Abstract
Listeria monocytogenes is a major foodborne pathogen causing increased morbidity worldwide. It forms resistant biofilm structures in food processing facilities after sanitization, consequently creating a public health concern. Many studies on the metabolism and transmission of L. monocytogenes has provided insights into its intracellular infection process, however there is limited understanding on the substrate utilization of the bacteria. Therefore, the main objective of this study was to investigate the carbon and nitrogen substrate catabolism and the biofilm forming potential of 3 Malaysian L. monocytogenes strains (LM41, LM92 and LM115) previously isolated from ready-to-eat foods. Biolog Phenotype Microarray (PM) system was used to study the catabolic activity of the foodborne strains in 190 carbon and 380 nitrogen sources. PM analysis showed that the carbon and nitrogen catabolic activity of L. monocytogenes strains were considerably limited and these strains utilised Tween 40 and Tween 80, which are commonly used for the sanitation in food and meat processing industries. Furthermore, all 3 strains showed strong biofilm forming potential in nutrient-rich and nutrient-limited media, irrespective of the serogroups. The data generated could be utilised to develop alternative measure to inhibit biofilm formation in L. monocytogenes in the food processing environment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Listeria monocytogenes is a facultative anaerobic, non-sporulating, Gram-positive rod shaped bacterium that is commonly found in the soil, plant material and raw and processed food products. It can survive and grow over a wide range of environmental conditions such as refrigeration temperatures (0–4 °C), low pH (2–4) and high salt concentration (10% NaCl), thus making them very hard to control (Doijad et al. 2015; Zhou et al. 2012). It is an emerging foodborne pathogen and the causative agent of human listeriosis. Infection is typically acquired through the ingestion of contaminated food products and the common site of infection is the intestinal epithelium. L. monocytogenes infection has several clinical conditions, including meningitis, encephalitis, gastroenteritis, septicemia, abortions, convulsions and causes high mortality (20–30%) rates among pregnant women, neonates, elderly and immune-compromised patients (Lomonaco et al. 2015). Listeria monocytogenes is accountable for the majority of deaths caused by food-borne epidemics in Europe and USA (Lomonaco et al. 2015). The largest listeriosis outbreak in U.S. history occurred in 2011, due to the consumption of cantaloupe from a single farm that caused 147 illnesses, 33 deaths, and 1 miscarriage in 28 states (CDC 2012). In Malaysia, foodborne L. monocytogenes had been detected in raw and ready-to-eat (RTE) foods and the majority of incidences resulted from the contamination of L. monocytogenes in various street-side foods, salads, vegetables, raw and processed deli meats and fish products (Jamali et al. 2013; Jeyaletchumi et al. 2012; Marian et al. 2012; Ponniah et al. 2010; Wong et al. 2011). Listeriosis is not a notifiable disease in Malaysia, hence official report on foodborne listeriosis is lacking.
Listeria monocytogenes has 4 evolutionary lineages (I, II, III and IV) containing 13 serotypes based on the subtyping of the somatic O and flagellar H antigens (Orsi et al. 2011). Lineage I consists of serotypes 1/2b, 3b, 4b, 4d and 4e (Liu 2008; Orsi et al. 2011). Lineage II contains serotypes 1/2a, 1/2c, 3a and 3c; and Lineage III includes serotypes 4a and 4c and some strains from serotype 4b (Kathariou 2002; Liu 2006, 2008). Both sporadic (serotype 1/2a, 1/2b and 4b) and outbreak cases (serotype 1/2a and 4b) of human listeriosis are mainly caused by L. monocytogenes strains belonging to Lineage I and II (Buchanan et al. 2017; Lomonaco et al. 2015; Orsi et al. 2011).
