke and Bowman (Mayer et al. 2012) corresponding to cDNAs tplb0013b07 and AK353650 were compared with the wheat A-, B-, and D-genome orthologous sequences from the flow-sorted wheat CS sequence database (Mayer et al. 2014) to reveal SNPs and develop KASP primers (Table 1). The KASP assays correctly discriminated between barley cv. Betzes, plants heterozygous for wheat and barley alleles (ditelosomic addition lines TA3588 for 7HS marker AK353650 and TA3587 for 7HL marker 13b07), and wheat CS (Fig. 2) and were used for further screenings.
Genotyping with chromosome 7 KASP markers. The co-dominant KASP assays correctly identified plants homozygous on barley alleles (cv Betzes, yellow circles), heterozygous on barley alleles (green triangles) and homozygous on wheat alleles (cv CS, blue squares). a 7HS marker AK353650, heterozygote—7HS ditelosomic addition line TA3588; b 7HL marker 13b07, heterozygote—7HL ditelosomic addition line TA3587. Black diamonds are blank controls (no call). c Physical positions of the markers on group 7 chromosomes are shown
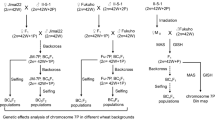
Development of wheat–barley translocation lines
The monosomic stocks CSM7A, CSM7B and CSM7D were crossed with the 7H addition line TA3697 and three sets of double monosomic lines with 2n = 42 chromosomes were selected from the F1 plants. The progenies of plants from three double monosomic populations were screened with KASP markers, and plants with a disassociation of barley 7HS and 7HL markers were analyzed by GISH. In total, 1281 seedlings from three populations were screened (supplemental Table S1). The number of seedlings without the barley chromosome (both 7HS and 7HL molecular markers were negative) and with a complete barley chromosome (both 7HS and 7HL molecular markers were positive) were similar in the three populations, ranging from 40 to 45% each. The remaining 15–20% of the seedlings had broken barley chromosome, represented by telosomes, isochromosomes, RobTs and dicentric chromosomes (supplemental Table S1). These data demonstrate that in 80–85% of the pollen mother cells of double monosomic plants, wheat and barley group-7 chromosomes segregate normally, and at least in 15–20% of the cells, the barley chromosome was broken at the centromere. Most of the broken 7H chromosomes were stabilized as telosomes and only 2–3% fused and formed RobTs or structures with two centromeres (dicentric chromosomes). The frequency of group-7 compensating translocation chromosomes was 0.3–0.9%.
To verify the structure of the translocation chromosomes, we combined molecular marker and cytogenetic analyses. The barley 7HS and 7HL chromosome arms were identified by KASP markers and confirmed cytologically by (GAA) n -banding to distinguish the short arm of barley chromosome 7H, which has a distinctive interstitial (GAA) n site (Figs. 1, 3). The wheat chromosome arms were identified using FISH with cDNA tplb0015e09 probe 7S-4 physically mapped on chromosome arms 7S, combined with probes to pAs1 and (GAA) n repeats (Fig. 3). The CS chromosome arm 7AL has a much weaker (GAA) n signal located more proximal than the (GAA) n signal on 7AS. The wheat 7DL arm has a distinctive interstitial (GAA) n band, that is absent in 7DS (Fig. 3). The wheat 7BL arm was also identified by its specific (GAA) n pattern. Compensating RobTs involving all six wheat group-7 chromosome arms and barley 7H arms and one non-compensating T7BS·7HS was recovered (Table 2, Fig. 3). Independent breakage-fusion events resulted in two T7HS·7DL and three T7DS·7HL RobTs.
Cytogenetic analysis of translocation chromosomes. a Short arms of wheat group-7 chromosomes in translocation chromosomes T7WS·7HL were identified by FISH with cDNA probe 7S-4 and probes to (GAA) n - and pAs1-repeats. FISH images are shown on the left, GISH images are on the right. b Long arms of wheat group-7 chromosomes in translocation T7HS·7WL were identified by their (GAA) n pattern (on the left). FISH: (GAA) n and pAs1 repeats are shown in green; cDNA probe 7S-4 is red; chromosomes counterstained with DAPI are blue. GISH: barley chromatin is labeled in green; (GAA) n -repeat is shown in white pseudocolor; chromosomes counterstained with PI are red
Plants with RobTs were self-pollinated and homozygous progenies were selected by KASP markers and confirmed by cytogenetically (Fig. 4). These plants were used for further analyses.
