Abstract
Objective
The molecular heterogeneity of prostate cancer (PCa) gives rise to distinct tumor subclasses based on epigenetic modification and gene expression signatures. Identification of clinically actionable molecular subtypes of PCa is key to improving patient outcome, and the balance between specific pathways may influence PCa outcome. It is also urgent to identify progression-related markers through cytosine-guanine (CpG) methylation in predicting metastasis for patients with PCa.
Methods
We performed bioinformatics analysis of transcriptomic, and clinical data in an integrated cohort of 551 prostate samples. The datasets included retrospective The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) cohorts. Two algorithms, Least Absolute Shrinkage and Selector Operation and Support Vector Machine-Recursive Feature Elimination, were used to select significant CpGs.
Results
We found that PCa progression is more likely to occur after the third year through conditional survival (CS) analysis, and prostate-specific antigen (PSA) was a better predictor of Progression-free survival (PFS) than Gleason score (GS). Our study first demonstrated that PCa tumors have distinct expression profiles based on the expression of genes involved in androgen receptor (AR) and PI3K-AKT, which influence disease outcome. Our results also indicated that there are multiple phenotypes relevant to the AR-PI3K axis in PCa, where tumors with mixed phenotype may be more aggressive or have worse outcome than quiescent phenotype. In terms of epigenetics, we obtained CpG sites and their corresponding genes which have a good predictive value of PFS. However, various evidences showed that the predictive value of CpGs corresponding genes was much lower than GpG sites in Overall survival (OS) and PFS.
Conclusions
PCa classification specific to AR and PI3K pathways provides novel biological insight into previously established PCa subtypes and may help develop personalized therapies. Our results support the potential clinical utility of DNA methylation signatures to distinguish tumor metastasis and to predict prognosis and outcomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prostate cancer (PCa) is the first common cancer of the genitourinary tract in the old man and the incidence rate is rising still now [1,2,3]. The diagnosis and treatment of prostate cancer have been continuously developed and improved in recent years. For example, prostate-specific antigen (PSA) is used for early screening of disease [4], and the decision on how to treat a patient depends on tumor TNM stage and Gleason score (GS). However, the majority of cancer-associated deaths including PCa are due to metastases rather than primary tumor burden [5]. Thus, decreasing mortality of PCa depends on understanding the biology that underlies metastasis such as the identification of biomarkers involved in cancer metastasis that would benefit the design of more effective clinical intervention strategies.
The androgen receptor (AR) is expressed in primary and metastatic PCa which regulates multiple cellular events, such as cell proliferation, migration, and invasion [6,7,8]. Tumors which could not be cured by surgery or radiotherapy are treated with therapies based on downregulation of androgen levels in the circulation or blockade of the androgen receptor (AR). Thus, AR downregulation is considered a chemopreventive strategy for PCa [9, 10]. The AR axis is not only the target of choice in androgen-sensitive PCa but also remains a valuable target in castration-resistant prostate cancer (CRPC). A number of novel experimental and clinical therapies take into account the resistance mechanisms leading to the reactivation of the AR pathway. Progression of PCa on these novel therapies targeting the AR pathway is often reflected by rising PSA levels, suggesting the persistence of the AR pathway as the biological driver of the disease [11]. Among other signalling pathways potentially contributing to progression, the role of the PI3K-AKT pathway may be particularly important in PCa due to the existence of signalling feedback between this pathway and the AR pathway [12,13,14]. Therefore, the strategy of clinical studies is either testing combinatory blockade of both pathways from the onset of treatment or evaluating the addition of a PI3K/AKT inhibitor when progressing to AR pathway modulation to counteract the eventual reciprocal feedback loops. Here, we stratify PCa into subgroups based on the genes expression patterns of PI3K and AR pathways and report their association with metastasis, and prognostic gene expression signatures.
DNA methylation abnormalities have been reported as a critical event for cancer initiation and development. The methylation pattern of CpG sites is an epigenetic regulator of gene expression. Several studies have compared DNA methylation patterns between primary PCa and metastatic prostate cancer [15,16,17]. The alterations of DNA methylation cause changes in gene expression that promote oncogenesis. Cancer cells are frequently less methylated at CpGs than normal cells, although hypermethylation at tumor CpG islands has also been reported [18,19,20]. Herein, we perform an integrative analysis aiming to uncover novel CpG sites and their matched genes that may contribute to predict PCa metastatic outcome.
Methods
Data selection and processing
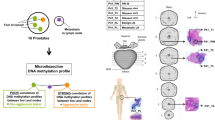
The RNA-Seq gene expression profiles of patients with PCa were downloaded from the cancer genome atlas (TCGA) portal (https://cancergenome.nih.gov/). It contains 551 prostate samples (499 prostate tumor samples and 52 normal samples). Clinical data on tumors were also downloaded from the TCGA database. The RNA expression data including 5 localized PCa and 4 metastatic PCa were downloaded from Gene Expression Omnibus (GEO, GSE27619) portal (https://www.ncbi.nlm.nih.gov/geo/). The RNA expression data of cells with PCa (including 2 PC-3 cells and 2 PC-3 + TGF-β cells) were downloaded from GSE159116. The DNA methylation level of CpG sites (including 8 aggressive prostate cancer tissues and 6 metastatic prostate cancer tissues) was downloaded from GSE157272. R software (4.0.5) was used for data extraction and sorting to obtain the gene expression matrices and clinical data.