Listeria monocytogenes is one of the leading foodborne pathogens in both developed and developing countries and able to form resistant biofilm structures in food processing environment (Kadam et al. 2013; Lomonaco et al. 2015). Biofilms are bacterial communities which are surrounded by extracellular polymeric substances attached to biotic or abiotic surfaces (Zhou et al. 2012). Listeria monocytogenes biofilms have been found to show increased resistance to cleaning, disinfectants, desiccation and UV exposure, leading to enhanced persistence to processing plants (Gandhi and Chikindas 2007). Biofilm structures can disperse and contaminate the food products during processing and packaging, thus making them a food safety concern (O’Toole et al. 2000; Zhou et al. 2012). Industrially processed foods, such as cheese and meats have often been reported to be contaminated with L. monocytogenes (Alonso et al. 2014; Doijad et al. 2015; Hain et al. 2007). Previous studies have shown that serotype 1/2a, 1/2b and 4b are responsible for 95% of the clinical cases of listeriosis worldwide (Kadam et al. 2013; Lomonaco et al. 2015). In the current study, 3 Malaysian foodborne L. monocytogenes strains belonging to the pathogenic serogroups were studied for their biofilm forming ability.
Phenotype Microarray is a well-established platform to determine the cellular phenotypes of various microorganisms under different growth conditions (Bochner 2001). Studies with other bacteria have successfully determined their metabolic activity in a variety of substrates (Chelvam et al. 2015; Chong et al. 2017; Farrugia et al. 2013; Tang et al. 2010). Although there is a relatively high incidence of foodborne L. monocytogenes in raw and RTE foods in Malaysia (Jamali et al. 2013; Kuan et al. 2013; Marian et al. 2012; Ponniah et al. 2010; Wong et al. 2011), studies regarding the catabolic activity of L. monocytogenes are lacking. Furthermore, biofilm formation is a major issue in the food industry since microbial biofilms are resistant to industrial chemicals and cleaning processes (Da Silva and De Martinis 2013; Kadam et al. 2013). While previous studies have shown the formation of different biofilm structures between different serotypes of this pathogen (Doijad et al. 2015; Stepanović et al. 2000), there are no reports about the importance of substrate utilization in biofilm formation. Therefore, the present study focuses on the carbon and nitrogen substrate utilization of 3 pathogenic L. monocytogenes strains using Biolog Phenotype Microarray system and their biofilm forming ability in different substrates.
2 Materials and methods
2.1 Background of bacterial strains
Malaysian L. monocytogenes strains isolated from RTE food samples (Jamali et al. 2013) were selected for this study. The strains belong to serogroups 1/2a, 3a (LM92), 1/2c, 3c (LM41) and 4b, 4d, 4e (LM115). Among them was LM115, a multiple-drug resistant strain (Jamali et al. 2013). The strains were maintained in LB broth with 50% glycerol stock and revived in LB broth to perform the individual assays.
The Phenotype MicroArray (PM) assay included 6 × 96-well PM panels (PM1, PM2A, PM3B, PM6, PM7 and PM8), which determined the ability of 3 L. monocytogenes strains to show responses in 190 carbon sources and 380 nitrogen compounds (Table S1). The principle of the test relies on the reduction of tetrazolium violet dye to detect respiration (NADH formation) in various compounds (Bochner 2001).
2.2 Analysis of phenotype microarray data
PM data of plates PM1, PM2A, PM3B, PM6, PM7 and PM8 were recorded using OmniLog OL_FM_12 kinetic software (Biolog, USA) and analyzed using Microsoft Excel. The OmniLog PM Software generated a time course curve for tetrazolium color development for 48 h. For each well, the average area under growth (AUG) and the average slope of time course data were used to measure the threshold value (maximum 100 OmniLog Biolog unit, OU) for growth in the PM plates. Phenotypes were determined based on the difference in average area under growth curve (AUG). The well was considered positive if the standardized average area is equal or exceeded the threshold value. Wells with a high average area value but with a slope close to zero were considered as less significant than wells with a smaller area but increasing signal over time. During data processing, the option of A1 zero (negative control) was selected to subtract the background from each wells. Plates were analyzed in duplicates and errors were excluded from the analysis (Kalai Chelvam et al. 2014).