Chromosomes of plants, homozygous on compensating RobTs. a, b, e, f GISH: barley chromatin is shown in green, (GAA) n -repeat is shown in white pseudocolor, chromosomes counterstained with PI are red; c, d FISH: BARE-repeat hybridized to barley chromatin is shown in green, pAs1-repeat is red, (GAA) n -repeat is shown in white pseudocolor
Characterization of wheat–barley translocation lines
To evaluate the ability of barley chromosome arms 7HS and 7HL to compensate for the loss of homoeologous wheat arms, the sporophytic compensation was determined as seed set per spikelet. The fertility of all homozygous translocation lines did not differ significantly from the CS wheat control (Table 2), except a non-compensating T7BS·7HS line, where it was very low. We observed that plants with RobTs involving 7HS tended to be more vigorous, have longer spikes and a slightly higher grain yield per plant than plants with 7HL (Table 2, Fig. 5).
The β-glucan content in grain from plants homozygous for the wheat–barley translocation chromosomes involving barley 7HL and grown under two temperature conditions was measured. Analysis of variance showed a significant influence of both genotype and temperature on β-glucan content and their interaction (p value < 0.01) with a stronger genotype influence (supplemental Table S2). In plants grown under higher temperatures, β-glucan increased by 14–40% above the CS wheat control, depending on the RobT involved. In plants grown under lower temperatures, the β-glucan content increased by ~ 30% above the wheat control in two lines and was the same as in wheat in line with T7DS·7HL (Table 3, Fig. 6). The TKW was higher in lines grown under lower temperatures and seemed to correlate negatively with β-glucan content (the correlation coefficient was − 0.43).
Discussion
Wheat is a substantial part of the human diet, as a source of starch, energy and essential nutrients, such as protein, vitamins, minerals and dietary fiber (Shewry and Hey 2015). The wheat grain dietary fiber includes mostly arabinoxylan, whereas the content of soluble β-glucan, the most health beneficial component, is very low. Barley has high content of soluble β-glucan, but is not a popular grain in the human diet (Fincher and Stone 1986; Cui et al. 2000; Collins et al. 2010). Introgression of desirable traits into wheat from wild and cultivated relatives, including barley is possible by interspecific hybridization followed by directed chromosome engineering, the strategy reducing the size of the introgressed alien chromatin by induced homoeologous chromosome recombination (reviewed in Sears 1981; Friebe et al. 1996; Qi et al. 2007; Molnár-Láng and Linc 2015).
We reduced the size of introgressed barley chromatin harboring a gene of interest, HvCslF6, to the size of a chromosome arm by producing the complete set of six compensating whole-arm translocation chromosomes. The physical position of HvCslF6 near the centromeric region of barley chromosome arm 7HL and three wheat orthologues on chromosome arms 7AL, 7BL, 7DL was shown by FISH and confirmed the substitution of one copy of wheat CslF6 by barley orthologue in each of the RobTs. The increase of β-glucan in grain of the translocation lines indicates that HvCslF6 is functional in a wheat background, which is in agreement with previous reports (Cseh et al. 2011, 2013). Though the grain β-glucan content substantially increased in some translocation lines, it was still much lower than in the barley parent. As previously reported, the grain β-glucan content in barley is controlled by several QTLs. Though the HvCslF6 was reported as a major QTL with 21–39% impact, the genetic control of β-glucan content and metabolic pathways are not fully understood (reviewed in Burton et al. 2008; Doblin et al. 2009) and other barley genes could be essential for the high accumulation of β-glucan in endosperm cell walls. The interreaction of the barley polysaccharide synthase with wheat enzymes involved in biosynthesis of endosperm cell wall polysacharides is not known. It was shown that the suppression of a TaCslF6 gene significantly reduces the β-glucan content in wheat grain (Nemeth et al. 2010), but it is still not known whether all three wheat copies are equally important for β-glucan biosynthesis. The difference in β-glucan content in the three translocation lines (Table 3) may indicate the possible unequal impact of the three wheat TaCslF6 genes.
