Abstract
Mercury (Hg) jeopardizes ecological balance and public health globally owing to its cytotoxicity, indegradability, coexistence in different chemical forms, uneven distribution, mobility, and long-lasting in the atmosphere. Therefore, the endeavors for feasible, economic, systematic, easily available, and sustainable approaches can be customized, which are conclusively enduring. Bioremediation, particularly microbial remediation means, as a greener and cheaper strategy has piqued the interest of biotechnologists and ecologists to mitigate Hg risk. By the virtue of versatile physiological performance, microorganism either prokaryotic or eukaryotic in planktonic or in the sessile-biofilm state, solitary or in consortium, indigenously occurred or intendedly introduced as GMO under aerobic or anaerobic circumstances and even extremophiles, have the ability to detoxify Hg and transform its phase to less-toxic valence through various metabolic scenarios like biosorption, bioprecipitation, bioleaching, bioaccumulation, and biovolatilization. Importantly, all previous mechanisms were catalyzed by different enzymatic systems including mercuric reductase and organomercurial lyase. However, their recruitment in in situ or ex situ technologies also implemented sensibly. Interestingly, the combined remediation technology boosted microbial-remediating efficiency and potentiality, especially upon hybridizing microbes with novel biosorbents such as carbon-based material, polymer, and nanoparticles. All preceding aspects were addressed in detail inclusively in this chapter to compensate the gap among knowledge, lab-scale practice, and field-scale application for alleviating Hg-toxicity.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Bioventing
- Biostimulation
- Composting
- Algal remediation
- Fungal remediation
- Extremophiles
- Demethylation
- Methylation
- Biochar
- MOFs
- Mer-A gene
- Metagenomics
1 Introduction
Mercury (Hg) is metallic element that was ranked among “big three” heavy metals in the list of hazardous materials according to the Agency for Toxic Substances and Disease Registry (ATSDR 2015). Owing to its inert nature, nonbiodegradability, toxicity, and long-lasting in the atmosphere, its presence represents a risky stalemate that jeopardizes all living creatures and their ambient ecosystem (Kim et al. 2015). Despite its release in the environment could be from natural roots (e.g., oceanic emission, volcanic eruption, photoreduction, and degassing from minerals (combustion of organic substances), the majority of its aquatic and terrestrial genesis was assigned to anthropogenic activities. As well known, Hg is the main ingredient in different medical devices (e.g., oesophageal dilators, sphygmomanometers, dental amalgams, etc.), electrical apparatus (e.g., batteries, switches, etc.), measuring tools (e.g., psychrometers, thermometers, flow meters, manometers, barometers, hydrometers, etc.), as insecticides, herbicides, fungicides. Besides, its role in paper pulps, painting industry, steel industry, and chloro-alkali process are documented. Hereby, all previous activities undoubtedly contribute to global Hg tainting besides mining and fossil fuel incineration (Balan et al. 2018; Amin et al. 2022).
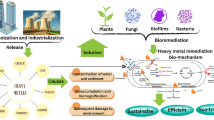
Mercury characterizes by its odorless nature, tolerance to a broad range of temperatures, and coexists in both liquid and vapor phases and also in different organic and inorganic states, namely, elemental (Hg0), mercurous (Hg22+), mercuric (Hg2+), monomethyl mercury, ethyl mercury or dimethyl mercury, etc. Each form of Hg possesses its own discerned physiochemical properties, environmental attitude, and biotoxicity (He et al. 2015). Remarkably, (Mahbub et al. 2017a) reported that alkylated compounds of mercury are severe neurotoxins; however, Amin et al. (2022) reported that the inorganic form of mercury (Hg2+), which is commonly present form in the environment, is the most toxic due to its superior affinity to cysteine moiety of protein, more soluble in lipids and highly accessible through biological membranes. On the other hand, Saranya et al. (2017) documented that the association of mercury with chloride, hydroxide, sulfide, and oxide groups plays a crucial role in elevating mercury poisoning symptoms. Generally, Minamata’s disease is considered the most popular disease caused by mercury besides other gastrointestinal, hematological, renal, cardiovascular, and neurological disorder, which had been detected (Amin et al. 2022) (Fig. 8.1).
Whatever the mercury state in the environment, US Environmental Protection Agency (EPA) determined the limits of mercury by 2.0 μg/L in water and the range of soil 6.6–3600 mg/kg according to the land use (Mahbub et al. 2017a). The bioaccumulative traits of mercury compounds through the food chain lead to biomagnifications and inherent toxicity; hereby, imperative necessity for scavenging such elements and reducing its pathogenicity via several remediation approaches. Conventionally, the hazardous wastes were manipulated through their digging up in isolated landfills or capped sites. Nonetheless, numerous shortcomings emerged beginning from handling/transportation and safety requirements, passing through precise finding of proper landfill sites, which are potentially cost processes, ending with the possibility for pollutant migration elsewhere, which is risky and entails monitoring and strict maintenance of the landfill barriers long into the future (Gong et al. 2018). Therefore, physicochemical techniques were developed, to compensate the drawbacks of isolation method, through degrading the pollutant completely or transforming it into innocuous matter. However, the type of contaminated site enforces technologists to employ specific approaches. Namely, in ground water and wastewater polluted with mercury, the precipitation method is the most commonly applied via utilizing coagulants such as sodium sulfide and lignin derivatives followed by filtration or clarification. Also, adsorption process proved its effectiveness in cleaning up Hg2+ contaminated water (O’rear et al. 2014). Whereas, soil washing, stabilization/solidification, vitrification, thermal treatment, and electrokinetic recovery are considered being the most efficient means of soil treatment via employing acid/alkali chelating agents/surfactants, stabilizing agents (e.g., lime, ceramics, zeolites, Portland cement, fly ashes, sulfur polymer, aluminosilicates, metal oxides, bentonites, biosolids and animal manure activated carbon, biochar, clay minerals, phosphates, etc.), high temperature with low pressure (350 °C/1 atm pressure), and a low intensity direct current (Gong et al. 2018).
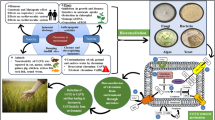
Unfortunately, the precipitated sludge that requires further multistage-treatment prior to disposal, the generation of fouling or plugging, the possibility for leachability, diminishing soil functionality/quality, and the capital cost of chemicals/energy are deemed the major obstacles that limit such physicochemical methods to be at the experimental phase for field application or even greenhouse studies (Gong et al. 2018; Taha et al. 2023). Arguably, such limitations symbolize the driving force that promoted researchers and biologists to harness green technology for removing or transforming mercury in natural bioremediation processes by plants and microbes. Interestingly, the phytoremediation technology found an ecologically sound in cleaning up of several contaminants, especially mercury, depending on biochemical, physical, biological, and microbial interactions of the plants. Via several mechanisms including, phytostimulation, phytostabilization, phytoextraction, rhizofiltration, phytovolatilization, and phytodegradation, the phytoremediation process could be implemented by various plant species (Verma 2021). Nevertheless, the type/physicochemical properties of contaminant, choice/bioavailability of hyperaccumulators phytoremediator plant species, the ingathering of contaminate in the edible parts of fruit and vegetable crop, the slow rate of growth process, plant seasonal variation and handling/disposing of contaminated plants are the substantial constraints that handicap the extended application of phytoremediation (Farraji et al. 2016). Building on this previous knowledge, the microbial manipulation of heavy metals has piqued the interest of technologists and researchers to find a cost-effective, sustainable, easy solution for mercury removal. Hence, in the current chapter, the microbial strategies are detoxifying mercury via different microbial groups and under various microbial growth conditions would be discussed. Besides, the remedy methods, genetic system, enzymatic pathways, and the hybridization of microbes with advanced approaches would be also addressed (Fig. 8.2).