2.3 Validation test for PM data
To confirm the data obtained in the PM analysis, the L. monocytogenes strains (LM41, LM92 and LM115) were grown in minimal medium supplemented with individual carbon and nitrogen substrates. Overnight cell cultures of L. monocytogenes strains were harvested at 6000 rpm for 5 min, washed with 1× phosphate buffered saline (PBS). Cellular turbidity was adjusted to McFarland standard No.5 which corresponds to 108 CFU-mL−1. Serial dilutions were carried out to ensure that the final concentration of cells in each well was standardized to 103 CFU-mL−1. An aliquot of 200 μL inoculum (M9 minimal medium) with individual carbon and nitrogen substrates were added into the wells of sterile 96-well microtiter plates and incubated at 37 °C for 24 h. After incubation, cellular turbidity was measured at O.D.600 nm wavelength using a microplate reader (Epoch, Germany). Negative control contained minimal medium and the substrates. This experiment was repeated 3 times and the results were averaged.
2.4 Biofilm assay
The biofilm assay was carried out according to the method described by Stepanović et al. (2000). The cell density of an overnight cell cultures of 3 L. monocytogenes strains (LM41, LM92 and LM115) were adjusted. An aliquot of 200 μL bacterial culture was added into the wells of microtiter plates and incubated at 37 °C for 24 and 48 h. After incubation, the unbound cells were removed and washed with 1x PBS. The adhered cells were heat-fixed at 80 °C for 30 min. Quantification of biofilm cells was performed by staining of adhered cells with Crystal Violet (0.5%), followed by washing and de-staining using Ethanol:Acetone (80:20%) solution. The absorption of the eluted stain was measured at O.D.590 nm wavelength.
Based the O.D590nm readings and calculation of cut-off O.D. (O.D.c), strains were classified as non-biofilm producer, weak, moderate and strong biofilm producer. The cut-off O.D. is defined as 3 standard deviations above the mean O.D. of negative control (Kalai Chelvam et al. 2014; Stepanović et al. 2000). Strains were:
-
non-biofilm producer (0), if O.D. ≤ O.D.c,
-
weak biofilm producer (+), if O.D.c < O.D. ≤ (2 × O.D.c),
-
moderate biofilm producer (+ +), if (2 × O.D.c) < O.D. ≤ (4 × O.D.c) and,
-
strong biofilm producer (+ + +), if O.D. > (4 × O.D.c).
Negative control contained sterile LB broth, and the value of negative control was deducted from the background and the experiment was performed in triplicates to ensure the reproducibility of the results.
Overnight cell cultures of 3 L. monocytogenes strains were harvested at 6000 rpm and washed with 1× PBS. The cell density of bacterial cultures was adjusted. Aliquots of 200 μL bacterial cultures in minimal medium were added to the wells of microtiter plates and incubated at 37 °C for 24 and 48 h. The adhered cells were heat-fixed, followed by staining, washing and de-staining. The absorbance (O.D.590 nm) was measured. Negative control contained minimal medium only. The results were averaged and the standard deviation of negative control was calculated for the biofilm classification (Stepanović et al. 2000).
Carbon and nitrogen substrates were selected and added to M9 minimal medium to induce biofilm formation in L. monocytogenes strains. 20 mM solutions of each carbon and nitrogen substrates were aseptically prepared. Aliquots of 200 μL bacterial cultures were added into the wells of microtiter plates containing 20 mM carbon or nitrogen substrates and then incubated at 37 °C for 24 and 48 h. Unbound cells were removed and washed with sterile dH2O. The adhered cells were heat-fixed, followed by staining, washing and destaining. Absorbance was measured at O.D.590 nm. Negative control contained minimal medium and the substrates. This assay was performed in triplicates and the results were averaged.
3 Results
The tested L. monocytogenes strains were able to catabolize different carbon and nitrogen sources. The preferred carbon substrates were: carbohydrates, polymers, nucleosides and the preferred nitrogen substrates were: amino acids, amines, nucleosides and dipeptide sources (Table 1).
