Abstract
Key message
A new genomic model that incorporates genotype × environment interaction gave increased prediction accuracy of untested hybrid response for traits such as percent starch content, percent dry matter content and silage yield of maize hybrids.
Abstract
The prediction of hybrid performance (HP) is very important in agricultural breeding programs. In plant breeding, multi-environment trials play an important role in the selection of important traits, such as stability across environments, grain yield and pest resistance. Environmental conditions modulate gene expression causing genotype × environment interaction (G × E), such that the estimated genetic correlations of the performance of individual lines across environments summarize the joint action of genes and environmental conditions. This article proposes a genomic statistical model that incorporates G × E for general and specific combining ability for predicting the performance of hybrids in environments. The proposed model can also be applied to any other hybrid species with distinct parental pools. In this study, we evaluated the predictive ability of two HP prediction models using a cross-validation approach applied in extensive maize hybrid data, comprising 2724 hybrids derived from 507 dent lines and 24 flint lines, which were evaluated for three traits in 58 environments over 12 years; analyses were performed for each year. On average, genomic models that include the interaction of general and specific combining ability with environments have greater predictive ability than genomic models without interaction with environments (ranging from 12 to 22%, depending on the trait). We concluded that including G × E in the prediction of untested maize hybrids increases the accuracy of genomic models.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Single-cross hybrids developed through the use of doubled-haploid technology have significantly increased the number of potential hybrids that can be tested in the field. Hybrid performance (HP) prediction is thus of fundamental importance for modern hybrid breeding programs. Van Eeuwijk et al. (2010) pointed out that while some researchers argue that the specific combining ability (SCA) of parental lines is the main factor that determines HP, other authors have concluded that the principal driving force of HP is additive gene action (Duvick et al. 2004; Bernardo 1996a, b). However, when studying HP, it is important to consider two sources of variation: general combining ability (GCA) (additive effects among lines), and SCA (non-additive effects among hybrids); such as dominance or epistatic deviations.
Applying linear mixed models for computing the best linear unbiased prediction (BLUP) of hybrids utilizing only field data was proposed and used by Bernardo (1994). Later, Bernardo (1996a, b, 1999), using the pedigree relationship matrix (obtained from the co-ancestry coefficient), showed promising results when predicting unobserved hybrids based on several observed hybrids. The BLUPs of unobserved hybrids can be computed using an additive relationship matrix derived from a pedigree or an additive genomic relationship matrix derived from markers; it has been shown that predictions based on markers have generally yielded higher accuracy than those based on pedigree (e.g., Rodríguez-Ramilo et al. 2015).
Based on the work originally developed by Bernardo (1994, 1996a, b, 1999) and on genomic prediction studies conducted in the last 15 years (Meuwissen et al. 2001), the BLUP type of prediction using the ridge regression BLUP (RR BLUP) or its equivalent model, the genomic relationship matrix (GBLUP) (de los Campos et al. 2013), has been employed extensively in HP prediction (Xu et al. 2014; Lehermeier et al. 2014; Schrag et al. 2010; Technow and Melchinger 2013; Technow et al. 2014; Piepho 2009; Zhao et al. 2013; Massman et al. 2013).
The genomic-enabled HP prediction models developed by Massman et al. (2013) consider the variance of the GCA of the parental lines, as well as the variance due to SCA of the crosses, and compare the HP prediction accuracy of the genomic RR BLUP with the prediction accuracy of BLUP. The authors found no improvement in the HP prediction accuracy of the genomic RR BLUP when compared with the standard BLUP; both were calculated across a large number of environments.
In plant breeding, multi-environment trials for assessing genotype × environment interactions (G × E) play an important role in selecting phenotypes with high performance and stability across environments. Environmental conditions modulate gene expression, which leads to G × E, such that the estimated genetic correlations of an individual line’s performance across environments summarize the joint action of genes and environmental conditions (López-Cruz et al. 2015). Recent studies have shown that multi-environment linear mixed models can account for the correlation between environments within the GBLUP framework and thus can predict performance of unobserved phenotypes using pedigree and molecular markers. Burgueño et al. (2012) were the first to use marker and pedigree GBLUP models to assess G × E under genomic prediction; they clearly showed that modeling G × E using both pedigree and markers increase prediction accuracy. Heslot et al. (2014) incorporated crop-modeling data for studying genomic G × E, and Jarquín et al. (2014) proposed a random effect GBLUP model where the main effects and interaction effects of markers and environmental covariates were introduced using highly dimensional random variance–covariance structures. Heslot et al. (2014) tested the proposed models using a large winter wheat data set and concluded that prediction accuracy increased, on average, by 11%. Jarquín et al. (2014) indicated that the prediction accuracy for grain yield in wheat using models that incorporate G × E was higher (17–34%) than the accuracy of models that did not include it. So, given the importance of G × E in plants and the empirical evidence of improved prediction accuracy when modeling G × E, the question is how to include G × E in genomic prediction models for HP.