Differential expression analysis and functional enrichment analysis
To identify differentially expressed genes (DEGs) or CpG sites between 2 groups, we used the R language “limma” package to screen them from GEO and TCGA transcriptome data. Mann–Whitney test was performed to determine differential expression levels of genes. Adjusted P value < 0.05 were considered to be statistically significant. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were conducted with R language “clusterProfiler”, “GOplot”, “AnnotationHub” and “enrichplot” package, false discovery rate (FDR) < 0.05 was set as the threshold. Bar chart was used to visualize the biological process (BP), cellular component (CC), and molecular function (MF) of GO enrichment. The bar chart was also used to visualize the pathways of KEGG. Functional similarity, which is defined as the geometric mean of their semantic similarities in the MF and CC aspect of GO, is designed for measuring the strength of the relationship between each protein and its partners by considering the function and location of proteins. we measure the functional similarity among proteins through the GOSemSim package. Gene Set Cancer Analysis (GSCA, http://bioinfo.life.hust.edu.cn/GSCA/#/) is an integrated database for genomic and immunogenomic gene set cancer analysis [21]. Pathway Activity module in this database presents the difference in genes expression between pathway activity groups (activation and inhibition) that defined by pathway scores. Methylation module in this database explores the differential methylation between tumor and paired normal, the correlation between methylation with expression and the survival affected by methylation level for selected cancer types. The linear correlation between the expression of 14 genes and the small molecules from Genomics of Drug Sensitivity in Cancer (GDSC) was analyzed using the Pearson correlation coefficient.
Consensus clustering for genes in PI3K and AR pathway
Gene sets PI3K (n = 105) and AR (n = 185) were belonged to the PI3K-AKT pathway and AR pathway (http://netpath.org/index.html), respectively. The unsupervised clustering “Pam” method based on Euclidean and Ward’s linkage was used in this analysis, executed by using the “ConsensuClusterPlus” R package, and repeated 1000 times to ensure the classification stability. Patients were grouped into gene clusters based on gene expression to identify genes associated with the pathways. DEGs among gene clusters were determined by setting significance cutoff criteria to P < 0.05, which was implemented by employing the limma R package. Median expression levels of coexpressed PI3K and AR genes were used to assign quiescent (PI3K ≤ 0, AR ≤ 0), PI3K (PI3K > 0, AR ≤ 0), AR (PI3K ≤ 0, AR > 0), and mixed (PI3K > 0, AR > 0) subgroups to each sample.
Two algorithms to select significant CpGs
We choose differentially methylated CpG sites for filtration, a Least Absolute Shrinkage and Selector Operation (LASSO) algorithm, with penalty parameter tuning conducted by ten-fold cross-validation, was built to select candidate CpGs. Simultaneously, another algorithm, Support Vector Machine-Recursive Feature Elimination (SVM-RFE), was also used for CpGs selection. Finally, we combined CpGs from either the LASSO or the SVM-RFE algorithms, and then performed L1 penalized Cox analysis to further narrow CpGs in the training cohort. SurvivalMeth (http://bio-bigdata.hrbmu.edu.cn/survivalmeth/) were used to investigate the effect of DNA methylation-related functional elements on prognosis.
Clinical-pathologic factors
Tumors were assessed by histological grading using the Gleason scoring system (6–10 score). PCa was further classified into a low aggressive tumor (Gleason score < 7), intermediate aggressive tumor (Gleason score = 7, 3 + 4 vs 4 + 3), and high aggressive tumor (Gleason score > 7). PSA level over 4.0 ng/ml as a threshold for screening for prostate cancer. PSA: G1: 0–4.0 ng/ml; G2: 4.1–10.0 ng/ml; G3:10.1–20.0 ng/m; G4: > 20.0 ng/ml. Progression-free survival (PFS) was defined as the first occurrence of a new tumor event. Overall survival (OS) was calculated from the date of diagnosis to the date of death or last follow-up.
Statistical analysis
The Kaplan–Meier plotter was employed to generate survival curves for the subgroups in each dataset. The log-rank test evaluated the statistically significant differences. Conditional survival (CS) was defined as the probability of surviving an additional number of y years, given that a patient had already survived for x years, and was calculated as CS(x|y) = S(x + y)/S(x), with S(x) representing the survival at x years estimated using the Kaplan–Meier method. Each column represents the years survived from therapy and each row represents the percentage to reach a certain total survival time from that point of survived years. The Kruskal–Wallis test was used to compare more than two groups, and the Wilcoxon test was used to compare two groups. ROC was used to find the best cutoff of continuous variables, and two groups were divided according to the cutoff for univariate logistic regression. Risk factor analysis was performed using univariate and multivariate logistic regression analyses. Variables showing statistical significance of the univariate analysis were included in the multivariate logistic regression analysis, and the forward stepwise method was used to select the variables that were eventually included in the model, all P < 0.05 was considered significant. Above all, ROC curve combined with log-rank test and logistic regression analyses confirmed the significant predictive value of genes for metastasis.