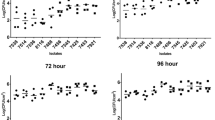
Among the 190 carbon substrates tested, only 38 (20%) were catabolized by the strains. The carbon substrates were classified into carbohydrates (n = 26), nucleosides (n = 5), polymers (n = 4), amino acids (n = 2) and amide (n = 1) (Fig. 1). Out of 38 carbon substrates, only 29 (15.3%) were catabolized by all 3 strains (Fig. 2). However, alcohol, carboxylic acid, fatty acid and esters were not catabolized by any of the 3 strains.
The catabolic phenome of Listeria monocytogenes strains. Strengths of carbon and nitrogen utilization phenotypes of L. monocytogenes strains LM41, LM92 and LM115 are determined using Biolog™ Phenotype Microarray plates PM1, PM2A, PM3B, PM6, PM7 and PM8. The maximal kinetic curve height is expressed as a color-scale ranging from 0 (green) to 200 (red) area under the curve (AUC) units. Substrates are considered as ‘strongly utilized’ if AUC ≥ 180, ‘weakly utilized’ if AUC ≤ 50, and ‘not utilized’ if AUC < 10 (color figure online)
Venn diagram showing the carbon catabolic activity of three L. monocytogenes strains. A total of 190 carbon substrates are tested. They are categorized as: A: Alcohol, B: Amide, C: Amine, D: Amino acid, E: Carbohydrate, F: Carboxylic acid, G: Ester, H: Fatty acid, I: Nucleotide and J: Polymer. Y-axis indicates the percentage of carbon utilised. X-axis shows the carbon category for each carbon substrate tested. The Venn diagram was obtained based on average growth curve area and the numerals indicate the carbon substrates utilized by three strains, utilized by two strains and each strain individually
A total of 380 nitrogen substrates were tested in PM3B, PM6, PM7 and PM8 microplates. Only 61 (16%) nitrogen substrates were catabolized by the strains. The substrates were categorized into amino acids (n = 6), amines (n = 5), nucleosides (n = 7), heterocyclic organic compounds (n = 3) and peptides (n = 10). Among them, only 7 substrates were actively metabolized by all 3 strains (Fig. 3). The substrates were: d-glucosamine, D-mannosamine, N-acetyl-d-glucosamine, cytidine, uridine, xanthosine and uric acid. Strain LM115 utilized only 5 peptide nitrogen sources, whereas strain LM92 utilized only 1 peptide in plate PM3B. Strain LM41 showed a higher respiration rate (10.5%) in plates PM6, PM7 and PM8, while strains LM92 and LM115 did not utilise in any of them (Figs. 1, 3).
Venn diagram showing the nitrogen catabolic activity of three L. monocytogenes strains. A total of 380 nitrogen substrates are tested. They are categorized as: A: Amino acid, B: Amine, C: Nucleoside, D: Heterocyclic Organic Compound and E: Dipeptide. Y-axis indicates the percentage of nitrogen utilised. X-axis shows the nitrogen category for each nitrogen substrate tested. The Venn diagram was obtained based on average growth curve area and the numerals indicate the nitrogen substrates utilized by three strains, utilized by two strains and each strain individually
To validate the PM analysis, 24 carbon and nitrogen substrates were selected based on the PM catabolic profile (Table 1). The validation experiment supported the PM analysis, with the exception of l-cysteine, D-xylose, riboflavin, thiamine, D-biotin, Tween 40 and l-glutamic acid. These substrates were not utilized in PM growth conditions; however they supported bacterial growth when added to the minimal medium (Table S2).