Schematic diagram summarizes Hg-bioremediation by various microbial groups through different remediating pathways that were catalyzed by different enzymatic systems expressed from varied genes and their recruitment in environmental sited via in situ and ex situ technologies, besides the advanced materials in combined approaches that would ameliorate the performance of microbes to guarantee the success of detoxification process
2 Microbial Pathways in Mercury Remediation
A plethora of microbial species possess the capability to detoxify a vast array of metal contaminants, by the virtue of their versatile metabolic activities. Remarkably, various species, either indigenous, genetically modified (GMOs), or exogenously introduced, could exert more than pathway in containment metals and metalloids simultaneously and restrict their availability in contamination site, even the dead cells could participate more or less in detoxification process. Generally, mercury-remediating microbes symbolize by their tolerance and low sensitivity to the toxicity of mercury ions. Remarkably, the binding of metals on the cell wall or internally by intracellular proteins (e.g., phytochelatins, metallothioneins, siderophores, etc.), enzymatic conversion of metals, reduced metal uptake, modifying uptake system and utilizing effective efflux systems are the common strategies by which microbes could resist heavy metals (Tarekegn et al. 2020; Tarfeen et al. 2022). Thereby, the biosorption/adsorption, bioprecipitation, biotransformation (bio-reduction, bio-oxidation, methylation, demethylation), bioaccumulation/sequestration, bioleaching, and biovolatilization are the most widely mechanisms utilized by microbes in bioremediation process.
2.1 Biosorption/Adsorption
As a surface phenomenon based mainly on the cellular surface traits, the adsorption of mercury takes place by both live and dead biomass. In fact, heavy metal ions like mercury were trapped passively on the microbial surface (i.e., avoiding energy requirement) and bound by different physical and chemical interactions (e.g., Van der Waals, electrostatic, covalent bonding, and ion exchange) to negatively charged surface groups of phosphates, carboxylates, sulfates, hydroxyl, and amides. Such functional groups constitute the main cell wall ingredients of proteins, polysaccharides, extracellular polymeric substances (EPS), and lipids, which allow external adsorption of mercury; however, in some resistant microbes, metal ions could pass through porins and reside in the periplasmic space creating a potent binding with the cellular membranes; facilitating by such way the activation of another remediating strategy such as internal sequestration or biotransformation. Interestingly, the dead biomass seemed being more effective in biosorption strategy, comparing to live biomass, due to insensitivity to higher concentrations, adaptability to alterations in environmental conditions, and the unnecessity for adjusting nutritional and growth conditions (Jin et al., 2018; Tarfeen et al. 2022).
In this context, a study conducted by Balan et al. (2018) reported that Hg-tolerant Pseudarthrobacter oxydans and Pseudomonas frederiksbergensis succeeded in removing 2 ppm of Hg under experimental conditions. By the aid of Fourier transform infrared analysis (FTIR), they explained that nitro compound, alkynes, alkenes, alcoholic alkyl halide, primary amines, aliphatic and aromatic amines, alkanes, carboxylic acid, and amide groups represented the active ligand to Hg that enable both bacterial species to tolerate and immobilize Hg. On the other hand, Rhizopus oryzae and Aspergillus niger removed about 90% of Hg at 10 and 100 ppm by their live and dead biomass; revealing that the higher sorption performance (up to 90.38%) was implemented by R. oryzae (dead cells) at 100 ppm. Besides, FTIR analysis and kinetic studies (Pseudo-second-order kinetic model and Langmuir isotherm) reflected that the chemisorption process happened on the homogenous surface (Anuar et al. 2020).
2.2 Bioaccumulation
The metabolic-dependent active absorption and infiltration of contaminants internally by living biomass to the middle of the cell is known as intracellular accumulation. It commences by adsorption of contaminant externally followed by its uptake through the phospholipid bilayers of living biomass. This process takes place with the aid of active transporters and protein channels in a process mimic that occurs for internalization of essential ions such as K+, Na2+, Mg+2, and Ca2+ using ion pumps and passive diffusion mechanisms (Tarfeen et al. 2022). Despite bioaccumulation process being time-consuming, relative to biosorption, the removal rate could be enhanced easily by adjusting the reaction conditions as revealed by (Jin et al. 2018; Tarekegn et al. 2020). The intracellular accumulation could be described as a toxicokinetic process that influenced by the sensitivity of living organisms to the contaminants and based on their concentrations and microbial physiology. However, the accumulative microbes characterized by their distinguished capability to transform and modify the toxicity of the sequestered contaminant to be less toxic, by other additional pathways while remaining inside the cellular compartments (Tarekegn et al. 2020). Notably, two mercury-tolerant bacterial strains isolated from gold mining tailings in Indonesia were identified as Fictibacillus nanhainensis and Bacillus toyonensis exhibited their potential accumulative performance for mercury by more than 81% removal capacity (Nurfitriani et al. 2020). In the same sense, Tazaki and Asada (2007) found that bacteria resident in Geita (small gold mine pond near Lake Victoria, Tanzania), accumulate mercury through EPS as visualized by transmission electron microscope (TEM). Whereas, a white rot fungus Phlebia floridensis trapped about 70–84% of mercury, which induced morphological and textural alterations in the bioremediated hyphae as depicted by scanning electron microscope (SEM) and energy dispersive X-ray (EDX) (Sharma et al. 2022).
2.3 Bioprecipitation
It could also be called biomineralization or biocrystallization (Tarekegn et al. 2020). It involves the conversion of heavy metals or metalloids from their soluble states to insoluble states such as sulfides, hydroxides, phosphates, and carbonates. The microbial growth, metabolic activity, and various enzymatic systems mediate such a process by liberating microbial metabolites such as organic acids, EPS, and electron donors, which thereafter change the surrounding environment chemistry to that favor the precipitation. Interestingly, bioprecipitation process relies fundamentally on the environmental changes generated by microbial activity like alterations in pH and redox potential changes (Jeyakumar et al. 2023). Undoubtedly, no one can deny the pivotal role of sulfate-reducing bacteria (SRB) in immobilizing heavy metals by producing their sulfides (Vitor et al. 2015; Zhang and Wang 2016). Wherein, Groudev et al. (2014) reported that indigenous SRB-dwelling cinnamonic forest soil stimulated the mobility of Zn, Cu, and Cd and precipitated them as insoluble metals sulfides; however, pertinent studies on mercury are scarce. Notwithstanding that, a study mediated by Pan-Hou and Imura (1981) found that Clostridium cochlearium was able to form HgS anaerobically. Besides, HgS was formed by Klebsiella aerogenes NCTC418 after its cultivation in continuous aerobic culture in the presence of HgCl2 (2 μg/mL) and the author confirmed the elevation of cellular sulfide upon the existence of mercuric ions (Aiking et al. 1985) On the other hand, Håkansson et al. (2008) hybridized electrokinetic remediation with the metabolic activity of SRB, in which contaminated soil from a chlor-alkali industry encompassed mercury (100 mg/kg) treated with iodide/iodine complexing agent and exposed to electric field. The complexes of mercury iodide reacted with H2S in water solution, which generated by the action of SRB and resulted in mercury precipitation in mercury sulfide crystals.