The significant influence of growing conditions on β-glucan content in barley grain has been well documented (Swanston et al. 1997; Molina-Cano et al. 2007; Li et al. 2008; Kim et al. 2011). Higher β-glucan content with an increased soluble portion was observed under higher temperature and lower precipitation conditions during grain filling (Swanston et al. 1997; Molina-Cano et al. 2007). The same conditions resulted in a decrease in 1000 kernel weight (Swanston et al. 1997). The β-glucan content in the wheat translocation lines in our contrasting temperature experiment was in agreement with these data. Plants grown under higher temperatures after spike emergence till maturity had a lower TKW and higher grain β-glucan content (Table 3). The difference in TKW may correlate with grain size, plumpness and chemical composition. In our experiment, grain with a lower TKW was either plump, but smaller in size, or shrunken. These two types of grain may have different endosperm/aleurone tissue ratio or different amount of cell wall components per grain. Considering that β-glucan content and solubility in wheat grain differ between the aleurone layer and endosperm, TKW may correlate with both β-glucan content and quality.
A significant increase in β-glucan content in barley grain after increasing the HvCslF6 copy number in plants, transformed with a construct containing HvCslF6 cDNA was shown (Burton et al. 2011). We are presently using ph1b-induced homoeologous recombination to shorten the barley chromatin in T7AS·7HL, T7BS·7HL, and T7DS·7HL lines. Because HvCslF6 is located close to the centromere, it should be feasible to produce wheat–barley recombinant chromosomes consisting of wheat goup-7 short arms, the proximal part of the long arm derived from barley with the HvCslF6 gene and the distal part of this arm derived from wheat of group-7 long arm (rec7AS·7HL-7AL, rec7BS·7HL-7BL and rec7DS·7HL-7DL). Because meiotic metaphase-I pairing in wheat is controlled by homology at the chromosome end, two or three recombinant chromosomes can be combined in one wheat line, which is expected to have a regular bivalent pairing. This will allow combining two or three copies of HvCslF6 and possibly double or triple the β-glucan content in wheat grain. We observed reduction of grain yield (Table 3) in lines with barley RobTs, which may be caused by incomplete compensation of a 7H chromosome arm for substituted wheat chromatin. Reduction of barley chromatin may reduce the plant yield penalty caused by incomplete compensation.
The lines with RobTs involving 7HS have longer spikes. QTLs, associated with spike length have been mapped on chromosomes 2H and 7H of barley (Wang et al. 2016) and a gene, involved in the determination of spike length was allocated to the genetic centromere of barley chromosome 7H (Shahinnia et al. 2012). Increased tillering capacity and earlier flowering time were observed in wheat–barley 7H addition lines (Farkas et al. 2014). A stipe rust-resistance gene was mapped on barley chromosome arm 7HL (Li et al. 2016). Thus, the lines produced during our experiment may carry valuable genes for agronomic performance and disease resistance, that can be screened using the stable homozygous RobT of this immortalized germplasm resource.
Author contribution statement
EJ, JP BF and BSG conceived the project. TVD and BF performed the experiments. TVD wrote the initial manuscript; all authors contributed to the final version.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1006/jmbi.1990.9999
Beresford G, Stone BA (1983) (1 → 3), (1 → 4)-beta-d-glucan content of triticum grains. J Cereal Sci 1:111–114
Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Stone BA, Newbigin EJ, Bacic A, Fincher GB (2006) Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3; 1,4)-beta-d-glucans. Science 311:1940–1942. https://doi.org/10.1126/science.1122975
Burton RA, Jobling SA, Harvey AJ, Shirley NJ, Mather DE, Bacic A, Fincher GB (2008) The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol 146:1821–1833. https://doi.org/10.1104/pp.107.114694