Results
Clinical-pathologic factors predict outcome
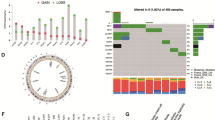
In our research, the 5-year OS of PCa patients is 98%, and the 5-year PFS is 73%. Kaplan–Meier estimates for conditional survival up to 6 years in 499 PCa patients given 0–5 years of survival were shown that there was no significant difference in OS (Fig. 1A). The survival probability increased per year already survived relative to the total survival time. The probability of achieving 5-year PFS after resection increased from 73 to 78%, 83%, 89%, and 96% per additional year survived (i.e. 1, 2, 3, and 4 years, respectively). The 1-year CS (i.e. probability of surviving the next year) decreased from 94 to 92% at 3 years after therapy, and then increased to 96% at 5 years (Fig. 1B). The chi-squared test showed that the progression group exhibited higher TN staging, PSA and GS than the group without disease progression, suggesting that highly malignant PCa is associated with disease progression (Fig. 1C). Then, we explored the relationship between PSA GS and TN stage, respectively. Among the 383 N0 samples, GS is highly correlated with PSA (r = 0.19, P < 0.001), and this correlation is also in 86 N1 samples (r = 0.20, P = 0.079), (Fig. 1D). Both GS (left) and PSA (right) in T3 and T4 groups with significantly higher scores than T1 and T2 groups, as shown in (Fig. 1E). Both GS and PSA models were shown a good AUC, and the PSA model was better than the GS model in predicting PFS (Fig. 1F). Our data indicate that clinical-pathologic factors have a good prediction effect on PFS of PCa.
Clinical-pathologic factors predict the outcome of PCa. A, B Kaplan–Meier estimates for conditional survival (CS) up to 6 years in 499 PCa patients given 0–5 years’ OS (A) and PFS (B). C Pie charts showing the Chi-squared test of clinic-pathological factors in PCa. D PSA predicts Gleason score (GS) with a decent correlation coefficient in both N0 and N1 stage. E The difference of GS (left) and PSA (right) among T1–T4 stages. F Time-dependent AUC of GS and PSA models
Identification of DEGs in metastatic PCa and enrichment analysis
The genes that met the cutoff criteria of an adjusted P-value < 0.05 were considered DEGs. Gene expression profiles of GSE27619 identified 4593 DEGs in PCa samples when compared with metastatic samples, GSE159116 identified 1264 DEGs in PC-3 cells when compared with PC-3 + TGF-β cells (Fig. 2A). The results of venn analysis showed that 257 genes were the common DEGs both in tissues and cells (Fig. 2B). We found DEGs of GSE27619 mainly enriched in these 4 tumor-associated pathways: EGFR tyrosine kinase inhibitor resistance, PI3K-Akt signaling pathway, TGF-beta signaling pathway, and TNF signaling pathway. However, DEGs of GSE159116 are mainly enriched in the pathways of HIF-1 signaling pathway, TNF signaling pathway, PI3K-Akt signaling pathway, Focal adhesion (Fig. 2C). To explore the biological functions of 257 DEGs, they were categorized into BP, CC, and MF. Under stringent threshold conditions (P-adjust < 0.05), we identified 136 specific BP, 2 CC of GO terms were enriched in these genes (Fig. 2D). Genes were enriched in the top 5 BP terms related to cell proliferation (Fig. 2E). On the basis of GO enrichment analysis, we found that these genes included in the DEGs were highly associated with two CC terms, such as caveola, plasma membrane raft (Fig. 2F). Thus, our findings suggest a specific association between DEGs and tumor metastasis.
Identification of DEGs and enrichment analysis. A Volcano plot of DEGs in PCa from GEO database. Red plots represent aberrantly expressed mRNAs with P < 0.05 and absolute log FC > 0.5. Black plots represent normally expressed mRNA. Green plots represent aberrantly expressed mRNAs with P < 0.05 and log FC < − 0.5. B Venn diagram summarize the common DEGs both in PCa cells and tissues. C GSEA of representative KEGG terms in tissues (left) and cells (right). D Bar-plot of GO enrichment in cellular component terms and biological process terms. E Hierarchical clustering of the gene expression profiles in each BP terms (left), chord plot displays the relationship between genes and BP terms (right). Fold enrichment of each GO term is indicated by the x-axis and bar color. F Circle plot displays the relationship between genes and CC terms
Gene characteristics and pathway activity of PI3K and AR pathways
The AR regulates multiple cellular events in PCa metastasis, such as cell proliferation, migration, and invasion. There is signal feedback between PI3K-Akt and the AR pathway, and PI3K-Akt signaling pathway is also considered as an important pathway related to metastasis in PCa cells and tissues. Therefore, we stratify PCa samples into subgroups based on the genes expression patterns of PI3K-Akt and AR pathways. We performed the unsupervised clustering of 276 pathway genes, which classified the TCGA cohort into three genomic clusters, namely, gene clusters A-C (Figure S1A). The 105 gene signatures that were in PI3K-AKT pathway were termed as the pathway gene signature A, and the 185 gene signatures that were in the AR pathway were termed as the pathway gene signature B. Concurrently, to reduce the noise or redundant genes, we used the Boruta algorithm to perform dimension reduction in the pathway gene signatures A and B. The heatmap delineated the transcriptomic profile of the 276 genes identified across the genomic clusters (Fig. 3A). Furthermore, we explored the prognostic implications of the gene clusters by integrating them with survival information, and we found that were no significant differences in OS (Figure S1C) and PFS (Figure S1D) among the three clusters. Then, we calculate the correlation between genes and clusters. Positive correlation is defined as direct A and negative correlation as direct B. The Kruskal–Wallis test was used to detect the significant differences among three clusters of top 10 related genes in direct A and direct B, respectively. The expression of all genes was significantly different among the three clusters (Fig. 3B, C). The results of venn analysis showed that 14 genes were co-existing in AR and PI3K pathways (Fig. 3D). Therefore, we considered that these 14 genes may play a key role in these two pathways.