Moreover, mercury precipitation in hydroxide form is also rare. Nonetheless, microbial-induced carbonate precipitation (MICP) seemed to be a promising tool in mercury remediation where, the activity of nitrate reductase enzyme of Proteus mirabilis 10B, either under aerobic or anaerobic conditions, entrapped about 322 and 309 of mercury, in their oxide forms (i.e., HgO and Hg2O), in calcite matrix during 168 and 186 hrs., respectively, in an investigation conducted by Eltarahony et al. (2020). On the other hand, the ureolytic strains of Metschnikowia pulcherrima and Raoultella planticola transformed the soluble form of mercury (350 ppm) completely into insoluble forms of CaHgO2, HgO and Hg2O within 102 hrs., which also encapsulated inside CaCO3 trap (Eltarahony et al. 2021); yet, the denitrification and ureolysis processes mediate the precipitation stage through elevating pH and alkalinity of solution. Strikingly, MICP is a proficient technique that remediates several heavy metals and nuclides in their carbonate form (Kim et al. 2021; Wang et al. 2023). Also, the precipitation of heavy metals in hydroxide form was also detected by several research groups (Chan et al. 2009; Li et al. 2019), but in case of mercury, it was not reported.
2.4 Bioleaching/Biomining
It contradicts the bioprecipitation, in which it involves the extraction of metals from their ores by dissolution of their insoluble minerals to soluble form by the catalysis of organic acids released by the acidophilic microbes (Gong et al. 2018; Jeyakumar et al. 2023). Broadly, as reported by Tarekegn et al. (2020), metals are present in the environment in sulfide and oxide forms, such processes could be catalyzed by various microbial genera such as Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans, Leptospirillum ferrooxidans, Aspergillus sp., Alternaria sp., Mucor sp., Penicillium sp., Rhizopus sp. and Cladosporium sp. (Jin et al. 2018). Bioleaching process or biological leaching was employed mainly in biohydrometallurgy and is implemented through one of three pathways, including acidolysis, complexolysis, and redoxolysis. Importantly, acidolysis entails the bioleaching microbe characterized by its ability to produce low molecular weight organic/inorganic acids (e.g., oxalic acid, gluconic acid, citric acid, sulfuric acid, etc.) during their metabolic activity to assure the recovery and mobilization of required metal. While redoxolysis could happen either directly, by direct microbial oxidation of metal sulfide with electrons gained directly from the reduced minerals, or indirectly through the use of an oxidant such as Ferric iron that is generated via microbial oxidation of ferrous iron, which coexists naturally in the minerals. Whereas, complexolysis encompasses the complexation of metal by organic acids or metabolites like siderophores in a slower rate process than acidolysis (Okoh et al. 2018). However, some bacterial strains like Citrobacter sp. could exert bioleaching process through excreting free inorganic phosphate. Although bioleaching is a cost-effective process, relative to other chemical approaches, its application is restricted to Cu, Ur, and Au (Tarekegn et al. 2020); the sluggish leaching kinetics are the main impediments in mercury remediation from contaminated solid waste as reported by Xie et al. (2020).
2.5 Biovolatilization
Via such a strategy, the enzymatic system of the microbes mediates the conversion of metals from the soluble phase to the volatile phase. By the virtue of the cytoplasmic flavoenzyme mercuric reductase (MerA) and mercurial lyase (MerB), Hg-resistant microorganisms reduce Hg2+ to its volatile state Hg0 (Gong et al. 2018; Tarfeen et al. 2022). As stated by Anthony (2014), Hg-resistance property is carried on transposons (Tn2) or conjugative plasmids (HgR), which enable the microbe to resist nor only mercury but also other heavy metals and even antibiotics. A broad spectrum of microorganism could eliminate the toxicity of organomercurials through volatilization after mercury uptake into cytoplasm through a specific transport system (MerT) or through auxiliary transporters MerC/MerF, followed by its reduction to elemental form, which volatilized immediately from the cells through passive diffusion system before re-oxidation to its divalent form occurs (Essa et al. 2002). In this regard, a mercury-resistant Bacillus sp. strain, which was isolated from molybdenum-lead mining soils in China, exhibited an appreciable resistance and removal rates to several heavy metals, especially mercury. The authors determined the Hg-adsorption rate and Hg-volatilization rate at 36 h, which reached 8.24% and 89.08%, respectively; reflecting the promising bioremediation potential of the strain in remediating 100 mg/L of mercury under nutrient availability (Yao et al. 2023). Whereas, a novel Hg(II)-volatilizing filamentous fungus Penicillium spp., DC-F11 was able to diminish mercury-phytotoxicity and total Hg in contaminated soil through multisystem collaborative process started with extracellular adsorption and precipitation followed by volatilization (Chang et al. 2020). Upon combining chemical extraction and microbial volatilization, Chen et al. (2018) got rid of 77% of Hg from field-tainted soil by using ammonium thiosulfate (0.5 M) as a first-step remediating process, thereafter, ≥81% of Hg2+ was reduced and volatilized by Enterobacter cloacae.
2.6 Biotransformation
2.6.1 Bioremediation via Redox State Change (Bioreduction/Biooxidation)
It describes the bioremediation through switching the oxidative state of heavy metal to a different state with entirely varied physicochemical traits. Such alteration in oxidative state may influence metal mobility, toxicity, and bioavailability (Gong et al. 2018; Tarfeen et al. 2022). This approach seems to be effective in the case of metals whose toxicity differs with varied redox states; therefore, intensive attention should be paid before employing this method, in particular, in field applications. That is for evading transformation to a more hazardous or more mobile phase, which would impact adversely on the ecosystem and public health (Gong et al. 2018). As highlighted by Colombo et al. (2014), gaseous mercury (Hg0) is highly mobile in groundwater and can accumulate easily in aquatic creatures. Hence, via such a study, the authors utilized anaerobic bacteria like Geothrix fermentans and facultative anaerobic bacteria such as Shewanella oneidensis and Cupriavidus metallidurans to oxidize dissolved mercury to its divalent state, which could be eliminated subsequently via complexation with sulfide and precipitation as insoluble HgS phase. Through X-ray absorption near edge structure (XANES) spectroscopy, the authors emphasized the covalently binding of Hg2+ with thiol moieties in both dead and live cells. In addition, Hg0 oxidation by anoxic is commonly observed in waterlogged soils and anoxygenic sediments (Bouffard and Amyot 2009; Poulin et al. 2016). Regarding mercury reduction, it could be executed under different aeration conditions and either solitary on in combination with other bioremediating pathways. Wherein, an investigation performed by Wu et al. (2022) declared that the plant symbiotic Metarhizium robertsii reduced Hg2+ to its gaseous form by the catalysis of mercury reductase as a second step after demethylating methylmercury by the activity of methylmercury demethylase, by such combinatory pathways, the fungus curtailed the accumulation of mercury in the plants and promoted their cultivation in contaminated soils. On the other hand, phototrophic non-sulfur purple bacteria (e.g., Rhodopseudomonas palustris, Rhodobacter capsulatus, and Rhodobacter sphaeroides) reduced HgII under anaerobic photoheterotrophic incubation (Grégoire and Poulain 2016), hence, participating in Hg redox cycling. Interestingly, Heliobacterium modesticaldum, which is a member of spore-forming fermentative photoheterotrophs, was reported as an effective Hg reducer anaerobically through the pathway cometabolized by ferredoxin (i.e., reduced redox cofactor) rather than MerA reductase, which was not detected in this strain (Grégoire et al. 2018).