We hypothesize that incorporating G × E into GBLUP models could increase the accuracy of HP prediction. However, none of the previous studies on HP prediction has explicitly incorporated G × E into hybrid prediction models. The main objective of this study was to elaborate on the models of Massman et al. (2013) and Technow et al. (2014) by including G × E within the framework of the reaction norm model proposed by Jarquín et al. (2014). The proposed models consider the interaction of SCA effects × environment and GCA × environment. To illustrate the proposed genomic G × E hybrid prediction model, we used an extensive data set including 2724 maize hybrids derived from 531 maize inbred lines, of which 507 are dent lines and 24 are flint lines (used as testers). The inbred lines were genotyped with Illumina 50K SNPs and evaluated for 12 years (2004–2015) at 58 different locations.
Materials and methods
Experimental data
Phenotypes
This data set was provided by the maize breeding program at RAGT (http://www.ragtsemences.com/). It included 2724 maize hybrids derived from 531 maize inbred lines, 507 of which were end-of-selection dent lines and 24 flint lines were used as testers. The hybrids were evaluated over the course of 12 years (2004–2015) in a total of 58 different locations. The following traits were analyzed in this study: adjusted percent starch content (SC%), percent dry matter content (DMC%) and silage yield (YLD) in kg/ha. The phenotypes were pre-adjusted for the field effects per trial, per year. Two field designs were used in the trials: (1) the standard augmented block design with check lines arranged in a randomized complete block design (Federer and Raghavarao 1975) (three controls were replicated three times each); and (2) a randomized complete block design with two replicates. Pre-adjusted phenotypes were computed using a full fixed effects model including individuals (1 and 2) and blocks (2), residuals were assumed to be independent and identically normally distributed with mean zero and homogeneous variance. Trials with coefficients of variation higher than 7% were discarded. Figure 1 gives a schematic representation of the tested hybrids obtained by crossing the dent and flint lines. Hybrids tested in the field are represented by black squares. Only about 22% of all possible hybrids were tested in the field.
Genotypes
The lines were genotyped using the 50K Illumina chip for maize (http://www.illumina.com), from which 49,013 SNPs were obtained. Standard quality controls were applied to the data, removing all non-bi-allelic markers and non-mapped markers. Beagle v3.2 software (Browning and Browning 2009; https://faculty.washington.edu/browning/beagle/b3.html) was used for imputing missing values in the genotypes. After editing, 22,690 markers were available to make the predictions. A genomic relationship matrix \(\bf {G} \) that includes dent and flint lines was built as follows: let \(\bf X\) be the matrix of markers for dent and flint lines, whose entries were set to 0 and 2 for recessive and dominant homozygous, respectively, and let W be the matrix of centered and standardized markers, that is, \({w_{{\text{ij}}}}=({x_{{\text{ij}}}} - 2{p_{\text{j}}})/\sqrt {4{p_{\text{j}}}(1 - {p_{\text{j}}})},\) where \(i\) indexes individuals and \(j\) the markers, and \({p_{\text{j}}}\) is the allele frequency of the reference allele in the population (see Technow et al. 2014); then \({\bf{G}}={\bf {W}}{{\bf {W}}^\prime}\!/p\) (López-Cruz et al. 2015). Further information about the population structure can be obtained by performing an eigenvalue decomposition of this matrix (Golob and Van Loan 1996), that is, \({\bf{G}}={\varvec{\Gamma}} {\varvec{\Lambda}} {{\varvec {\Gamma}} ^\prime },\) where \(\varvec{\Gamma}\) is the square matrix whose columns correspond to the eigenvectors of \({\bf {G}},\) and \(\varvec \Lambda\) is the square diagonal matrix whose diagonal elements are the corresponding eigenvalues. Figure 2 depicts the first two eigenvectors of the genomic relationship matrix for dent and flint lines, which are clearly distinguished in the graph. The proportion of variance explained by the first two principal components was 20% and just 90 principal components were necessary to explain 80% of the sum of the eigenvalues of the genomic relationship matrix (data not shown).
Statistical models
Technow et al. (2012) compared the prediction accuracy of GBLUP and BayesB models when predicting hybrid performance using simulated data based on marker genotypes of Central European flint and dent inbred lines from a maize breeding program. Massman et al. (2013) proposed a model for predicting hybrid performance using genotypic (genomic) information from the parents. The genotypic information was used to build relationship matrices for both the parents and the hybrids. The proposed model is a linear mixed model that includes random effects due to the parents’ general combining ability, and the hybrids’ specific combining ability, whose variance–covariance matrices are built based on markers. Technow et al. (2014) compared the prediction accuracy achieved by the GBLUP (Bernardo 1996a; Massman et al. 2013) and BayesB prediction methods using data from 1254 single-cross maize hybrids that were generated by the University of Hohenheim breeding program and concluded that the prediction accuracies of both methods were about the same.