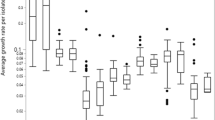
All 3 strains were able to produce strong biofilm in both LB broth and minimal medium after 24 and 48 h incubation. To induce biofilm formation in the studied strains, 23 carbon and nitrogen substrates were chosen based on the PM catabolic profile (Table 1). Out of 23 substrates tested, only 15 substrates induced biofilm formation in M9 medium. The substrates were: adenosine, cytidine, uridine, glutamic acid, rhamnose, Tween 40, riboflavin, thiamine, biotin, salicin, fructose, maltose, galactose, trehalose and xylose (Table 2). All 3 strains formed weak biofilm in threonine and glycerol. Strain LM92 formed moderate biofilm in glycine, lysine, cysteine and glucose; whereas LM41 and LM115 formed weaker biofilm. Moreover, glucose did not induce strong biofilm formation in any of the strains, instead it produced moderate to weak biofilms. All 3 strains formed strong biofilm in minimal medium when supplemented with Tween 40 and Tween 80, although, they did not use them as sole carbon sources in PM growth conditions (Table 2). On the contrary, none of the strains formed biofilm when thymidine was added to the medium. The PM analysis indicated growth of three strains in thymidine, yet it failed to initiate biofilm formation in minimal medium.
4 Discussion
According to the PM analysis, the carbon catabolism capacity of the L. monocytogenes strains was extremely low, only 29–38% out of 190 carbon substrates tested (Fig. 1). Compared to other pathogenic bacteria, i.e. Acinetobacter baumannii, Bacillus subtilis and Escherichia coli were able to catabolize a variety of carbon and nitrogen sources (Bren et al. 2016; Farrugia et al. 2013), thus signifying that L. monocytogenes strains are metabolically less competent. The foodborne strains mainly utilized carbohydrates, polymers and nucleotides and the catabolism rate and choice of carbon sources were rather similar between the 3 strains (Fig. 2). Moreover, the nitrogen substrate catabolism was even lower than carbon catabolism. The strains utilized only 16% out of 380 nitrogen substrates tested, wherein 13% was utilized by strain LM41 only (Fig. 3). All 3 strains catabolized a higher number of nitrogen substrates individually and their preference for nitrogen sources were relatively extensive. For example, 7 (2%) out of 380 nitrogen sources were utilized by 3 strains, while strain LM41 utilized 10% of the dipeptide nitrogen sources and other 2 strains were unable to utilize peptides (Figs. 1, 3). The genomic analysis of two L. monocytogenes strains (Lim et al. 2016) showed that the strains carry a number of carbohydrate, amino acid, fatty acid and nucleotide metabolizing genes and therefore, this foodborne pathogen should be able to metabolize a wide range of carbohydrate and amino acid compounds. The strains were unable to utilize alcohol, carboxylic acid, ester and fatty acid sources and the limited carbon and nitrogen catabolic activity of the strains indicates an incomplete or restrictive metabolic pathway involved in their metabolism.
Furthermore, L. monocytogenes strains showed limited growth activity in d-glucose, while an increased growth rate was observed in l-glutamic acid, fructose, maltose, galactose, salicin and plant-derived carbon sources, arbutin, D-xylose and sorbitol (Fig. 1). Conceivably, glucose is not the sole carbon source for their optimum growth. L. monocytogenes has a complex phosphotransferase system mediated glucose transport system, where it catabolizes fructose, mannose, cellobiose as sole carbon sources when grown in a defined minimal liquid medium and were able to utilize branched-chain amino acids (Tsai and Hodgson 2003). In this study, none of the strains were able to grow on ammonia, nitrate, nitrite and urea since they lack the genes for nitrate and nitrite reductases (Haber et al. 2017); as a result, they only catabolized organic nitrogen sources (Fig. 1). Additionally, glycine, xylose, riboflavin, thiamine, biotin and Tween 40 supported bacterial growth in the validation experiment, while they did not represent growth in PM analysis (Table S2). This could be due to the low concentration of substrates in the Biolog PM plates. Moreover, riboflavin, biotin and thiamine may act as cofactors in minimal medium since they did not require them for growth.