2.6.2 Mercury Methylation
It is a process dedicated to transfer a methyl group to mercury and the formation of methymercury (MeHg+). Although MeHg+ is more toxic than other Hg forms, but in this case, it may be considered as less toxic to the manipulating microorganism. Such process was executed predominantly by SRB such as Desulfovibrio desulfuricans under low availability of sulfate ions and anoxic incubation (Barkay et al. 2003; Wagner-Döbler, 2003). Yet, other microbial groups of methanogens (Gilmour et al. 2018), iron-reducing bacteria (Fleming et al. 2006), and some members of Chloroflexi and Firmicutes phyla also recently recognized as Hg methylators. Notably, hgcAB genes encode the corrinoid protein, which commences the initial stage in methylation process and was utilized to identify Hg methylators among a broad microbial spectrum in any contaminated habitats. As noticed by Lin et al. (2014), Geobacter sulfurreducens, which is metal dissimilating anaerobic bacteria, had the ability to reduce mercury when hgcAB gene was deleted; reflecting the presence of a physiological link between two pathways of Hg transformations. Interestingly, the investigations concerning mercury methylation by phototrophs are scarce and more/deep mechanistic studies are required (Grégoire and Poulain 2014). Nevertheless, (Franco et al. 2018) studied the potential role of Nostoc paludosum in detoxifying mercury by methylation and they found that the cyanobacterium removes mercury through reduction and volatilization in lieu of methylation.
2.6.3 Demethylation of Methylmercury
It is called also MeHg degradation, which describes the removal of methyl group from organomercurial compounds; utterly forming insoluble mercuric sulfide in the presence of hydrogen sulfide. Two main scenarios addressed demethylation process according to their final byproducts. Namely, in reductive demethylation, methane (CH4) and elemental Hg(0) are generated; yet, oxidative demethylation produces CO2 and Hg(II). Under aerobic conditions and availability of mercury (μM), the reductive demethylation is preferential, while under anoxic circumstances and low existence of mercury (nM), oxidative is favored. Broadly, demethylation process is catalyzed by two successive enzymatic machines. It begins with the catalysis of organomercurial lyase (MerB) that cleaves the C–Hg bond generating CH4 and Hg2+, which is less toxic than methyl mercury by 100-times, followed by mercuric reductase (MerA), which yielding volatile Hg0 after reduction (Lu et al. 2016).
Intriguingly, diverse microbial species among both prokaryotes and eukaryotes exerted their best performance in detoxification of MeHg via demethylation accompanying by other pathways till reaching the safest state. Lu et al. (2016) recruited Geobacter bemidjiensis in Hg detoxification. The data revealed that such iron-reducing bacterium mediated Hg transformations anaerobically through simultaneous accompanying strategies of MeHg yielding, degradation, Hg(II) reduction, and Hg(0) oxidation. The authors proposed that G. bemidjiensis employed a reductive demethylation strategy to degrade MeHg and transform it to a volatile phase by the virtue of MerB and MerA. Meanwhile, under phototrophic conditions, Kritee et al. (2017) studied thoroughly and deeply the demethylation process by Isochrysis galbana using Hg stable isotope. The data highlighted the production of a pool of isotopically HgII confirming the demethylation capacity of algal cells. In this context, Li et al. (2022a, b) examined the capacity of 15 marine microalgae (Diatoms (8 species), Dinoflagellates (4 species), Chlorophyta (2 species), and Chrysophyte) in mercury methylation and demethylation potential in natural environments. The authors found that all examined microalgae lack the ability to methylate inorganic Hg, while six species induced MeHg demethylation at an equivalent level with photodemethylation. Besides, they suggested that demethylation ability could be attributed to the extracellular phyco-secretions (i.e., photo-induce demethylation and thiol biomolecules) in association with bacterial activity.
3 Molecular Aspects-Mediating Microbial Remediation
The versatile resistance mechanisms mediated by microbes, as described in detail in the previous section, are attributed to their enzymatic systems expressed from specific genes (Christakis et al. 2021; Li et al. 2022a, b; Yu and Barkay 2022; Yadav et al. 2023).
3.1 mer Operon-Mediated Inorganic Hg Reduction and Volatilization
Hg(II) and organomercury compounds are detoxified to a volatile less-toxic form (Hg0) by Hg resistance-mediated system (mer) (Priyadarshanee et al. 2022; Yu and Barkay 2022).
The mer operon is distributed widely among bacteria, archaea, and integrated into chromosomal DNA or on mobile genetic elements such as plasmids, transposons, and integrons (Krout et al. 2022; Yu and Barkay 2022). It consists of regulatory proteins such as MerR and MerD, inner membrane-spanning transporter proteins (e.g., MerC, MerE, MerF, MerG, and MerT) that transport Hg2+ to the cytoplasm for reduction by MerA, a protein with reductase activity and MerP (periplasmic Hg(II) scavenging protein) (Agarwal et al. 2019; Li et al. 2022a, b). Priyadarshanee et al. (2022) depicted a schematic representation of bacterial mer operon-mediated Hg detoxification system. MerR and MerD are dual-function transcriptional regulators tightly regulating the mer operon expression through binding to the mer operator/promoter (O/P) region. They function as either activators or repressors in the absence or presence of Hg2+.
Mercury resistance has been classified into two categories according to the mer determinants: narrow-spectrum and broad-spectrum. The broad-spectrum mer determinants (merA and merB genes) resist both organic and inorganic mercury compounds, in contrast to the narrow-spectrum mer determinants (merA), which only resist inorganic mercury (Agarwal et al. 2019; Priyadarshanee et al. 2022).
Upon exposure to ionic Hg2+, the toxic heavy metal (Hg2+) binds to MerP cysteine residues at positions 14 and 17, thus transferring the Hg2+ to the mercury-specific transporter MerT. Consecutively the Hg2+ bounds to MerT, and it is transferred directly to the MerA amino-terminal domain cysteine residues. Thenceforth, the Hg2+ is conveyed to the MerA (mercuric reductase, an NAD(P)H dependent flavin disulfide oxidoreductase) cysteine residues located in the active site then the Hg2+ is reduced into volatile less-toxic form (Hg0). Eventually, the Hg0 passively diffuses from the cellular environment (Zheng et al. 2018; Agarwal et al. 2019; Zhang et al. 2020; Priyadarshanee et al. 2022).