Identification of gene subtypes by genes in PI3K and AR pathways. A The expression levels of the genes in different clusters were presented in the heatmap with clinic-pathological variables as the annotations. B The statistical differentially expression in three clusters of top 10 genes in pathway gene A. C. The statistical differentially expression in three clusters of top 10 genes in pathway gene B. *P < 0.05; **P < 0.01; ***P < 0.001. D Venn diagram summarize the common genes both in PI3K-AKT and AR pathways. E Graphical summary of the 14 venn genes and their pathways, direct groups distribution. F The statistical differentially expression in three clusters of 14 venn genes
As we all know that these 14 genes were co-expressed in AR and PI3K-AKT pathways, and most genes were negatively correlated with clusters (Fig. 3E). Then, 14 genes of Venn diagram both in PI3K and AR pathway were obvious differentially expressed among 3 clusters. (Fig. 3F). Kaplan–Meier progression-free survival analysis of best separation was performed based on TCGA survival data, and only log-rank P value < 0.05 was shown in Figure S1B. Genes expression of SRC, AKT2, PIK3R1, FOXO1, and MDM2 were significantly related to the PFS of PCa patients. Statistically significant variables screened from the univariate analysis were included in the non-conditional binary multivariate logistic regression. AKT2 was the only independent risk gene of PFS in PCa (Table 1). We divided the PCa patients into the progression group and the non-progression group, and to find the best cutoff of classification variables used ROC curve, so that it has the minimum P value when predicting the tumor progression, all AUC > 0.5 of 14 genes, (Figure S2).
To better illustrate the characteristics of 14 co-expressed genes, we also tested the correlation between them. GSK3B and CREB1 had the highest positive correlation (r = 0.78), while CREB1 and PIK3R2 had the highest negative correlation in Fig. 4A (r = − 0.52). The distributions of functional similarities were summarized as boxplots, the lines in the boxes indicate the mean of the functional similarities. MDM2, GSK3B, AKT1 were the three top-ranked proteins potentially playing central roles in the 14 genes. MDM2 was the only protein with a cutoff value > 0.65 which is widely used to separate significant and non-significant correlation (Fig. 4B). Next, we analyzed RNA sequencing data for the 499 PCa patient samples, and found that 14 genes directly associated with the AR and PI3K-AKT pathways were recurrently dysregulated (ie, either elevated or suppressed compared with normal, AKT2 (61%), AKT1 (31%), GSK3B (25%). Cluster 3 demonstrate up-regulation of 14 pathway genes (activated) to varying extents, whereas clusters 1 and 4 shows down-regulation of the pathway genes (inactivated) and cluster 2 exhibits the least disruption of the AR and PI3K-AKT pathways (Fig. 4C). Figure 4D displayed global activity of 14 genes in PCa. The results showed that CREB1, PIK3R2, and GSK3B genes were not in the TSC/mTOR pathway. The pie of gene in cor-response pathways means the activity/inhibition/non-significant effect contributes to the pathways in PCa. Pathway analysis validated that these genes were mainly associated with the activation of the RTK pathway, and disruption of cell cycle (Fig. 4E). The link between gene and pathway shows that most genes activate EMT (epithelial-mesenchymal transition) pathway and inhibit the Hormone AR pathway (Fig. 4F). Because the nature of genetic alternations influences clinical treatment intervention. Drug-sensitivity analysis in our study demonstrated that the expression of GSK3B, PIK3R2, EGFR, AKT1, RAC1, and SRC were negatively correlated with most drug resistance, and others were positively correlated with most drug resistance (Figure S3A). Then, we found that genes expression of SRC, FOXO1, PIK3R1 and genes methylation of FOXO1, MDM2, PIK3R1, PTEN, CREB1, EGFR were significantly related to the PFS of PCa (Figure S3B-C). The methylation level of 14 genes was negatively correlated with the expression level except SRC (Figure S3D). Methylation analysis revealed that AKT2 was not methylated in PCa tissues (Figure S3E). The above results in our research suggest that these 14 Venn genes play an important role in different processes of prostate cancer.
The characteristics and pathway activity in cancers of 14 co-expressed genes. A Correlations among 14 gene expression levels in human PCa tissues. B Summary of functional similarities of the 14 genes. The dashed line represents the cutoff value. C Unsupervised hierarchical clustering of the data revealed 4 distinct clusters (1–4). D The global activity of 14 genes in cancer-related pathways. E The percentage of cancers in which a gene has an effect (activation or inhibition) on the pathway. F The profiles in cancer-related pathways across cancer types
Dual analysis of PI3K and AR pathways identifies four distinct subgroups of PCa
To stratify PCa tumors based on their relative expression levels of AR and PI3K-AKT pathway genes, we utilized RNA-seq data from PCa patient tumors of TCGA. Genes belonging to gene sets AR (n = 185) and PI3K-AKT (n = 105) were selected for analysis. To aid in selecting genes co-regulated within each pathway and relevant to PCa biology, we used consensus clustering (Ward. D2 and Euclidean distances were used as the clustering algorithm and distance metric, respectively) to identify robustly co-expressed AR and PI3K pathway genes to be used for subtyping, and three clusters were the optimal choice (Fig. 5A). Median expression levels of co-expressed AR and PI3K genes were calculated for each sample and used in assigning one of four profiles specifically relevant to these two pathways: quiescent, AR, PI3K and mixed (Fig. 5B). Expression levels of AR and PI3K genes across the subgroups are visualized in Fig. 5C. Furthermore, we found that was no significant differences in OS (Fig. 5D) among the four subgroups, and mixed cases had a significantly worse outcome than cases in the other three groups (log-rank test P = 0.009, Fig. 5E). Our results indicate that there are multiple phenotypes relevant to the AR-PI3K axis in PCa, where tumors with mixed phenotypes may be a more aggressive or have worse outcome than quiescent phenotype.