Burton RA, Collins HM, Kibble NAJ, Smith JA, Shirley NJ, Jobling SA, Henderson M, Singh RR, Pettolino F, Wilson SM, Bird AR, Topping DL, Bacic A, Fincher GB (2011) Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3; 1,4)-beta-d-glucans and alters their fine structure. Plant Biotechnol J 9:117–135. https://doi.org/10.1111/j.1467-7652.2010.00532.x
Collins HM, Burton RA, Topping DL, Liao ML, Bacic A, Fincher GB (2010) Variability in fine structures of noncellulosic cell wall polysaccharides from cereal grains: potential importance in human health and nutrition. Cereal Chem 87:272–282. https://doi.org/10.1094/Cchem-87-4-0272
Cseh A, Kruppa K, Molnar I, Rakszegi M, Dolezel J, Molnar-Lang M (2011) Characterization of a new 4BS.7HL wheat–barley translocation line using GISH, FISH, and SSR markers and its effect on the beta-glucan content of wheat. Genome 54:795–804. https://doi.org/10.1139/011-044
Cseh A, Soos V, Rakszegi M, Turkoesi E, Balazs E, Molnar-Lang M (2013) Expression of HvCslF9 and HvCslF6 barley genes in the genetic background of wheat and their influence on the wheat beta-glucan content. Ann Appl Biol 163:142–150. https://doi.org/10.1111/aab.12043
Cui W, Wood PJ, Blackwell B, Nikiforuk J (2000) Physicochemical properties and structural characterization by two-dimensional NMR spectroscopy of wheat beta-d-glucan—comparison with other cereal beta-d-glucans. Carbohydr Polym 41:249–258. https://doi.org/10.1016/S0144-8617(99)00143-5
Cui SW, Wu Y, Ding H (2013) The range of dietary fibre ingredients and a comparison of their technical functionality. In: Delcour JA, Poutanen K (eds) Fibre-rich and wholegrain foods. Elsevier Science, ProQuest Ebook Central, pp 96–119. doi:https://doi.org/10.1533/9780857095787.1.96
Danilova TV, Friebe B, Gill BS (2012) Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma 121:597–611. https://doi.org/10.1007/s00412-012-0384-7
Danilova TV, Friebe B, Gill BS (2014) Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor Appl Genet 127:715–730. https://doi.org/10.1007/s00122-013-2253-z
Danilova TV, Zhang G, Liu W, Friebe B, Gill BS (2017) Homoeologous recombination-based transfer and molecular cytogenetic mapping of a wheat streak mosaic virus and Triticum mosaic virus resistance gene Wsm3 from Thinopyrum intermedium to wheat. Theor Appl Genet 130:549–556. https://doi.org/10.1007/s00122-016-2834-8
Devos KM, Dubcovsky J, Dvorak J, Chinoy CN, Gale MD (1995) Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor Appl Genet 91:282–288. https://doi.org/10.1007/BF00220890
Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A (2009) A barley cellulose synthase-like CslH gene mediates (1,3; 1,4)-beta-d-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci USA 106:5996–6001. https://doi.org/10.1073/pnas.0902019106
EFSA Panel on Dietetic Products Nutrition, Allergies (2011) Scientific opinion on the substantiation of health claims related to beta-glucans from oats and barley and maintenance of normal blood LDL-cholesterol concentrations (ID 1236, 1299), increase in satiety leading to a reduction in energy intake (ID 851, 852), reduction of post-prandial glycaemic responses (ID 821, 824), and “digestive function” (ID 850) pursuant to Article 13(1) of Regulation (EC) No 1924/2006 EFSA J 9:2207. doi:https://doi.org/10.2903/j.efsa.2011.2207
Farkas A, Molnar I, Kiss T, Karsai I, Molnar-Lang M (2014) Effect of added barley chromosomes on the flowering time of new wheat/winter barley addition lines in various environments. Euphytica 195:45–55. https://doi.org/10.1007/s10681-013-0970-7
Fincher GB, Stone BA (1986) Cell walls and their components in cereal grain technology. In: Pomeranz Y (ed) Advances in cereal science and technology, vol 8. American Assoc of Cereal Chemists, St Paul, MN, USA, pp 207–296
Food, Drug Administration HHS (2006) Food labeling: health claims; soluble dietary fiber from certain foods and coronary heart disease. Final Rule Fed Regist 71:29248–29250
Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS (1996) Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91:59–87. https://doi.org/10.1007/BF00035277
Han F, Ullrich SE, Chirat S, Menteur S, Jestin L, Sarrafi A, Hayes PM, Jones BL, Blake TK, Wesenberg DM, Kleinhofs A, Kilian A (1995) Mapping of beta-glucan content and beta-glucanase activity loci in barley-grain and malt. Theor Appl Genet 91:921–927. https://doi.org/10.1007/BF00223901
Havrlentova M, Kraic J (2006) Content of beta-d-glucan in cereal grains. J Food Nutr Res 45:97–103
Houston K, Russell J, Schreiber M, Halpin C, Oakey H, Washington JM, Booth A, Shirley N, Burton RA, Fincher GB, Waugh R (2014) A genome wide association scan for (1,3; 1,4)-beta-glucan content in the grain of contemporary 2-row spring and binter barleys. BMC Genom 15:907. https://doi.org/10.1186/1471-2164-15-907
Islam AKMR (1983) Ditelosomic additions of barley chromosomes to wheat. In: Proceedings of the sixth International Wheat Genetics Symposium, Kyoto, Japan, pp 233–238
Islam AKMR, Shepherd KW, Sparrow DHB (1978) Production and characterization of wheat–barley addition lines. In: Proceedings of the fifth International Wheat Genetics Symposium, New Delhi, pp 365–371
Islam AKMR, Shepherd KW, Sparrow DHB (1981) Isolation and characterization of euplasmic wheat–barley chromosome addition lines. Heredity 46:161–174. https://doi.org/10.1038/hdy.1981.24
Izydorczyk MS, Dexter JE (2008) Barley beta-glucans and arabinoxylans: molecular structure, physicochemical properties, and uses in food products—a review. Food Res Int 41:850–868. https://doi.org/10.1016/j.foodres.2008.04.001
Jaaskelainen AS, Holopainen-Mantila U, Tamminen T, Vuorinen T (2013) Endosperm and aleurone cell structure in barley and wheat as studied by optical and Raman microscopy. J Cereal Sci 57:543–550. https://doi.org/10.1016/j.jcs.2012.05.002
Kato A, Lamb JC, Birchler JA (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA 101:13554–13559. https://doi.org/10.1073/pnas.0403659101
Kato A, Albert PS, Vega JM, Birchler JA (2006) Sensitive fluorescence in situ hybridization signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech Histochem 81:71–78. https://doi.org/10.1080/10520290600643677
Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Falin LJ, Grabmueller C et al (2016) Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acid Res 44:D574–D580. https://doi.org/10.1093/nar/gkv1209
Kim H-S, Park K-G, Baek S-B, Kim J-G (2011) Inheritance of (1–3)(1–4)-beta-d-glucan content in barley (Hordeum vulgare L.). J Crop Sci Biotechnol 14:239–245. https://doi.org/10.1007/s12892-011-0045-z
Law Library of Congress (U.S.). Global Legal Research Directorate (2014) Restrictions on genetically modified organisms. Global Legal Research Center, Washington, D.C
Li JZ, Baga M, Rossnagel BG, Legge WG, Chibbar RN (2008) Identification of quantitative trait loci for beta-glucan concentration in barley grain. J Cereal Sci 48:647–655. https://doi.org/10.1016/j.jcs.2008.02.004
Li K, Hegarty J, Zhang CZ, Wan AM, Wu JJ, Guedira G, Chen XM, Munoz-Amatriain M, Fu DL, Dubcovsky J (2016) Fine mapping of barley locus Rps6 conferring resistance to wheat stripe rust. Theor Appl Genet 129:845–859. https://doi.org/10.1007/s00122-015-2663-1
Limberger-Bayer VM, de Francisco A, Chan A, Oro T, Ogliari PJ, Barreto PL (2014) Barley beta-glucans extraction and partial characterization. Food Chem 154:84–89. https://doi.org/10.1016/j.foodchem.2013.12.104
Manninen I, Schulman AH (1993) BARE-1, a copia-like retroelement in barley (Hordeum vulgare L). Plant Mol Biol 22:829–846. https://doi.org/10.1007/Bf00027369
Marcotuli I, Houston K, Schwerdt JG, Waugh R, Fincher GB, Burton RA, Blanco A, Gadaleta A (2016) Genetic diversity and genome wide association study of beta-glucan content in tetraploid wheat grains. PLoS One. https://doi.org/10.1371/journal.pone.0152590