3.2 hgcA Gene-Mediated Methylation
Net production and the tremendous bioaccumulative nature of neurotoxic methylmercury (MeHg) in terrestrial and marine food webs are regulated by microbial processes of methylation and demethylation (Lin et al. 2021; Gionfriddo et al. 2023; Luo et al. 2023). Iron-reducing bacteria, sulfate-reducing bacteria, methanogenic archaea, and fermentative bacteria play a role in the conversion of inorganic mercury into MeHg, which primarily takes place under anaerobic conditions (Christakis et al. 2021; Cardona et al. 2022; Frey et al. 2022). hgcAB is a gene cluster that encodes the proteins, HgcA and HgcB, which are crucial for the methylation process. First, the HgcA, a corrinoid methyltransferase (encoded by the hgcA gene), is a member of the carbon monoxide dehydrogenase/acetyl-CoA synthase delta subunit family, which is involved notably in the methyl transfer reactions. It has a cytosolic corrinoid binding domain (CBD) which transfers the methyl group to Hg(II), and a transmembrane domain (TMD) for the Hg uptake and cellular MeHg efflux. As well, the HgcB, a dicluster ferredoxin (iron-sulfur cluster protein encoded by the hgcB gene) that contains three conserved cysteine residues at the C-terminus. It plays a pivotal role in methylation as an electron donor, thereby reducing the cobalt ion of HgcA besides binding and delivering the Hg(II) to HgcA (Yu and Barkay 2022; Gionfriddo et al. 2023; Lin et al. 2023; Luo et al. 2023).
Apart from Hg methylation, reductive demethylation, and oxidative demethylation are two mechanisms involved in biotic MeHg demethylation. The reductive demethylation occurs under oxic conditions, whereby the mer operon (merB gene) encodes the organomercurial lyase (MerB), which cleaves the C-Hg bond of organomercurials by protonolysis resulting in Hg(II), which is further reduced to generate methane and volatile elemental Hg. As for oxidative demethylation, it takes place mainly in anaerobes lacking mer operon whereas, the Hg2+, CO2, and CH4 are the end products. However, further research is required to identify the genes mediating oxidative demethylation (Tiodar et al. 2021; Yu and Barkay 2022; Luo et al. 2023; Tada et al. 2023).
In addition to the above mechanisms, other pathways assigned for other alternative enzymatic systems were detected to detoxify mercury, especially in transgenic bacteria. As documented by (Shahpiri and Mohammadzadeh 2018), mt-1 and ppk genes encoding metal-scavenging agents (i.e., metallothionein) and polyphosphate kinase adopt an intrinsic role in Hg resistance and accumulation. Interestingly, Ruiz et al. (2011) described that Hg sequestration is governed by metallothionein (mt-1) and polyphosphate kinase (ppk) genes, which are expressed in transgenic bacteria (Escherichia coli/pBSK-P16S-mt1-rpsT and pBSK-P16S-g10-ppk-rpsT). Similarly, Deng and Jia (2011) and Alcántara et al. (2018) reported that the expression of the metallothionein gene and polyphosphate synthesis aided in the Hg removal efficiency of the recombinant strain Rhodopseudomonas palustris and Lactobacillus sp. respectively.
4 Microbial Paradigms of Mercury Bioremediation
Irrespective of whether aerobic or anaerobic conditions, planktonic or aggregated biofilm, the remediating microbes are able to decontaminate Hg pollution using one or more of the previously mentioned pathways either sequentially or simultaneously run. The bacterial remediation, mycoremediation, and even phycoremediation were effectively achieved by a wide range of bacteria, fungi (unicellular or filamentous), and algae (microalgae or macroalgae) as summarized in Table 8.1.
4.1 Extremophiles as Mercury Bioremediators
Extremophiles are microorganisms that possess attractive skills to tolerate high Metals and radionuclides levels, extreme physical (e.g., radiation, temperature), chemical (e.g., acidic/alkaline pH, salinity), and other climate-changing conditions. They can act as bioremediation minor factories (micro factories), by which their performance can even be enhanced and customized for metals and radionuclides elimination (Marques 2018). Extremophilic bacteria and Achaea can simultaneously evolve defense mechanisms against multiple and concurrent extrema (Rothschild and Mancinelli 2001). The secret of their advantageous traits lies behind their speedy-adapting transcriptional and translational scenarios that modulate, either by inhibition or activation, many responses such as anti-oxidative stress, metal-binding/transport, and membrane-permeability (Mukherjee et al. 2012; Dekker et al. 2016). Remarkably, extremophiles cell membranes possess a distinct structure/composition with an inner layer carrying a positive charge that regulates the function of metal transporters and minimizes the entrance of metals and protons, which ultimately manage acidity and metal toxicity (Zhang et al. 2016; Singh and Singh 2017).
Thus, thermophiles, halophiles, radiophiles, and polyextremophiles have been described as important microbial resources for metal bioremediation. Genome sequencing of extremophilic microorganisms such as Sulfolobus solfataricus (Schelert et al. 2013), Laptospirillum ferriphilum (Mi et al. 2011), and the extremely thermoacidophilic Metallosphaera sedula (Auernik et al. 2008) has harbored clusters coding the mercury resistance gene merA. Meanwhile, Acidithiubacillus ferroxidans SUG 2–2 strain was examined for its ability to volatilize mercury in acidic soils contaminated with this metal (pH = 2.5) (Takeuchi et al. 2001; Giovanella et al. 2020). Strikingly, the metallophilic Cupriavidus metallidurans is able to evacuate heavy metals like Cd, Hg, and Cu; concomitantly degrading the poisonous organic contaminant toluene aerobically and anaerobically as being facultative anaerobe (Rojas et al. 2011; Bravo et al. 2020a; Millacura et al. 2018; Alviz-Gazitua et al. 2019).
On the other hand, C. metallidurans strain MSR33, which is a transconjugant derivative of metallophilic C. metallidurans CH34, encoded the environmental plasmid pTP6 that authorized it not only to expand mercury resistance from 2 to 4-folds, relative to the pristine strain, but also reducing inorganic and organic mercurial compounds into Hg(0) under aerobic and anaerobic conditions (Rojas et al. 2011; Bravoa et al. 2020a). Consequently, it was utilized by (Bravo et al. 2020b) in remediating mercury-contaminated agricultural soil via a rotary drum bioreactor as ex situ technology. The data of this study recorded 82% elimination potential and revealed the fostering of nitrogen-fixing and nitrification processes by endogenous communities in the remediated soil. In the same sense, Haloferax sp. HA1, Haloferax sp. HA2, Halobacterium sp. HA3, and Halococcus sp. HA4 are hydrocarbon-utilizing halophilic archaea strains, isolated from a hypersaline coastal area of the Arabian Gulf, they showed resistance to mercury and were able to volatize 42.6, 46.2, 50.8, and 51.6% of Hg (II) (100 ppm), respectively after 8 days, under 45 °C as incubation temperature and 4 M solution (Al-Mailem et al. 2011).
4.2 Plant-Microbes Interaction in Mercury Remediation
The soil is inhabited by various microbial groups; remarkably, the mycorrhizal fungi that occupy rhizosphere are the most distinguished dwellers. Their role in absorption or adsorption of heavy metals triggered through their extending mycelia in the soil, which fosters the increment of plant roots surface area profoundly (Jinet al. 2018; Singh et al. 2021, 2022). Such endophytic mycorrhiza increases the plants’ capability to withstand heavy metal ions through the production of siderophores, organic acids, and chelating agents. However, their ability to acidify the ambient medium and activate metal phosphates is also accounted as another means for fungi to synergize plants. Furthermore, the exopolysaccharides secreted by fungi, upon increasing heavy metal levels, could capture heavy metals on the surface of fungal cell walls, blocking by such way the mobility and bioavailability of heavy metals in the plant.