Stratification of PRAD tumors based on the expression of AR and PI3K genes. A Heatmap depicting consensus clustering solution (k = 3) for AR and PI3K genes in PRAD samples. B Scatter plot showing median expression levels of coexpressed AR (x-axis) and PI3K (y-axis) genes in each PRAD sample. Subgroups were assigned on the basis of the relative expression levels of AR and PI3K genes. C Heatmap depicting expression levels of coexpressed AR and PI3K genes across each subgroup. D Kaplan–Meier overall survival analysis of patients stratified by subgroup. E Kaplan–Meier progression-free survival analysis of patients stratified by subgroup
Selection of candidate CpGs and building a metastatic predictive signature
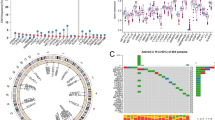
In our study, we compared the methylation values between 8 aggressive prostate cancer samples and 6 metastatic prostate cancer samples from GSE157272 datasets. Using the primary filter criteria (adjusted P-value < 0.05 and |logFC|> 1), we derived a list of 2824 differential CpGs, and the differential CpG sites between these 2 groups were shown in Figure S4A. Next, we used two different algorithms to select the most significant CpGs for classifying with and without metastatic patients. First, we used the LASSO algorithm to identify a set of 9 CpGs (Fig. 6A). Second, we performed the SVM-RFE algorithm and selected a set of 19 CpGs (Fig. 6B). Heatmap analysis showed the differential CpGs level between aggressive PCa samples and metastatic PCa samples (Fig. 6C). Gene set analysis demonstrated that the 23 global CpGs methylation profiles were highly correlated with PCa metastasis (Fig. 6D). After combining the CpGs selected by the LASSO and SVMRFE algorithms, 23 CpGs were identified, with 5 CpGs selected simultaneously by the two algorithms (Fig. 6E). These data verify that methylation values of CpGs might have an important value in the prediction of prostate cancer metastasis.
Two algorithms were used for metastatic feature selection. A LASSO and B SVM-RFE algorithms in the discovery cohort. C Heatmap analysis of the 23 differential CpGs level between 2 groups D Gene set analysis barcode plot, with differential CpGs shown as a shaded rectangle with CpGs horizontally ranked by moderated t-statistic. E Venn diagram summarize 5 CpGs were selected simultaneously by the two algorithms
The value of CpGs and their corresponding genes in predicting PCa metastasis
To better illustrate the characteristics of 5 CpGs selected simultaneously by the above two algorithms from GSE157272 datasets, we also tested the correlation between them. There was a high correlation of methylation levels among the five CpGs, either positive or negative relationship (all |r|> 0.75, Fig. 7A). Subsequent ROC Curve analysis used by GSE157272 datasets showed that the above 5 CpGs could perfectly distinguish patients with or without PCa metastasis (Fig. 7B). Then, we retested the AUC value with TCGA data to avoid overfitting, and the results of AUC were shown in Figure S5 (all AUC > 0.63). The results also showed that the above 5 CpGs could perfectly distinguish patients with or without PCa metastasis. We analyzed the clinicopathological characteristics of these CpGs, and found the methylation levels of 4 CpGs were significantly correlated with OS (Fig. 7C). We compared the DNA methylation levels of 5 CpGs in different T, N and M stages and found that the CpGs methylation levels of N1 and M1 were lower than that of N0 and M0, but there was little change of DNA methylation levels in different T stages (Figure S6A-C). To investigate the prognostic value of these 5 CpGs, we built the risk prediction model which uses these 5 CpGs. All samples were divided into two (high and low) groups according to the risk score of our model (Table S1), and the methylation levels were significantly lower in the low-risk group than in the high-risk group (Figure S7A). Then, we found that the samples of the prognostic index were significantly higher in the high-risk group compared to the low-risk group (Figure S7B), and there were also significant differences between the high-risk and low-risk groups in terms of survival (P < 0.001), (Figure S7C). Finally, the heatmap shows the methylation levels of CpGs between the high-risk and low-risk groups (Figure S7D). The above results indicated that these 5 CpGs play an important role in predicting PCa metastasis.