Mayer KFX, Waugh R, Langridge P, Close TJ, Wise RP, Graner A, Matsumoto T, Sato K, Schulman A, Muehlbauer GJ et al (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491:711–717. https://doi.org/10.1038/nature11543
Mayer KFX, Rogers J, Dolezel J, Pozniak C, Eversole K, Feuillet C, Gill B, Friebe B, Lukaszewski AJ, Sourdille P et al (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. https://doi.org/10.1126/science.1251788
McGuire PE, Qualset CQ (1990) Transfer of the Yd2 barley yellow dwarf virus resistance gene from barley to wheat. In: Burnett PA (ed) World perspectives on barley yellow dwarf. CIMMYT, Mexico, pp 476–481
Mickelson-Young L, Endo TR, Gill BS (1995) A cytogenetic ladder-map of the wheat homoeologous group-4 chromosomes. Theor Appl Genet 90:1007–1011. https://doi.org/10.1007/BF00222914
Miftahudin Ross K, Ma XF, Mahmoud AA, Layton J, Mahmoud AA, Layton J, Milla MAR, Chikmawati T, Ramalingam J, Feril O, Pathan MS et al (2004) Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics 168:651–663. https://doi.org/10.1534/genetics.104.034827
Molina-Cano JL, Moralejo M, Elia M, Munoz P, Russell JR, Perez-Vendrell AM, Ciudad F, Swanston JS (2007) QTL analysis of a cross between European and North American malting barleys reveals a putative candidate gene for beta-glucan content on chromosome 1H. Mol Breed 19:275–284. https://doi.org/10.1007/s11032-006-9075-5
Molnár-Láng M, Linc G (2015) Wheat–barley hybrids and introgression lines. In: Molnár-Láng M, Ceoloni C, Doležel J (eds) Alien introgression in wheat. Cytogenetics, molecular biology, and genomics. Springer, Cham, pp 315–346. doi:https://doi.org/10.1007/978-3-319-23494-6_12
Molnar-Lang M, Kruppa K, Cseh A, Bucsi J, Linc G (2012) Identification and phenotypic description of new wheat—six-rowed winter barley disomic additions. Genome 55:302–311. https://doi.org/10.1139/G2012-013
Nagaki K, Tsujimoto H, Isono K, Sasakuma T (1995) Molecular characterization of a tandem repeat, Afa family, and distribution among Triticeae. Genome 38:479–486. https://doi.org/10.1139/g95-063
Naranjo T, Roca A, Goicoechea PG, Giraldez R (1987) Arm homoeology of wheat and rye chromosomes. Genome 29:873–882. https://doi.org/10.1139/g87-149
Nemeth C, Freeman J, Jones HD, Sparks C, Pellny TK, Wilkinson MD, Dunwell J, Andersson AA, Aman P, Guillon F, Saulnier L, Mitchell RA, Shewry PR (2010) Down-regulation of the CslF6 gene results in decreased (1,3; 1,4)-beta-d-glucan in endosperm of wheat. Plant Physiol 152:1209–1218. https://doi.org/10.1104/pp.109.151712
Panato A, Antonini E, Bortolotti F, Ninfali P (2017) The histology of grain caryopses for nutrient location: a comparative study of six cereals. Int J Food Sci Technol 52:1238–1245. https://doi.org/10.1111/ijfs.13390
Pauli D, Brown-Guedira G, Blake TK (2015) Identification of malting quality QTLs in advanced generation breeding germplasm. J Am Soc Brew Chem 73:29–40. https://doi.org/10.1094/Asbcj-2015-0129-01
Qi L, Friebe B, Zhang P, Gill BS (2007) Homoeologous recombination, chromosome engineering and crop improvement. Chrom Res 15:3–19. https://doi.org/10.1007/s10577-006-1108-8
Reeves A, Tear J (2000) MicroMeasure for Windows, version 3.3
Schreiber M, Wright F, MacKenzie K, Hedley PE, Schwerdt JG, Little A, Burton RA, Fincher GB, Marshall D, Waugh R, Halpin C (2014) The barley genome sequence assembly reveals three additional members of the CslF (1,3; 1,4)-beta-glucan synthase gene family. PLoS One. https://doi.org/10.1371/journal.pone.0090888
Sears ER (1981) Transfer of alien genetic material to wheat. In: Evans IT, Peacock WJ (eds) Wheat science—today and tomorrow. Cambridge University Press, Cambridge
Shahinnia F, Druka A, Franckowiak J, Morgante M, Waugh R, Stein N (2012) High resolution mapping of dense spike-ar (dsp.ar) to the genetic centromere of barley chromosome 7H. Theor Appl Genet 124:373–384. https://doi.org/10.1007/s00122-011-1712-7
Shewry PR (2013) Improving the content and composition of dietary fibre in wheat. In: Delcour JA, Poutanen K (eds) Fibre-rich and wholegrain foods. Elsevier Science, ProQuest Ebook Central, pp 153–168. doi:https://doi.org/10.1533/9780857095787.2.153