It is worth mentioning the pivotal role of arbuscular mycorrhizal fungi (AMF), which are mutualistic symbionts colonized in plant roots, in enhancing the uptake of water, micro, and macronutrients from the soil to the plants. Besides, AMF ooze a glycoprotein, called glomalin that consolidate the structure and physicochemical properties of the soil, which increases soil fertility (Herath et al. 2021). Remarkably, in a study conducted by Li et al. (2023), the phytoremediating role of AMF in the mercury (Hg) uptake by rice plants was investigated by using 199Hg isotope. The results of pot trials highlighted the Hg content in rice associated with AMF group ranged from 52.82 to 96.42% lower than that in control group (without AMF inoculation). Let alone the tendency of Hg to accumulate in non-edible parts of the plant like the stems and leaves. Additionally, the accumulated Hg in the grains inoculated by AMF recorded only 20.19%, while AMF non-inoculated grains contained 48.07% of Hg. That resulted in ameliorating in the growth of rice indicated by increasing in some growth indices like biomass and antioxidant enzyme activities.
Interestingly, plant growth-promoting (PGP) bacteria also assist in phytoremediation process through two steps. Firstly, metals chelation and transformation in the soil and subsequently quenching their availability and facilitating the uptake of necessary soil-bound metals. Secondly, enhancing plant growth and further plant biomass by improving vital processes such as nitrogen fixation, phosphorus solubilization, iron sequestration, and production of 1-aminocyclopropane 1-carboxylic acid deaminase and other phytohormones (Gong et al. 2018). In this concern, Rafique et al. (2015) utilized a nitrogen-fixing bacterium, Cronobacter sp., which is root nodules symbiont, as a simultaneous biofertilizer and mercury bioremediator. Moreover, Enterobacter. aerogenes was used to bioremediate mercury and zinc (Ravikumar et al. 2007), chromium (Panda and Sarkar 2012), cadmium, and copper (Huang et al. 2005). This bacterium was found to have a symbiotic relationship with legume plants of Vicia faba, Phaseolus vulgaris, Pisum sativum, and non-legume plants of Cucumis sativus and Lycopersicon esculentum and help the plants to remediate the mercury in the soil (Sorkhoh et al. 2010).
5 Microbial Bioremediation Strategies of Hg
Through utilizing the previous microbial remediation mechanisms, which was achieved by various microbial groups as listed formerly, two distinct strategies can be applied in real environmental locations depending on the site characteristics, at which the process of bioremediation would be executed, including in situ and ex situ.
5.1 In-Situ Bioremediation of Hg
In this strategy, the decontamination process occurred at the polluted place/site itself by using the biological agent in the contaminated site. It encompasses the detoxification of sorbed and dissolved Hg in different places such as saturated soil, unsaturated soil, and groundwater using indigenous microbes either solitary or in a consortium. For example, in aquatic environments, Hg methylation can occur by microbial communities at anoxic–oxic conditions in the soils and sediments, which generally contain organic matter (OM) that is considered a main vector of MeHg and Hg transport from the catchments to the surface of the water. Different microorganisms have been identified as Hg methylators in these environments as mentioned by Schaefer et al. (2014), who detected Hg methylating microbial communities in the tropical swamp at southern Sweden and Florida. In addition, in situ technology involves the acceleration of detoxification process through adjustment and modifying the ambient conditions in place to be more appropriate for wild microorganisms to maximize their performance in the least period. As referred by He et al. (2015), this strategy is predominantly preferred due to its practical easiness, low possibility of pollutant transfer, less expensive and less destructive/intrusive to on-site ecological operations. Nonetheless, the depth of the contaminated site and pollutant concentration could configure one of this strategy’s limitations (Kulshreshtha et al. 2014). Remarkably, in situ strategy could be fulfilled through several types as follows.
5.1.1 Bio-sparing Process
The bio-sparing process occurs by air injection through a pipe found below the table of water to enhance the indigenous microorganisms’ growth by elevating oxygen concentration. Also, it is completely different from the bio-venting process in mixing both the groundwater and soil by air injection in the saturated area, which leads to the movement of volatile organic compounds from the saturated to the unsaturated area, this process is affected by the pollutant biodegradability and characteristics of the soil. It characterizes by easiness and flexibility in designing and constructing the system of air injection points (Jain et al. 2012).
5.1.2 Bio-venting Process
Bio-venting process in which the indigenous soil microorganisms can be stimulated to degrade the targeted contaminant by injecting a small amount of oxygen to increase microbial activity. The air injection occurred in the unsaturated area and also supplemented it with moisture and nutrients. This process could be more efficient in the case of anaerobic bioremediation, also mixing both oxygen and nitrogen will increase the remediation potency (Jain et al. 2012).
5.1.3 Bio-augmentation Process (Bio-vagnification)
Bio-augmentation process aims to increase the native microbiota either by introducing naturally occurring or genetically engineered microorganisms (GMO) to decontaminate the polluted site. This treatment usually uses a microbial consortium that has the ability to produce all the required degradative enzymes and pathways. This process is used to treat soil, ground, and wastewater (Jain et al. 2012). In this sense, the microbial biomass could be used as immobilized material in matrices like silica and alginate to be a suitable biosorbent with suitable porosity and strength. Intriguingly, utilizing Hg-resistant strains in immobilized form to decontaminate polluted sites is evident intensively in numerous laboratory-scale pilot studies (Pepi et al. 2013; Jafari et al. 2015).
However, Vidali (2001) declared the necessity of well-competing ability of exogenous microbes with indigenous populations to ensure the sustainability and successfulness of the bioaugmentation process. Notably, Mahbub et al. (2017a) removed about 60% of soil-bound Hg via bioaugmentation with improved growth of cucumber and lettuce in the bio-augmented soils. He and coworkers demonstrated that insufficient application of bioaugmentation in the soil could be attributed to several reasons including, poor abundancy of Hg in soil, coexistence of mixed contaminants, and improper nutrients supplements, which collectively interfere with biochemical potential and metabolic activity of Hg-remediating microorganisms. Meanwhile, the combination remediating therapy seems to be influential. Nakamura et al. (1999) merged both chemical leaching processes with seeding by Pseudoalteromonas haloplaktis for removing 85% of Hg content found in Minamata Bay sediments.
5.1.4 Bio-stimulation Process (or Accelerated Natural Attenuation)
This process modifies the polluted environment to stimulate indigenous microorganisms for enhancing the bioremediation. This can occur via circulating an inflow of extra nutrients and electron acceptors (e.g., nitrogen, carbon, oxygen, and phosphorus) through contaminated areas (Riseh et al. 2022). In this regard, Feng et al. (2014) demonstrated that uplifting the concentration of sulfate to 59.9 mg/L in inflow water promoted sulfate-reducing bacteria to enhance Hg methylation in the wetland ecosystem. Meanwhile, Winardi et al. (2020) performed a comparative study to remediate Hg from the soil in Ka-limantan Barat-Indonesia. The different groups of sampling plots were exposed to different in situ bioremediating technology. The design included aeration (bioventing), while biostimulation was implemented by nutrient addition and pH flocculation. The experiment was conducted during rainy and dry seasons to detect the seasonal variation effect. The finding of this comparative study unveiled the effective Hg-remediation accounted by 89% within 90 days under rainy conditions, neutral pH with nutrients addition.