In this study, we extended the GBLUP-type models to take into account the effect of the environment and the effect of G × E. We consider two models: (1) GBLUP + Env model (M1); and (2) GBLUP + Env + G × E model (M2). The first model is the one discussed in Massman et al. (2013) and Technow et al. (2014). The second model builds on model 1 by including the effect of G × E interaction. The proposed models are described below. Both models assume a homogeneous error variance across environments.
GBLUP + Env model (M1)
The linear model for hybrid performance that includes the effect of the environments is given by Technow et al. (2014):
where y is the response vector (i.e., the adjusted hybrids’ phenotypic information), \({{\bf{Z}}_{\text{E}}}\) is the design matrix for environments (location within year), \({{\boldsymbol{\beta}}_{\text{E}}}\) is the vector of environmental effects, \({{\boldsymbol \beta} _{\text{E}}} \sim N({\bf {0}}, \,\sigma _{\text{E}}^2{\bf {I}})\), \({{\bf{g}}_{\text{D}}}\) is the vector of random effects due to the GCA of dent lines, \({{\bf {g}}_{\text{F}}}\) is the vector of random effects due to the GCA of markers for flint lines and h is the vector that includes SCA random effects and denotes the interaction effects between flint and dent parental lines for the hybrids. \({{\bf{Z}}_{\text{D}}},~{{\bf {Z}}_{\text{F}}},~{{\bf {Z}}_{\text{H}}}\) are incidence matrices that relate \(\bf y\) to \({{\bf{g}}_{\text{D}}},\,~{{\bf{g}}_{\text{F}}},~\, {\bf{h}},~{\text{with}}~~{{\bf{g}}_D} \sim N({{\bf 0}},\sigma _{\text{D}}^2{{\bf{G}}_{\text{D}}}),\) \({{\bf{g}}_{\text{F}}} \sim N({{\bf 0}},\sigma _{\text{F}}^2 {\bf{G}}_{\text{F}})\), \({\bf{h}} \sim N({\bf{0}},\sigma _{\text{H}}^2 {\bf{H}})\), where \(\sigma _{\text{D}}^2\), \(\sigma _{\text{F}}^2\) and \(\sigma _{\text{H}}^2\) are variance components associated with general and specific combining abilities, and \({{\bf{G}}_{\text{D}}},~\;{{\bf{G}}_{\text{F}}}\) and \(\bf H\) are relationship matrices for dent and flint lines and hybrids, respectively. Finally, \({\boldsymbol{\varepsilon}} \sim N({\bf{0}},\sigma _\varepsilon ^2{\bf{I}})\), where \(\sigma _\varepsilon ^2\) is the variance associated with the residuals.
The relationship matrices \({{\bf{G}}_{\text{D}}}\) and \({{\bf G}_{\text{F}}}\) were computed using the markers (VanRaden 2008). Let \({{\bf {X}}_{\text{m}}}\), \(m \in \left\{ {{\text{Dent}},{\text{ Flint}}} \right\}\) be the matrix of markers whose entries were set to 0 and 2 for recessive and dominant homozygous, respectively. Let \({{\bf W}_{\text{m}}}\) be the matrix of centered and standardized markers, that is, \({w_{{\text{ijm}}}}=({x_{{\text{ijm}}}} - 2{p_{{\text{jm}}}})/\sqrt {4{p_{{\text{jm}}}}(1 - {p_{{\text{jm}}}})},\) where \(i\) indexes individuals and \(j\) the markers, and \({p_{{\text{jm}}}}\) is the allele frequency of the reference allele in the population of flint or dent lines (see Technow et al. 2014). Then \({{\bf G}_{\text{m}}}={{\bf W}_{\text{m}}}{\bf W}_{\text{m}}^\prime /p\) (Technow et al. 2014; López-Cruz et al. 2015), where \(p\) is the number of markers. This gives an average diagonal \({{\bf G}_{\text{m}}}\) value of around one; therefore, \(\sigma _{\text{m}}^2\) is defined on the same scale as \(\sigma _\varepsilon ^2\).