Then, we identified the corresponding genes (only mRNA) based on the locations of 23 CpGs screened out by two algorithms, and the results were shown in Table S2. Heatmap (Fig. 8A) and boxplot (Fig. 8B) analysis showed that the most corresponding genes of CpGs have differential expression profiles between normal and PCa samples. To better illustrate the characteristics of 13 corresponding genes, we also tested the correlation between them, and the expression levels of most genes were positively correlated (Fig. 8C). To find the best cutoff of progression variables used the ROC curve, so that it has the minimum P value when predicting the tumor progression, all AUC > 0.5 of 13 corresponding genes, (Fig. 8D). After that Kaplan–Meier progression-free survival analysis of best separation was performed based on TCGA survival data, and 8 genes in which log-rank P value < 0.05 were shown in Fig. 8E. Univariate and multivariate logistic regression analysis of PFS was performed based on the expression levels of the genes from TCGA. ALG10, SKOR2, and WDR63 were the independent risk genes of PFS in PCa (Table 2). The pie of the CpGs corresponding genes in cor-response pathways means the activity/inhibition/ non-significant effect contributes to the pathways in PCa (Figure S4B). Pathway analysis validated that 7 genes were mainly associated with the activation/inhibition of AR and RTK pathway, activation of EMT (Figure S4C). The above suggested that although the corresponding genes of CpGs showed a good predictive value of PFS, the AUC values and their correlation of expression were lower than the predictive value of methylation of CpGs.
The characteristics of 13 corresponding genes in PCa. A Heatmap analysis of expression profiles between normal and PCa samples from TCGA. B Boxplot analysis of expression profiles between normal and PCa samples from TCGA. C Correlations among 13 genes expression levels in human PCa tissues. D ROC curve of 13 genes to divided PCa progression group and the non-progression group. E Kaplan–Meier progression-free survival analysis of best separation was performed based on TCGA
Discussion
As we all know that metastasis is the main cause of death in patients with PCa. Survival estimates are traditionally calculated from the time of diagnosis or surgery [22]. PFS is an effective reference standard for predicting OS of PCa patients. However, in patients for PCa, predicted PFS changes considerably during follow-up. CS is the survival probability after already surviving a predefined time period, may, therefore, provide better insight [23]. CS is usually used in cancers with a poor survival prognosis as the survival estimates change considerably after surviving the first year in previous studies. In our study, CS was not only used to evaluate the OS of PCa patients, but also to predicted PFS, and the 1-year CS was equivalent to the 3 years after therapy (94% vs 94%). This indicates that tumor progression in the second and third years after therapy is equal to the first year after therapy. This also suggests that prostate cancer progression is more likely to occur after the third year. The following results we found that although prostate cancer had a better prognosis, its progression was still affected by clinicopathological factors (including TN stage, PSA, and GS). We also found that PSA and GS were associated with tumor size and lymph node metastasis, and PSA was a better predictor of PFS than GS. Current evidence suggests that clinicopathological factors can predict the progression of PCa. Many biomarkers have been evaluated for their ability to discriminate between metastasis and non-metastasis conditions. However, which angle to explore meaningful biomarkers is the focus of this study.
In addition to surgery and radiotherapy, downregulation of androgen levels in the circulation or blockade of the AR is a common therapy for PCa [24]. Continuous progress in the understanding of metastatic pathways is needed to inhibit the development of PCa metastasis. In our study, we found DEGs of tumors and cells both enriched in PI3K-AKT pathway. Two landmark papers defined the interplay between PTEN loss/PI3K activation and AR signaling in the development of prostate cancer [25, 26]. Carver et al. demonstrated the inverse relationship with AR inhibition being associated with upregulated AKT signaling as a result of increased phosphorylation of AKT target genes such as GSK-alpha and PRAS40 [25]. Our study first demonstrated that PCa tumors have distinct expression profiles based on the expression of genes involved in AR and PI3K-AKT, which influence disease outcomes and provide functional context to previously identified gene expression subtypes. Therefore, our study is the first to explore the regulation of AR and PI3K-Akt pathways on PCa metastasis from different perspectives.
In the current study, we hypothesized that the comprehensive characterization of the gene clusters and pathway-related gene expression patterns would be an approach in predicting the risk of PCa metastasis. Within these gene clusters, we found that the expression levels of all top 10 genes in both direct A and B were significantly different in the three clusters. It was clear that our clusters were useful for distinguishing genes expression, but not for evaluating prognosis. Then, our primary concern was the molecular characterization of co-expressed genes, and so we first fetched the venn genes based on AR and PI3K-AKT pathways. The results showed that there were significant differences in the expression of all co-expressed genes in three clusters. The pathway analysis of 14 co-expressed genes validated that these genes were mainly associated with the activation of RTK pathway, and disruption of cell cycle. The link between gene and pathway shows that most genes activate EMT pathway and inhibit the Hormone AR pathway. These results suggested that most of the 14 co-expressed genes may be related to the metastasis of PCa.
There is significant molecular heterogeneity in PCa giving rise to distinct tumor subclasses based on epigenetic modification and gene expression signatures [27, 28], leading to a growing interest in translating this information into clinical practice for outcome prognostication and the development of targets prediction based on each tumor's unique molecular signature. Aberrations in PI3K/AKT/mTOR signaling have been identified in approximately 40% of early prostate cancer cases and 70–100% of advanced disease [29]. Therefore, the PI3K Pathway is indispensable for in-deep exploration of PCa metastasis, which is consistent with our results obtained by KEGG analysis. Activation of the PI3K pathway is associated with resistance to androgen deprivation therapy, disease progression and poor outcomes in PCa. Given the complexity of the AR and PI3K pathways, they likely interact at numerous levels. Mulholland et al. [30] found that dual pathway inhibition with androgen deprivation and a PI3K-AKT inhibitor could lead to significant tumor regression as compared to single pathway inhibition. Our results indicated that there are multiple phenotypes relevant to the AR-PI3K axis in PCa, where tumors with mixed phenotypes may be a more aggressive or have worse outcome than quiescent phenotype. Our results also confirmed previous studies from another perspective. Furthermore, the correlation of gene expression heterogeneity along the AR-PI3K axis and prognostic subtypes of PCa indicates that different therapeutic strategies targeting tumor dependency on AR or PI3K could have clinical benefit in subsets of PCa patients.