5.1.5 Bio-attenuation Process (Natural Process)
Bio-attenuation process involves naturally occurring physical, chemical, and biological processes that decrease the toxicity, volume, mass, and contaminant concentration (Riseh et al. 2022). It could be implemented aerobically, anaerobically, and under simultaneous or sequential or both conditions. Despite its simplicity and lower cost, it suffers from some drawbacks like slow rate and effectiveness only in the case of simple or less complex contaminates. Nevertheless, its combination with other techniques (e.g., biostimulation and bioaugmentation) would boost its efficacy (Goswami et al. 2018).
5.2 Ex-Situ Bioremediation of Hg
The main concept of this strategy depends on the treatment of the contaminated site by soil excavation followed by transferring it away to another place to be remediated. As many biogenic processes that were mediated by microbial activity, the efficiency of this strategy count on different variables like pH, temperature, salinity, pollutants overload, and microbial biomass. This strategy includes five pathways as demonstrating:
5.2.1 Slurry-Phase Bioremediation
This technique depends on contaminated soil excavation and mixing it with water and then transporting the mixed soil to a bioreactor, followed by rubble and stone removal. The used water amount depends on the contamination concentration, type, biodegradation rate, and soil nature. Then, the soil can be separated by centrifugation and filtration, followed by soil drying and transferred to the original site (EPA 2003). In this context, Azoddein (2013) employed Pseudomonas putida (ATCC 49,128) in a field study using petroleum industrial plants (two different locations) in Peninsular, Malaysia contained 1000 ppm of Hg. The results revealed efficient removal recorded 90.5%, 97.27%, and after 96 h for point-1 and point-2, respectively; reflecting by such way the potentiality of such strain in remediating Hg from actual petroleum wastes. In a similar study conducted by Deckwer et al. (2004), mercury-contaminated wastewater was treated by Hg-resistant bacterial biomass in an aerated bioreactor and the data indicated the reduction of Hg2+ to volatile Hg0 gas that was constrained in an activated carbon filter.
5.2.2 Solid Phase Bioremediation
This process includes three steps: soil excavation, followed by transferring the soil to piles, sometimes the soil contains agricultural, organic, and municipal wastes, followed by the biodegradation process stimulation by oxygen supplying through a pipes network to enhance the respiration of microorganisms; subsequently increase microbial activity. This technique requires a large area and takes a long time to be done and complete (Hyman and Dupont 2001).
5.2.3 Land-Farming
This technique focuses on indigenous microbe stimulation and their aerobic manipulation towards contaminants. It mediates by spreading on the soil surface by excavated soil supplementation with minerals and nutrients to stimulate the biodegradation process. It could be described as a superficial process that is restricted to the treatment of the top 10–35 cm of soil (Vidali 2001).
5.2.4 Biopiles
This technique is similar to the land-farming technique but differs in using above-ground piles and used pipes for air injection through the soil; thereby, it could be considered as a merge between composting and landfilling. This process is characterized by its low cost and complete control of aeration, temperature, and nutritional feed, this technique is applied in treating surface-contaminated environments and limiting volatilization of low molecular weight compounds (Verma 2022).
5.2.5 Composting Bioremediation
Composting bioremediation process is similar to land farming in employing contaminated soil excavation and indigenous microorganisms that were stimulated through nutrients feeding and air injection. The main difference lies behind the soil supplementation, which is nonhazardous additives such as animal manure and agricultural residues (e.g., hay, straw, corncobs, etc.). Such organic supplements aid in the eventual distribution of the oxygen through the soil, maintain the moisture content, enrich microbial populations and raise up the compost temperature (Vidali 2001). However, this technique is not suitable for volatile pollutants due to the periodic turning through the process (Hobson et al. 2005). Recently, the combination between composting and carbon-based materials like biochar could ameliorate the bioremediation process by expanding the surface area that is supported by various functional groups and also extending more nutrients and organic matter that facilitate and expedite the metabolic activities of microbes (Gong et al. 2018).
Albeit the suitability of ex-situ technology to scavenge the toxicity of various pollutants within a suitable time frame, excavation and pollutant transfer process remains the major obstacle, which thereafter increases both transfer cost and probability of cross-contamination. Anyway, Mahbub et al. (2017b, c) stated that both methods, namely, in situ and ex situ, are still in the experimental phase of field studies. As the overall process entails accurate knowledge about the nature/concentration of contaminant and perceiving the physicochemical/local biogeochemical features of the contaminated sites and also appraise the multitasking functions of microbes that could be easily harmonized with any modification to achieve their goals. Hence, more investigations and researches are going on in this aspect (abo-Alkasem et al. 2023).
6 Advanced Approaches in Mercury Remediation
As a natural process, bioremediation process gains a lot of attention owing to several merits, including, safety, economic, easiness, and appreciable efficiency. That’s besides the possibility to recover heavy metals, its low requirement of energy/temperature, comparing to other physicochemical means, and so less expensive operation cost. Additionally, the feasibility to be executed on site avoids by such a way disrupting of normal activities and transportation step that consequently leads to additional risk (Gupta et al. 2016; Volarić et al. 2021). But the geochemical conditions, nutrients availability, physicochemical properties of contaminant, and contaminant concentration are collectively controlling the microbial performance and facilitate/retard the clean-up process (Jeyakumar et al. 2023). Meanwhile, the slow growth rate of microbes, longer growth time, the potential of more persistent/toxic byproduct, and regulatory uncertainty are habitually the main drawbacks of microbial remediation, which triggers researchers and technologists to adopt modern and advanced tools to speed up bioremediation and augment its efficacy as referred by Vidali (2001) and Tripathi and Ram (2018).
6.1 Synthetic Biology and Genetically Engineered Microorganisms (GMOs)
As a naturally inspired process rather than artificially designed, the genetic exchange among microorganisms promoted the researchers to invest recombinant DNA technology in bioremediation. Microbes are engineered with intendedly inserted desired traits such as metal homeostasis, higher metabolic rate, tolerance of biotic/abiotic stressors and overexpression of meta-chelators, uptake regulator, transport, and degradative genes. Thereby, GMOs act as smart cell factories that utilize risky unwanted wastes in an enhanced manner in contaminated groundwater, soil, and active sludge (Volarić et al. 2021). Hence, Tay et al. (2017) cloned MerR promotor of Shigella flexneri plasmid to bacterial biofilm. Interestingly, MerR is responsible for curli nanofibers synthesis that facilitates sequestration of mercury. Nonetheless, the safe release of foreign modified organisms in the ecosystem still symbolizes a cryptic matter and may cause unmeasurable, unaccounted, and unreacted adverse impacts on the natural structure including functional microbial community composition and diversity alterations as highlighted by Sarao and Kaur (2021) Volarić et al. (2021). In this regard, Xue et al. (2022a, b) designed and developed a self-controlled genetic circuit of Pseudomonas putida KT2440 and Escherichia coli cells, respectively, that exhibit superior performance in mercury sensing and adsorption, followed by programmable killing stage by utilizing a cell suicide module.