The elements of matrix \({\bf H}\) can be obtained directly from matrices \({{\bf G}_{\text{D}}}\) and \({{\bf{G}}_{\text{F}}}\) (see Bernardo 2002, p. 231–232, and; Technow et al. 2014). The derivation can be performed easily based on the fact that the SCA of hybrids can be represented as a first-order interaction between paternal and maternal lines. Here we include the derivation of the result, because the technique used to establish this is also the technique used to introduce G × E interaction. Let \({h_{{\text{ij}}}}\) represent the interaction effect of a hybrid obtained from a single cross between individual \(i\) in the dent population and individual \(j\) in the flint population. Assuming that \({h_{{\text{ij}}}}={g_{{{\text{D}}_{\text{i}}}}} \times {g_{{{\text{F}}_{\text{j}}}}},\) where \({g_{{{\text{D}}_{\text{i}}}}}~\) is the ith entry of \({{\bf{g}}_{\text D}}\) and \({g_{{{\text{F}}_{\text{j}}}}}\) is the jth entry of \({{\bf {g}}_{\text{F}}},\) then the expectation and the covariance function are as follows. \(E\left[ {{h_{{\text{ij}}}}} \right]=E\left[ {{g_{{{\text{D}}_{\text{i}}}}} \times {g_{{{\text{F}}_{\text{j}}}}}} \right]=E\left[ {{g_{{{\text{D}}_{\text{i}}}}}} \right] \times E\left[ {{g_{{{\text{F}}_{\text{j}}}}}} \right]=0.\) The covariance between two hybrids, one obtained from a single cross between individual \(i\) in the dent population and individual \(j\) in the flint population, and the other obtained by crossing individual \({i^\prime }\) in the dent population and \({j^\prime }\) in the flint population, is given by:
where \({{\bf {G}}_{{{\text{D}}_{{\text{i}}{{\text{i}}^{\prime}}}}}}\) and \({{\bf {G}}_{\text{F}}}_{_{{\text{j}}{{\text{j}}^{\prime}}}}\) are entries from \({{\bf {G}}_{\text{D}}}\) and \({{\bf{G}}_{\text{F}}}\), respectively. In compact notation, matrix \(\bf H\) for all possible crosses is obtained as the Kronecker product of \({{\bf{G}}_{\text{D}}}\) and \({{\bf{G}}_{\text{F}}}\), that is, \({\bf{H}}={{\bf{G}}_{\text{D}}} \otimes {{\bf{G}}_{\text{F}}}\) (e.g., Covarrubias-Pazaran 2016).
GBLUP + Env + GE model (M2)
Jarquín et al. (2014) suggested modeling the interaction between markers and environmental covariates using a Gaussian process with a specific type of covariance function that is induced by a reaction norm model. These authors showed that if the covariance function generated by the interaction terms is obtained using a first-order multiplicative model, then the covariance function is the Hadamard (cell-by-cell) product of two covariance structures, one describing genetic information, and the other environmental effects. Using this approach, we extended model (1) to include GCA and SCA × environment interaction. The model is as follows:
where \({{\bf{u}}_{\text{H}}} \sim N({\bf{0}},\sigma _{{\text{hE}}}^2{{\bf{V}}_{\text{H}}}),\) \({{\bf{u}}_{\text{D}}} \sim N({\bf{0}},\sigma _{{\text{DE}}}^2{{\bf{V}}_{\text{D}}}),\) \({{\bf{u}}_{\text{F}}} \sim N({\bf{0}},\sigma _{{\text{FE}}}^2{{\bf{V}}_{\text{F}}}),\) \(\sigma _{{\text{hE}}}^2,\) \(\sigma _{{\text{DE}}}^2\) \(\sigma _{{\text{FE}}}^2\) are variance components associated with hybrid × environment, dent × environment, and flint × environment interactions, respectively, and \({{\bf{V}}_{\text{H}}},~{{\bf{V}}_{\text{D}}}\) and \(~{{\bf{V}}_{\text{F}}}\) are their associated variance–covariance matrices. The elements of matrix \({{\bf{V}}_{\text{H}}}\) can be obtained following the approach of Jarquín et al. (2014). Assuming that the interaction between hybrids and environments \(E{h_{{\text{ijk}}}}\) can be represented as \({h_{{\text{ij}}}} \times {E_{\text{k}}},\) where \({E_{\text{k}}}={\beta _{{\text{E}_\text{k}}}},\quad {\text{for}}~k=1, \ldots ,E\) (environments), then the expected value and the covariance function are as follows: \(\mathbb{E}\left[ {{h_{{\text{ij}}}} \times {E_k}} \right]=\mathbb{E}\left[ {{h_{{\text{ij}}}}} \right] \times \mathbb{E}\left[ {{E_{\text{k}}}} \right]=0 \times 0=0,\)
Note that \({\text{Cov}}\left[ {{E_{\text{k}}},{E_{{{\text{k}}^{\prime}}}}} \right] \ne 0\) only if \(k={k^{\prime}}\). So using these results, the variance–covariance matrix is given by \({{\bf{V}}_{\text{H}}}={{\bf {Z}}_{\text{H}}}{\bf{H}}{\bf{Z}}_{\text{H}}^\prime \# {{\bf{Z}}_{\text{E}}}{\bf {Z}}_{\text{E}}^\prime ,\) where # stands for the Hadamard product. Note that if observations are sorted by environment, then V is a block diagonal matrix whose structure is similar to the marker × environment model of López Cruz et al. (2015). However, it is not necessary to sort the observations by environment to fit the model. The variance–covariance matrices \({{\bf{V}}_{\text{D}}}\) and \({{\bf{V}}_{\text{F}}}\) can be similarly derived and are as follows: \({{\bf{V}}_{\text{D}}}={{\bf{Z}}_{\text{D}}}{{\bf{G}}_{\text{D}}}{\bf{Z}}_{\text{D}}^\prime \# {{\bf{Z}}_{\text{E}}}{\bf{Z}}_{\text{E}}^\prime,\) \({{\bf {V}}_{\text{F}}}={{\bf{Z}}_{\text{F}}}{{\bf{G}}_{\text{F}}}{\bf{Z}}_{\text{F}}^\prime \# {{\bf{Z}}_{\text{E}}}{\bf{Z}}_{\text{E}}^\prime\) (see Jarquín et al. 2014, for more details).