Biofilm formation is a major nuisance in food manufacturing environment. Foodborne pathogens, namely Listeria monocytogenes, Salmonella spp., pathogenic Escherichia coli, Bacillus cereus, Campylobacter spp., Staphylococcus aureus have the capability to adhere and create biofilms on various surfaces in food processing industries (Giaouris and Simões 2018). Previous studies have shown that the foodborne L. monocytogenes strains are resistant to cleaning and sanitization and can survive after the disinfection process (Gandhi and Chikindas 2007; Mertins et al. 2007). Studies by Møretrø et al. (2017) and Dutta et al. (2013) demonstrated that L. monocytogenes showed increased resistance towards quaternary ammonium compounds (QAC), such as Benzalkonium chloride due to the presence of QAC resistance genes, qacH and bcrABC cassettes in their genome. Polysorbates are nonionic surfactants often emulsified in the sanitizers to clean food and meat processing plants. Møretrø et al. (2017) demonstrated that L. monocytogenes can survive as biofilms in the surface water residues containing residual QAC after disinfection, therefore conferring growth advantage to the bacteria. In this study, both Tween 40 and Tween 80 induced strong biofilm formation in the selected strains in nutrient limited conditions.
Many studies indicated that L. monocytogenes strains produce more biofilm in nutrient limited medium; while they have increased growth rate in nutrient-rich medium (Doijad et al. 2015; Zhou et al. 2012). In this study, all 3 strains showed enhanced growth activity and formed strong biofilms in both LB and minimal media (Table 2), suggesting that this foodborne pathogen employs an alternative mechanism to proliferate and produce biofilm in nutrient-starved conditions. This study also showed that the L. monocytogenes strains were able to attach and form biofilm on polystyrene microtiter plates. Polystyrene is one of the most widely used plastics, especially in commercial food packaging and the equipment used in food production facility. Several studies reported that persistent biofilm structures are readily formed on plastic surfaces and niches that are difficult to clean (Borucki et al. 2003; Doijad et al. 2015). On the contrary, thymidine did not induce biofilm formation in any of the strains, although increased growth rate was observed when supplemented in minimal medium (Table 2). Therefore, thymidine may not be the ideal carbon source to initiate the biofilm formation in L. monocytogenes, since it inhibited the biofilm-forming process in a nutrient limited condition. Further experimental analysis should be taken into consideration regarding the impact of thymidine in biofilm control and prevention.
5 Conclusion
In conclusion, this study revealed that the foodborne L. monocytogenes strains were strong biofilm producers even in the absence of nutrients. The biofilm-forming strains thus facilitate their survival and persistence in food processing environment which may lead to the subsequent contamination of food products. Additionally, the phenotypic analysis showed that the carbon and nitrogen catabolic activity of three strains were considerably limited. Further genomic analysis is required to investigate the biosynthetic pathways involved in their metabolism and identify the genes for the enzymes responsible for the catabolism of various substrates. Lastly, alternative approaches must be implemented to prevent biofilm formation in food processing environment to improve the food quality and ensure the welfare of human health.