As mentioned earlier, the molecular heterogeneity of PCa is not only related to gene expression characteristics but also may be regulated by epigenetic modifications. Locus-specific DNA methylation alterations in PCa have been known for decades, and these events impact gene expression potentially[31]. Combined panels of candidate DNA methylation markers have been shown to have high sensitivity and specificity for the discrimination of prostate cancers from benign tissue [32], and few studies for the metastasis of PCa. In addition, metastatic sites have been reported to show greater divergence for DNA hypomethylation within some patients [33]. Studies showing clonal stability of DNA hypermethylation, and evidence of a subset of clones that are more closely related to metastatic disease, support the idea of using DNA methylation markers for prostate cancer detection and prognosis. Although genomic and transcriptomic subtypes of PCa have been described in some studies [34, 35], we have identified a new epigenetic marker of PCa that was distinguishing primary PCa and PCa metastasis by methylation level of CpGs islands. Unlike previous studies, our study used a combination strategy that incorporated CpGs from two distinct algorithms to minimize the possibility of losing or ignoring important markers. In this study, we used a methylation microarray to screen differential CpGs in a discovery cohort and selected 23 significant CpGs by using two algorithms. Subsequently, we narrowed the selection to 5 candidate CpGs and built a signature. We found that the methylation levels of these 5 CpGs not only had a high correlation but also had a very high area under ROC curve, which could perfectly distinguish primary PCa and PCa metastasis by global methylome changes. We compared the DNA methylation levels of 5 CpGs in different T, N and M stages and found that the CpGs methylation levels of N1 and M1 were lower than that of N0 and M0, but there was little change in DNA methylation levels in different T stages. The above results indicated that the 5 CpGs, which were selected by integrating the union of features from LASSO and SVM-RFE, were reliable in further validations in this study, suggesting that the integration strategy was feasible.
Then, we identified the corresponding genes based on the locations of 23 CpGs screened out by two algorithms, and the results were shown that most corresponding genes of CpGs were the DEGs of normal and PCa samples. Kaplan–Meier progression-free survival analysis of best separation was performed based on TCGA survival data, and 8 genes were P value < 0.05. Univariate and multivariate logistic regression analysis of PFS was performed also based on the expression levels of the genes from TCGA. ALG10, SKOR2, and WDR63 was the independent risk genes of PFS in PCa. Combined with our objectives, we obtained CpG sites and their corresponding genes which have a good predictive value of PFS. However, various evidences showed that the predictive value of CpGs corresponding genes was much lower than GpG sites in OS and PFS.
Conclusions
In summary, we found that PCa progression is more likely to occur after the third year through CS analysis. The molecular heterogeneity of PCa giving rise to distinct tumor subclasses based on epigenetic modification and gene expression signatures. PCa-specific pathways analysis demonstrates that some reprogrammed pathways are less detrimental than others to clinical outcomes depending on the cancer type, which may be exploited for the development of more precise therapeutic strategies targeting unique pathways dependencies. We also identified a CpG methylation panel for the prognosis of PCa with high sensitivity and specificity. Our results support the potential clinical utility of DNA methylation signatures to distinguish tumor metastasis and to predict prognosis and outcomes.
Availability of data and materials
All data obtained and/or analyzed in this study were available from the corresponding authors in a reasonable request.
Abbreviations
- PCa:
-
Prostate cancer
- TCGA:
-
The Cancer Genome Atlas
- GEO:
-
Gene Expression Omnibus
- CpG:
-
Cytosine-guanine
- CS:
-
Conditional survival
- PSA:
-
Prostate-specific antigen
- GS:
-
Gleason score
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- AR:
-
Androgen receptor
- CRPC:
-
Castration-resistant prostate cancer
- DEGs:
-
Differentially expressed genes
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular function
- GSCA:
-
Gene Set Cancer Analysis
- GDSC:
-
Genomics of Drug Sensitivity in Cancer
- LASSO:
-
Least Absolute Shrinkage and Selector Operation
- SVM-RFE:
-
Support Vector Machine-Recursive Feature Elimination
- EMT:
-
Epithelial-mesenchymal transition
References
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68:7–30
Lo UG, Lee CF, Lee MS, Hsieh JT (2017) The role and mechanism of epithelial-to-mesenchymal transition in prostate cancer progression. Int J Mol Sci 18(10):E2079
Hayes JH, Barry MJ (2014) Screening for prostate cancer with the prostate-specific antigen test: a review of current evidence. JAMA 311(11):1143–1149
Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM (2012) Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med 4:127
Damodaran S, Kyriakopoulos CE, Jarrard DF (2017) Newly diagnosed metastatic prostate cancer: has the paradigm changed? Urol Clin North Am 44(4):611–621