On the covariance equation display above, note that \(E{h_{{\text{ijk}}}}\) represents the interaction effect for a hybrid obtained from a single cross between individual i in the dent population and individual j in the flint population tested in environment k. Likewise, \(E{h_{{{\text{i}}^{\prime}}{{\text{j}}^{\prime}}{{\text{k}}^{\prime}}}}\) represents the interaction effect for a hybrid obtained from a single cross between individual i in the dent population and individual \(j'\) in the flint population tested in environment \(k'.\)
Model assessment
Models 1 and 2 were first fitted using the full data in each year, and estimates of variance components were obtained from this analysis. To test the prediction ability of the proposed model, we mimicked a common problem breeder’s face when testing new hybrids using incomplete field trials: how to predict the performance of newly developed hybrids. The hybrids were evaluated in some environments but not in others, and their performance had to be predicted in environments where they were not evaluated. To mimic this problem, we performed a cross-validation analysis using a scheme that is known as cross-validation 2 (CV2), which considers some lines being observed in some environments, while missing in others; the problem consists of predicting the lines that were not observed. In CV2, the individual plot records are assigned to folds, so that individual records of a hybrid are potentially assigned to different folds (see Jarquín et al. 2014, Table 2). We performed a fivefold cross-validation for each year.
Note that we have a relatively small number of folds. We selected only fivefolds to be able to make the computations feasible, since with 12 years of data, two models and fivefolds, it is necessary to fit 12 × 2 × 5 = 120 models using the Markov Chain Monte Carlo technique, which is computationally expensive. The fivefold cross-validation has been used in many other studies. Under this validation scheme, 80% of the records are used in the training set and 20% in the validation set; thus the ratio between the training and validation populations is ¼. Models 1 and 2 were fitted using the records in the training set and predictions in the testing set were obtained to estimate prediction accuracy. We computed Pearson’s correlation coefficient for observations in the testing set for each year and we computed an average correlation by weighting the individual correlation in each site according to the number of hybrids predicted. The R code (R Core Team 2016) used to generate partitions for this type of cross-validation is included in the Supplementary materials.
Software
The models described above were fitted using the R package (R Core Team 2016) Bayesian generalized linear regression (BGLR) (de los Campos and Pérez-Rodríguez 2016). The software can be downloaded free of charge from https://cran.r-project.org/web/packages/BGLR/index.html. For more details, see Pérez-Rodríguez and de los Campos (2014). The R codes used to fit the two models are included in the Supplementary materials. Inferences were based on 30,000 iterations of the Gibbs sampler (Geman and Geman 1984), 5000 of which were taken as burn-in. For the hyper-parameters for the prior distributions, we used the default values provided by the BGLR package, which were set according to the rules given in the supplementary materials in Pérez-Rodríguez and de los Campos (2014).
Results
Estimated variance components
Estimated variance components of the parameters of the two fitted models (M1–M2) were described to determine how much of the total variance is explained by each component. The variance components of the two models are: environments (E), general combining ability of dent lines (D), general combining ability of flint lines (F), specific combining ability of hybrids (H), the interaction between hybrids and environment (H × E), interaction of dent lines × environment (D × E), interaction of flint lines × environment (F × E), and residual (Res). These components were obtained from the full data analysis performed when fitting models 1 and 2. The results for YLD are shown in Tables 1 and 2; in the case of SC% and DMC% for models 1–2, the results are included in the Supplementary materials (Tables B.1–B.4). In general, the variance associated with the main effects of environments (locations) accounts for a large proportion of the total variance; therefore, component E is usually not considered with the purpose of distinguishing the differences among the other components, which are small compared to the magnitude of E. When model M2 is fitted, the residual variances decrease consistently, compared with model M1. Depending on the trait, the average decrease in the residual variance is about 10–16%.