References
Alonso AN, Perry KJ, Regeimbal JM et al (2014) Identification of Listeria monocytogenes determinants required for biofilm formation. PLoS ONE 9(12):1–16. https://doi.org/10.1371/journal.pone.0113696
Bochner BR (2001) Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res 11(7):1246–1255. https://doi.org/10.1101/gr.186501
Borucki MK, Peppin JD, White D, Loge F, Call DR (2003) Variation in biofilm formation among strains of Listeria monocytogenes. Appl Environ Microbiol 69(12):7336–7342. https://doi.org/10.1128/AEM.69.12.7336
Bren A, Park JO, Towbin BD et al (2016) Glucose becomes one of the worst carbon sources for E.coli on poor nitrogen sources due to suboptimal levels of cAMP. Sci Rep 6:2–11. https://doi.org/10.1038/srep24834
Buchanan RL, Gorris LGM, Hayman MM et al (2017) A review of Listeria monocytogenes: an update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 75:1–13. https://doi.org/10.1016/j.foodcont.2016.12.016
CDC (2012) Multistate outbreak of listeriosis linked to whole cantaloupes from Jensen Farms, Colorado| Listeria| CDC. https://www.cdc.gov/listeria/outbreaks/cantaloupes-jensen-farms/index.html. Accessed 4 May 2018
Chelvam KK, Yap KP, Chai LC, Thong KL (2015) Variable responses to carbon utilization between planktonic and biofilm cells of a human carrier strain of Salmonella enterica serovar Typhi. PLoS ONE 10(5):1–11. https://doi.org/10.1371/journal.pone.0126207
Chong TM, Chen JW, See-Too WS et al (2017) Phenotypic and genomic survey on organic acid utilization profile of Pseudomonas mendocina strain S5.2, a vineyard soil isolate. AMB Express. https://doi.org/10.1186/s13568-017-0437-7
Da Silva EP, De Martinis ECP (2013) Current knowledge and perspectives on biofilm formation: the case of Listeria monocytogenes. Appl Microbiol Biotechnol 97(3):957–968. https://doi.org/10.1007/s00253-012-4611-1
Doijad SP, Barbuddhe SB, Garg S et al (2015) Biofilm-forming abilities of listeria monocytogenes serotypes isolated from different sources. PLoS ONE 10(9):1–14. https://doi.org/10.1371/journal.pone.0137046
Dutta V, Elhanafi D, Kathariou S (2013) Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl Environ Microbiol 79(19):6067–6074. https://doi.org/10.1128/AEM.01751-13
Farrugia DN, Elbourne LDH, Hassan KA et al (2013) The complete genome and phenome of a community-acquired Acinetobacter baumannii. PLoS ONE 8(3):e58628. https://doi.org/10.1371/journal.pone.0058628
Gandhi M, Chikindas ML (2007) Listeria: a foodborne pathogen that knows how to survive. Int J Food Microbiol 113(1):1–15. https://doi.org/10.1016/j.ijfoodmicro.2006.07.008
Giaouris EE, Simões MV (2018) Pathogenic biofilm formation in the food industry and alternative control strategies. In: Holban MA, Grumezescu MA (eds) Foodborne diseases: handbook of food bioengineering, 1st edn. Academic Press, Cambridge, Massachusetts, US, p 309–377. https://doi.org/10.1016/B978-0-12-811444-5.00011-7
Haber A, Friedman S, Lobel L et al (2017) l-Glutamine induces expression of Listeria monocytogenes virulence genes. PLoS Pathog 13(1):1–25. https://doi.org/10.1371/journal.ppat.1006161
Hain T, Chatterjee SS, Ghai R et al (2007) Pathogenomics of Listeria spp. Int J Med Microbiol 297(7–8):541–557. https://doi.org/10.1016/j.ijmm.2007.03.016
Jamali H, Chai LC, Thong KL (2013) Detection and isolation of Listeria spp. and Listeria monocytogenes in ready-to-eat foods with various selective culture media. Food Control 32(1):19–24. https://doi.org/10.1016/j.foodcont.2012.11.033
Jeyaletchumi P, Tunung R, Selina PM et al (2012) Assessment of Listeria monocytogenes in salad vegetables through kitchen simulation study. J Trop Agric Food Sci 40(1):55–62
Kadam SR, den Besten HMW, van der Veen S et al (2013) Diversity assessment of Listeria monocytogenes biofilm formation: impact of growth condition, serotype and strain origin. Int J Food Microbiol 165(3):259–264. https://doi.org/10.1016/j.ijfoodmicro.2013.05.025