Waltering KK, Helenius MA, Sahu B, Manni V, Linja MJ, Jänne OA et al (2009) Increased expression of androgen receptor sensitizes prostate cancer cells to low levels of androgens. Can Res 69:8141–8149
Zoran C, Frédéric RS (2014) Androgen receptor signaling in prostate cancer. Cancer Metastasis Rev 33:413–427
Mateo J, Smith A, Ong M, de Bono JS (2014) Novel drugs targeting the androgen receptor pathway in prostate cancer. Cancer Metastasis Rev 33(2–3):567–579
Lin Y, Lu Z, Kokontis J, Xiang J (2013) Androgen receptor primes prostate cancer cells to apoptosis through down-regulation of basal p21 expression. Biochem Biophys Res Commun 430:289–293
Cunha GR, Ricke W, Thomson A, Marker PC, Risbridger G, Hayward SW et al (2004) Hormonal, cellular, and molecular regulation of normal and neoplastic prostatic development. J Steroid Biochem Mol Biol 92:221–236
Attard G, Cooper CS, De Bono JS (2009) Steroid hormone receptors in prostate cancer: a hard habit to break? Cancer Cell 16(8):458–462
Manning BD, Cantley LC (2007) AKT/PKB signaling: navigating downstream. Cell 129:1261–1274
Mediwala SN, Sun H, Szafran AT, Hartig SM, Sonpavde G, Hayes TG et al (2013) The activity of the androgen receptor variant AR-V7 is regulated by FOXO1 in a PTEN-PI3K-AKTdependent way. Prostate 73:267–277
Trotta AP, Need EF, Selth LA, Chopra S, Pinnock CB, Leach DA et al (2013) Knockdown of the co-chaperone SGTA results in the suppression of androgen and PI3K/AKT signaling and inhibition of prostate cancer cell proliferation. Int J Cancer 133(12):2812–2823
Gerhauser C, Favero F, Risch T, Weischenfeldt J et al (2018) Molecular evolution of early-onset prostate cancer identifes molecular risk markers and clinical trajectories. Cancer Cell 34(6):996–1011
Abeshouse A et al (2005) Cancer Genome Atlas Research Network Te molecular taxonomy of primary prostate cancer. Cell 163:1011–1025
Bhasin JM, Lee BH, Matkin L, Ting AH et al (2015) Methylome-wide sequencing detects DNA hypermethylation distinguishing indolent from aggressive prostate Cancer. Cell Rep 13:2135–2146
Yegnasubramanian S, Kowalski J, Gonzalgo ML et al (2004) Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res 64:1975–1986
Herman JG, Baylin SB (2003) Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349:2042–2054
Skvortsova K, Masle-Farquhar E, Luu PL, Clark SJ et al (2019) DNA hypermethylation encroachment at CpG island borders in cancer is predisposed by H3K4 monomethylation patterns. Cancer Cell 35(2):297–314
Liu CJ, Hu FF, Xia MX, Guo AY et al (2018) GSCALite: a web server for gene set cancer analysis. Bioinformatics 34(21):3771–3772
Katz MHG, Hu CY, Fleming JB, Pisters PWT, Lee JE, Chang GJ (2012) Clinical calculator of conditional survival estimates for resected and unresected survivors of pancreatic cancer. Arch Surg 147(6):513–519
Kent TS, Sachs TE, Sanchez N, Vollmer CM, Callery MP (2011) Conditional survival in pancreatic cancer: better than expected. HPB 13(12):876–880
Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H et al (2009) A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Can Res 69:2305–2313
Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S et al (2011) Reciprocal feedback regulation of PI3k and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19:575–586
Crumbaker M, Khoja L, Joshua AM (2017) AR signaling and the PI3K pathway in prostate cancer. Cancers (Basel) 9(4):1–15
Chau CH, Figg WD (2005) Molecular and phenotypic heterogeneity of metastatic prostate cancer. Cancer Biol Ther 4(2):166–177
Roudier MP, True LD, Higano CS, Vesselle H, Ellis W, Lange P, Vessella RL (2003) Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol 34:646–653
Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C et al (2011) Reciprocal feedback regulation of PI3k and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19(5):575–586
Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, Plaisier S, Garraway IP et al (2011) Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 19(6):792–804
Bedford MT, Van Helden PD (1987) Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res 47:5274–5276
Paziewska A, Dabrowska M, Goryca K et al (2014) DNA methylation status is more reliable than gene expression at detecting cancer in prostate biopsy. Br J Cancer 111:781–789
Yegnasubramanian S, Háner MC, Zhang Y et al (2008) DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res 68:8954–8967
Fraser M, Sabelnykova VY, Yamaguchi TN, Boutros PC et al (2017) Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541(7637):359–364
Zhao SG, Chang SL, Erho N, Feng FY et al (2017) Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol 3(12):1663–1672
Acknowlegements
Not applicable.
Funding
This work was supported by the Natural Science Foundation of China (No. 81972373) and Scientific Research Foundation for Advanced Talents, Xiang’an Hospital of Xiamen University (NO. PM201809170001).
Author information
Authors and Affiliations
Contributions
Conception and design: YZ, CS; provision of study materials: XH, HRY, XL, and HMS; collection and assembly of data: YZ, XH, HRY; data analysis and interpretation: YZ, XH, XL; manuscript writing: YZ, CS. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, Y., Hu, X., Yu, H. et al. Alternations of gene expression in PI3K and AR pathways and DNA methylation features contribute to metastasis of prostate cancer. Cell. Mol. Life Sci. 79, 436 (2022). https://doi.org/10.1007/s00018-022-04456-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04456-2