For trait YLD, results from all models and years indicated that GCA of the flint component (F) accounted for much more variability than that explained by the GCA of the dent component (D). In general, the SCA explained the least variability of the three genetic components. In terms of the variance explained by the three interaction components when fitting M2, F × E explained the most variability, followed by components D × E and H × E. These patterns are very consistent across years, although there were a few exceptions. The pattern of results of the variance components for traits SC% and DMC% for all years and models is similar to the pattern for trait YLD (see Supplementary materials, Tables B.1–B.4 for traits SC% and DMC% for models 1–2).
Prediction accuracy of M1 vs M2
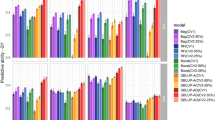
The average correlations between phenotypes and predictive values obtained from random cross-validation CV2 are reported for each model in Tables 3, 4 and 5 for traits YLD, SC% and DMC%, respectively. The results were obtained using Pearson’s correlation coefficient, and ranged from 0.42 to 0.50 for M1 and from 0.48 to 0.60 for M2, depending on the trait. These results clearly indicate that M2 had better prediction ability than M1. Tables 3, 4 and 5 also report the changes in percent prediction accuracy of M2 over M1. For YLD, in regards to percent change for M2 vs M1, the average increase in prediction accuracy was 16.73% (Table 3). In two years (2005 and 2008), the predictive ability superiority of M2 over M1 reached 20% and in 1 year (2008), it even increased to 50%. For trait SC%, the average percent change for M2 vs M1 was 12.30% (Table 4). Finally, for trait DMC%, the average percent increase in prediction accuracy of M2 vs M1 was 21.74% (Table 5). In summary, in most cases, the average percent change in the predictive ability of M2 over M1 is positive, which indicates that M2 has better predictive ability than M1.
These results (higher prediction accuracy of M2 over M1 for all three traits) are also depicted in Fig. 3, which shows the distribution of the percent change for M2 vs M1 for traits SC%, DMC% and YLD. Results indicate the importance of incorporating G × E into modeling to increase hybrid prediction accuracy of the three traits analyzed in this study.
Model comparison. Boxplot of percent change in prediction accuracy calculated by taking model 1 (M1) as a base. Percent change, M1 vs M2 \(=({r_{{\text{M2}}}} - {r_{{\text{M1}}}})/{r_{{\text{M1}}}} \times 100\), where \({r_{{\text{M1}}}}\), \({r_{{\text{M2}}}}\) are the Pearson’s correlations for models 1 and 2 (M1–M2), respectively
Discussion
Several studies have documented the benefits of using genomic multi-environment models for assessing the performance of genotypes across different environmental conditions (Burgueño et al. 2012; Dawson et al. 2013; Jarquín et al. 2014). Analyses of multi-environment trials can include G × E interactions using genomic covariance functions (Burgueño et al. 2012). The main objective of this study was to demonstrate that including the G × E term increases the predictive ability of a genomic-enabled prediction model used to predict hybrid performance based on genotypic information from parents only. In this article, we used the covariance functions proposed by Jarquín et al. (2014), who defined the covariance function based on the Hadamard product (cell-by-cell) of the matrix of genotypes with the design matrix associated with the environments. Using this approach, the authors showed that a large percentage of the phenotypic variance is explained by the main effect of environments, which agrees with the results of the analysis where we obtained variance components for environments (Tables 1, 2 and Tables B.1–B.4 in Supplementary materials).
The importance of the parents’ general combining ability, the specific combining ability of hybrids and the interaction between parental lines and the environment varies from trait to trait, but in general, these terms explain a sizable proportion of the total variance, and when included in the complete model, the prediction accuracy of the unobserved hybrids did increase. Results of our study in maize show the importance of genomic prediction accuracy of HP based on both general and specific combining ability. Results of this study are in agreement with other researchers that consider that HP is determined not only by additive effects due to male and female GCA but also by the intra gene dominance interaction producing the SCA effects (van Eeuwijk et al. 2010). Also, the effect of G × E is considered an important non-genetic factor affecting heterosis. This is the first study showing how the intra gene interaction due to dominance and its interaction with environments can be modeled to exploit these positive interaction genetic effects with environments (SCA × E). The model proposed in this study is essentially similar to the model of Massman et al. (2013) and Technow et al. (2014) except that it includes modeling the interaction term (SCA × E or/and GCA × E). These interaction effects positively affect prediction accuracy. The residual error decreased consistently when fitting model (2), which indicates that the G × E interaction term accounts for a considerable portion of the variance between environments. This is consistent with the findings of Jarquín et al. (2014), who reported that including the interaction term significantly reduced the error variance and increased the predictive ability of the model.