Kalai Chelvam K, Chai LC, Thong KL (2014) Variations in motility and biofilm formation of Salmonella enterica serovar Typhi. Gut Pathogens 6(1):1–10. https://doi.org/10.1186/1757-4749-6-2
Kathariou S (2002) Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J Food Prot 65(11):1811–1829. https://doi.org/10.4315/0362-028X-65.11.1811
Kuan CH, Wong WC, Pui CF et al (2013) Prevalence and quantification of Listeria monocytogenes in beef offal at retail level in Selangor, Malaysia. Brazil J Microbiol 44(4):1169–1172. https://doi.org/10.1590/S1517-83822014005000002
Lim SY, Yap KP, Thong KL (2016) Comparative genomics analyses revealed two virulent Listeria monocytogenes strains isolated from ready-to-eat food. Gut Pathogens 8(1):1–8. https://doi.org/10.1186/s13099-016-0147-8
Liu D (2006) Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J Med Microbiol 55(6):645–659. https://doi.org/10.1099/jmm.0.46495-0
Liu D (2008) Epidemiology. In: Liu D (ed) Handbook of Listeria monocytogenes. CRC Press, Florida, p 27–60
Lomonaco S, Nucera D, Filipello V (2015) The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect Genet Evol 35:172–183. https://doi.org/10.1016/j.meegid.2015.08.008
Marian MN, Sharifah Aminah SM, Zuraini MI et al (2012) MPN-PCR detection and antimicrobial resistance of Listeria monocytogenes isolated from raw and ready-to-eat foods in Malaysia. Food Control 28(2):309–314. https://doi.org/10.1016/j.foodcont.2012.05.030
Mertins S, Joseph B, Goetz M et al (2007) Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes. J Bacteriol 189(2):473–490. https://doi.org/10.1128/JB.00972-06
Møretrø T, Schirmer BCT, Heir E et al (2017) Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry. Int J Food Microbiol 241:215–224. https://doi.org/10.1016/j.ijfoodmicro.2016.10.025
Orsi RH, de Bakker HC, Wiedmann M (2011) Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301(2):79–96. https://doi.org/10.1016/j.ijmm.2010.05.002
O’Toole G, Kaplan HB, Kolter R (2000) Biofilm formation as microbial development. Annu Rev Microbiol 54(1):49–79
Ponniah J, Robin T, Paie MS et al (2010) Listeria monocytogenes in raw salad vegetables sold at retail level in Malaysia. Food Control 21(5):774–778. https://doi.org/10.1016/j.foodcont.2009.09.008
Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M (2000) A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 40(2):175–179. https://doi.org/10.1016/S0167-7012(00)00122-6
Tang JYH, Carlson J, Mohamad Ghazali F et al (2010) Phenotypic MicroArray (PM) profiles (carbon sources and sensitivity to osmolytes and pH) of Campylobacter jejuni ATCC 33560 in response to temperature. Int Food Res J 17(4):837–844. https://doi.org/10.1007/BF00441757
Tsai H, Hodgson DA (2003) Development of a synthetic minimal medium for Listeria monocytogenes. Appl Environ Microbiol 69(11):6943–6945. https://doi.org/10.1128/AEM.69.11.6943
Wong WC, Pui CF, Chai LC et al (2011) Biosafety assessment of Listeria monocytogenes in vegetarian burger patties in Malaysia. Int Food Res J 18(1):459–463
Zhou Q, Feng X, Zhang Q et al (2012) Carbon catabolite control is important for Listeria monocytogenes biofilm formation in response to nutrient availability. Curr Microbiol 65(1):35–43. https://doi.org/10.1007/s00284-012-0125-4
Acknowledgements
We thank University of Malaya for the financial support and research facilities. This study was supported by the High Impact Research Grant UM.C/625/1/HIR/MOE/CHAN/01/02 from University of Malaya. The funders had no role in project design, data collection and analysis, decision to publish or preparation of the manuscript. This work was performed at the Laboratory of Biomedical Science and Molecular Microbiology, Institute of Graduate Studies, University of Malaya.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharar, N.S., Chai, L.C. & Thong, K.L. Catabolic activity and biofilm formation of foodborne Listeria monocytogenes strains. J Consum Prot Food Saf 13, 289–298 (2018). https://doi.org/10.1007/s00003-018-1169-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00003-018-1169-6