Shewry PR, Hey SJ (2015) The contribution of wheat to human diet and health. Food Energy Secur 4:178–202. https://doi.org/10.1002/fes3.64
Shu XL, Rasmussen SK (2014) Quantification of amylose, amylopectin, and beta-glucan in search for genes controlling the three major quality traits in barley by genome-wide association studies. Front Plant Sci 5:197. https://doi.org/10.3389/fpls.2014.00197
Sibakov J, Lehtinen P, Poutanen K (2013) Cereal brans as dietary fibre ingredients. In: Delcour, JA, Poutanen K (eds) Fibre-rich and wholegrain foods. Elsevier Science, ProQuest Ebook Central, pp 153–168. doi:https://doi.org/10.1533/9780857095787.2.170
Swanston JS, Ellis RP, PerezVendrell A, Voltas J, MolinaCano JL (1997) Patterns of barley grain development in Spain and Scotland and their implications for malting quality. Cereal Chem 74:456–461. https://doi.org/10.1094/Cchem.1997.74.4.456
Szucs P, Blake VC, Bhat PR, Chao SAM, Close TJ, Cuesta-Marcos A, Muehlbauer GJ, Ramsay L, Waugh R, Hayes PM (2009) An integrated resource for barley linkage map and malting quality QTL alignment. Plant Genome 2:134–140. https://doi.org/10.3835/plantgenome2008.01.005
Tonooka T, Aoki E, Yoshioka T, Taketa S (2009) A novel mutant gene for (1-3, 1-4)-beta-d-glucanless grain on barley (Hordeum vulgare L.) chromosome 7H. Breed Sci 59:47–54. https://doi.org/10.1270/jsbbs.59.47
Turkosi E, Cseh A, Darko E, Molnar-Lang M (2016) Addition of Manas barley chromosome arms to the hexaploid wheat genome. BMC Genet. https://doi.org/10.1186/s12863-016-0393-2
Wang JB, Sun GL, Ren XF, Li CD, Liu LP, Wang QF, Du BB, Sun DF (2016) QTL underlying some agronomic traits in barley detected by SNP markers. BMC Genet 17:1–13. https://doi.org/10.1186/s12863-016-0409-y
Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BBT, Powell W (1997) Genetic distribution of bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol Gen Genet 253:687–694
Wicker T, Taudien S, Houben A, Keller B, Graner A, Platzer M, Stein N (2009) A whole-genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J 59:712–722. https://doi.org/10.1111/j.1365-313X.2009.03911.x
Wirkijowska A, Rzedzicki Z, Kasprzak M, Blaszczak W (2012) Distribution of (1-3)(1-4)-beta-d-glucans in kernels of selected cultivars of naked and hulled barley. J Cereal Sci 56:496–503. https://doi.org/10.1016/j.jcs.2012.05.002
Zhang P, Friebe B, Lukaszewski AJ, Gill BS (2001) The centromere structure in Robertsonian wheat-rye translocation chromosomes indicates that centric breakage-fusion can occur at different positions within the primary constriction. Chromosoma 110:335–344. https://doi.org/10.1007/s004120100159
Acknowledgements
We thank W. John Raupp for critical editorial review of the manuscript and Duane Wilson for technical assistance; Paul St Amand for consultations on KASP; Rebecca Miller, for expertise and providing equipment for grain moisture measurement and milling; Alina Akhunova, for expertise and lab equipment for spectrophotometry and real time PCR; Anita Dille and Krishna S.V. Jagadish for providing growth chambers. This work was supported through sponsored research agreement between Kansas State University and General Mills Inc. EJ is an employee and stakeholder of General Mills Inc. This is Contribution number 18-078-J from the Kansas Agricultural Experiment Station, Kansas State University, Manhattan, KS 66506-5502, USA. Funding was provided by General Mills (US) (Grant nos. BG4412 and BG3641).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have conflict of interest.
Additional information
Communicated by P. Heslop-Harrison.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Danilova, T.V., Friebe, B., Gill, B.S. et al. Development of a complete set of wheat–barley group-7 Robertsonian translocation chromosomes conferring an increased content of β-glucan. Theor Appl Genet 131, 377–388 (2018). https://doi.org/10.1007/s00122-017-3008-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-3008-z