The genetic variance was small when compared to the variance of environments, although predictions based on markers worked very well. Bernardo (1994) notes that accurate estimates of genetic variances are not necessary for effectively predicting the performance of single crosses and that approximations of their values are sufficient. In this study, we assumed homogeneous error variance across environments, although this is not ideal, the comparison between models is fair because in both of them the same assumption holds. We evaluated the accuracy of prediction using the cross-validation scheme, which quantifies the prediction accuracy of yield under conditions in the particular year-environment combinations included in the data set (Pérez-Rodríguez et al. 2015). Crossa et al. (2011) mentioned that a simple approach for evaluating predictive ability consists of dividing the data into a training sample and a validation sample, or testing set. Models are fitted using the training sample, and the fitted models are then used to predict outcomes in the validation sample. This approach is appropriate for large data sets as is the case in our study (e.g., Hastie et al. 2009). The random cross-validation just mimics the reality the researcher might face when predicting HP without knowing the actual observed values.
The gains in prediction accuracy obtained when G × E was included in the model are also consistent with the results presented in other studies (Burgueño et al. 2012; Jarquín et al. 2014; López-Cruz et al. 2015). The results confirmed the superior predictive ability of model 2. However, as mentioned in López-Cruz et al. (2015), interaction models are subject to the structure of the covariance matrix, i.e., the covariance between environments must be positive and constant. Thus, the interaction model is better suited to environments that are positively correlated.
In a study on the genomic prediction of HP for identifying superior single crosses early in a maize hybrid breeding program, Kadam et al. (2016) used an initial model that included the parents’ general combining ability and specific combining ability and their interactions with environments. Although the prediction of the single-cross performance was done using parental combining ability and covariance among single crosses for grain yield for different testing/training single-cross schemes, the authors did not model the parents’ general combining ability × environment and/or the specific combining ability × environment interaction terms, and therefore did not quantify their impact on the prediction accuracy of the HP. Results of our study clearly indicates the benefit of including and modeling the various interactions terms using appropriate variance–covariance structures given by the Hadamard product of the proposed model. The model used in this study allows borrowing information from correlated environments such that hybrids (or parents) observed in some environments can be predicted in others where they were not observed.
In general, incorporating G × E models in the genomic prediction of HP, as presented in this study, can be applied to any crop and adapted to most of the GBLUP models that have been used in recent genomic studies for assessing the genomic prediction of HP in different environments. Further research using non-linear kernel methods should be conducted to assess the possible increase in HP prediction accuracy due to a kernel method that is different from the linear kernel used in the GBLUP.
Author contribution statement
RAP, JC, GC and PPR conceived the idea and designed the study; RAP, JC and PPR developed the statistical models; RAP wrote the R scripts; RAP and PP carried out the statistical analysis; ST and BC were involved in the design of the field trials, data collection and edition; RAP, JC and PPR wrote the first version of the manuscript; JC, GC, ST, BC, SPE and PPR contributed critical comments and to the writing of the final manuscript.
References
Bernardo R (1994) Prediction of maize single-cross performance using RFLPs and information from related hybrids. Crop Sci 34:20–25
Bernardo R (1996a) Best linear unbiased prediction of maize single-cross performance. Crop Sci 36:50–56. doi:10.2135/cropsci1996.00111183X003600010009x
Bernardo R (1996b) Best linear unbiased prediction of the performance of crosses in maize. Crop Sci 41:68–71. doi:10.2135/cropsci2001.41168x
Bernardo R (1999) Marker-assisted best linear unbiased prediction of single-cross performance. Crop Sci 39:1277–1282
Bernardo R (2002) Breeding for quantitative traits in plants. Stemma Press, Woodbury
Browning BL, Browning SR (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84(2):210–223. doi:10.1016/j.ajhg.2009.01.005
Burgueño J, de los Campos G, Weigel K, Crossa J (2012) Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Sci 52:707–719
Covarrubias-Pazaran G (2016) Genome-assisted prediction of quantitative traits using the R Package Sommer. Plos One 11(6):e0156744. doi:10.1371/journal.pone.0156744
Crossa J, Pérez-Rodríguez P, de los Campos G, Mahuku G, Dreisigacker D, Magorokosho C (2011) Genomic selection and prediction in plant breeding. J Crop Improv 25:239–261
Dawson JC, Endelman JB, Heslot N, Crossa J, Poland J et al (2013) The use of unbalanced historical data for genomic selection in an international wheat breeding program. Field Crops Res 154:12–22
de los Campos G, Pérez-Rodríguez P (2016) BGLR: Bayesian generalized linear regression. R package v. 1.0.5
de los Campos G, Hickey JM, Pong-Wong R, Daetwyler HD, Calus MPL (2013) Whole genome regression and prediction methods applied to plant and animal breeding. Genetics 193:327–345
Duvick DN, Smith JSC, Cooper M (2004) Long-term selection in a commercial hybrid maize breeding program. Plant Breed Rev Part 2(24):109–152
Federer WT, Raghavarao D (1975) On augmented designs. Biometrics 31:29–35
Geman S, Geman D (1984) Stochastic relaxation, Gibbs distributions and the Bayesian restoration of images. IEEE Trans Pattern Anal Mach Intell 6(6):721–741
Golob GH, Van Loan CF (1996) Matrix computations, 3rd edn. Johns Hopkins University Press, Baltimore
Hastie T, Tibshirani R, Friedman J (2009) The elements of statistical learning: data mining, inference and prediction, 2nd edn. Springer, New York
Heslot N, Akdemir D, Sorrells ME, Jannink JL (2014) Integrating environmental covariates and crop modeling into the genomic selection framework to predict genotype by environment interactions. Theor App Genet 127(2):463–480
Jarquín D, Crossa J, Lacaze X, Cheyron PD, Daucourt J et al (2014) A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor Appl Genet 127:595–607
Kadam DC, Potts SM, Bohn MO, Lipka A, Lorenz AJ (2016) Genomic prediction of single crosses in the early stages of a maize hybrid breeding pipeline. G3 Genes Genom Genet 6:3443–3453. doi:10.1534/g3.116.031286
Lehermeier C, Krämer N, Bauer E, Bauland C, Camisan C et al (2014) Usefulness of multiparental populations of maize (Zea mays L.) for genome-based prediction. Genetics 198:3–16
López-Cruz M, Crossa J, Bonnet D, Dreisigacker S, Poland J, Jannink L-L, Singh RP, Autrique E, de los Campos G (2015) Increased prediction accuracy in wheat breeding trials using a marker × environment interaction genomic selection model. G3 Genes Genom Genet. doi:10.1534/g3.114.016097
Massman J, Gordillo A, Lorenzana RE, Bernardo R (2013) Genome-wide predictions from maize single-cross data. Theor Appl Genet 126:13–22. doi:10.1007/s00122-012-1955
Meuwissen, THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Pérez-Rodríguez P, de los Campos G (2014) Genome-wide regression & prediction with the BGLR statistical package. Genetics 198:483–495
Pérez-Rodríguez P, Crossa J, Bondalapati K, De Meyer G, Pita F, de los Campos G (2015) A pedigree-based reaction norm model for prediction of cotton yield in multi-environment trials. Crop Sci 55(3):1143–1151
Piepho HP (2009) Ridge regression and extensions for genome-wide selection in maize. Crop Sci 49(4):1165–1176
R Core Team (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org. Accessed 4 Apr 2017
Rodríguez-Ramilo ST, García-Cortés LA, Rodríguez de Cara MA (2015) Artificial selection with traditional of genomic relationships: consequences in coancestry and genetic diversity. Front Genet 6:127. doi:10.3389/fgene.2015.00127
Schrag TA, Möhring J, Melchinger AE, Kusterer B, Dhillon BS, Piepho HP, Frisch M (2010) Prediction of hybrid performance in maize using molecular markers and joint analyses of hybrids and parental inbreds. Theor Appl Genet 120(2):451–461
Technow F, Melchinger AE (2013) Genomic prediction of dichotomous traits with Bayesian logistic models. Theor Appl Genet 125(6):1133–1143
Technow F, Riedelsheimer E, Schrag TA, Melchinger AE (2012) Genomic prediction of hybrid performance in maize with models incorporating dominance and population specific marker effects. Theor Appl Genet 125(6):1181–1194
Technow F, Schrag A, Schipprack W, Bauer E, Simianer H, Melchinger AE (2014) Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics 197:1343–1355
van Eeuwijk FA, Boer MP et al (2010) Mixed model approaches for the identification of QTLs within a maize hybrid breeding program. Theor Appl Genet 120:429–440
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Xu S, Zhu D, Zhang Q (2014) Predicting hybrid performance in rice using genomic best linear unbiased prediction. Proc Natl Acad Sci 11(34):12456–12461. doi:10.1073/pnas.1413750111
Zhao Y, Zeng J, Fernando R, Reif J (2013) Genomic prediction of hybrid wheat performance. Crop Sci 53:1–9. doi:10.2135/cropsci2012.08.0463
Acknowledgements
The authors appreciate the positive and detailed comments from two anonymous reviewers and the time invested by the Associated Editor handling the manuscript. These contributions significantly improved the quality and clarity of the article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Matthias Frisch.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Acosta-Pech, R., Crossa, J., de los Campos, G. et al. Genomic models with genotype × environment interaction for predicting hybrid performance: an application in maize hybrids. Theor Appl Genet 130, 1431–1440 (2017). https://doi.org/10.1007/s00122-017-2898-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2898-0