6.2 Metagenomics
It is a technical term used to characterize the genetic profile of microorganisms in any ecosystem. It gives detailed information about the response of ecosystems members against environmental changes induced by any pollution by bestowing the sequence and functions of genomes concerning adaptive microorganisms in the site community (Malla et al. 2018; Jaiswal and Shukla 2020) via such advanced technology, the total DNA of any examined site (soil, wastewater, sludge, etc.) was extracted, which serve as DNA of all indigenous microbes collectively present in the examined site that terms site metagenome. Once, the extraction step was fulfilled, the construction of the DNA library followed to facilitate the screening of the target genes, which finalized by the intense expression of the target gene product (Volarić et al. 2021). In fact, Jaiswal et al. (2019) constructed mer operon metagenomic library of Panipat, which is one of the well-known sites contaminated with mercury, by utilizing E. coli as a host. The promising results indicated that the clones displayed the potentiality for mercury tolerance and volatilization by 90 ppm and 91.89%, respectively. Additionally, the efficiency of mercury remediation could be elevated by encapsulating the clones in polyacrylamide gel and alginate microspheres, which also enable their reusability.
6.3 Biosurfactants
Biosurfactants are surface-active compounds produced microbially and characterized by their amphiphilic nature (i.e., encompass both hydrophobic and hydrophilic moieties). They have been utilized recently as alternatives to synthetic surfactants by the dint of their biodegradability, biocompatibility, biosafety, bioavailability, specificity, withstanding extreme conditions, and higher surface and interfacial activity. Biosurfactants possess variable chemical structures; exhibiting a broad range of chelating capabilities with different metals (Jeyakumar et al. 2023). The biosurfactant produced by Bacillus sp. MSI 54 was characterized and its chemical structure of anionic nature lipopeptide was identified by FTIR and nuclear magnetic resonance spectroscopy (NMR). Its chelating capacity to mercury from fresh vegetables and wastewater was detected by atomic absorption spectroscopy, which was assessed by 75.5% (Ravindran et al. 2020).
6.4 Combined Remediation
To compensate the limitations of each sole method, it is recommended to hybridize two or more approaches together. Namely, utilize a consortium of different microorganisms, wherein each can remediate by a pathway differs from the pathway used by the others. Additionally, the amalgament between physical/chemical, physical/biological, or chemical/biological remediation approaches could be also grouped into the same concept (Gong et al. 2018). Interestingly, phyoextraction combined with electrokinetic remediation (Mao et al. 2016), chemical stabilization-assisted soil washing (Zhai et al. 2018), thermal treatment combined with a chelating agent, Ma et al. (2015) are promising paradigms on such combined biotechnology.
In this context, innumerable materials, fabricated either chemically or physically, were employed as immobilizing matrices for entrapping mercury-remediating microbes. In recent studies, innovative matrices varied in its chemical structure and physical properties, oscillated in its origin from natural to synthetic, and are exploited as ecofriendly, cost-effective, high surface area, porous-structure adsorbents that impregnated with microbial cells with degradative/remediating traits (Gong et al. 2018).
6.4.1 Carbon-Based Materials
Waste-derived materials, which represent an environmental burden, were directed to adopt bioremediation purposes (Beckers et al. 2019; Gong et al. 2018; O'Connor et al. 2018; Liu et al. 2022). However, such materials could be classified according to their feedstock into industrial waste-derived substance and biomass-derived materials (e.g., biochar, activated carbon, graphene, graphene oxide, etc.), while coal fly ash (CFA) is categorized among the most common industrial waste-derived substance (Wang et al. 2020; Liu et al. 2022). Strikingly, mercury‐volatilizing bacteria like Pseudomonas sp. DC‐B1 and Bacillus sp. DC‐B2 immobilized on 4% sawdust biochar diminished mercury phytoavailability in lettuce shoots, roots, and in soil by 2.0–48.6%, 12.3–27.4%, and 24.8–57.8%, respectively, within 56 days without changing community compositions of the soil microbial ecosystem; reflecting the successful hybridization of bacteria-biochar as green’ additives (Chang et al. 2019). Moreover, Yan et al. (2018) used graphene oxide as a carrying matrix for Enterococcus avium and the data revealed the improvement of remediating potential of this sulfate-reducing bacterium by accelerating the growth rate and maximizing the removal rate of sulfate and metal.
6.4.2 Polymers
Polymers, especially those that exhibit adsorptive capacity, also gained colossal popularity in remediation technology owing to their chemical stability, pore size, and considerable surface area. Acrylamide is one of the most common synthetic polymers that distinguished by its higher potential in adsorption of Hg(II); however, its microbial toxicity and environment-unfriendly restrict its utilization in immobilization of microbes (Wang et al. 2020). As a consequence, the attention was directed to employ natural polymers to guarantee the effectiveness of the remediation process in a sustainable manner. Wherein, chitosan polymer and its functionalized/modified co-polymer show multidimensional properties with tunable adsorptive capability, in particular to Hg as revealed by Goci et al. (2023). Upon synthesizing microbeads of chitosan/algal (Cladophora sp.) composite, the metals sorption capacity elevated more than each ingredient individually (Sargın et al. 2016). More so, McCarthy et al. (2017) entrapped the cells of Pseudomonas veronii in a xanthan gum-based biopolymer and coated them with zeolite granules. Such an innovative method employed combined remediation in a tripartite way (i.e., carbon-based material, polymer, and degradative bacteria), which exerted superior performance in response to mercury volatilization with increased viability for 16 weeks at least.
6.4.3 Nanomaterials
The miniaturization in dimensions and increasing the surface area of materials elevate their functionality, mechanical, electrical, chemical, and adsorptive features, which trigger the utilization of materials in nanoscale dimension sign for innovative products with promising applications. A vast array of nanomaterials (e.g., nanoparticles, nanocomposites, nanosheets, carbon nanotubes, etc.) emphasized their efficiency in Hg scavenging as stated by Wang et al. (2020). The recruitment of nanoadsorbents (e.g., porous silica, titania, etc.) as carriers to immobilize microorganisms proved its efficiency in recent years (Velkova et al. 2018). In this regard, the chitosan‐coated Fe3O4 nanoparticles and TiO2 nanoparticles were immobilized with Saccharomyces cerevisiae biomass to mitigate the toxicity of heavy metals (Choudhury et al. 2017; Peng et al. 2010). Similarly, Akhtar et al. (2021) hybridized the cells of Bacillus cereus and Lysinibacillus macrolides with ZnO nanoparticles on a rice crop irrigated with heavy metals contaminated water in a synergistic manner. Such hybridization maximized significantly the removal efficacy via the synergistic mechanism of both remediating bacterial consortium and nanoparticles. Furthermore, their hybridization enhanced the plant growth and its tolerance index, while lessened the bioaccumulation index and metallothionine content. On the other hand, Ozdemir et al. (2017) used nanodiamond as biosorbent carriers for thermophilic Bacillus altitudinis to eliminate Hg2+ along with other metals from food sample. The data unveiled the simultaneous preconcentration-separation of examined metals with 0.3 ml/min as an optimum flow rate under pH 6 and biosorption capacity assessed by 19.5 mg/g.
7 Concluding Remarks and Future Outlook
The continuous and regular disposing of various effluents containing mercury into water bodies increases the likelihood of their access and persistence in the food chain through agricultural crops and aquatic animals, leading then to the bioaccumulating and bioaugmentation in human bodies. However, traditional remediation technologies displayed some significant disadvantages, bioremediation techniques could compensate them in an environmentally friendly, least destructive, biosafe, and cost-efficient way. Bioremediation strategies seemed to be convenient to diverse environmental circumstances, via both in situ and ex situ approaches as explained herein. This chapter addressed the main principles, strategies, effectiveness of different microbial forms, and advanced tools of mercury microbial remediation, which had been studied. However, various microbial genera possess varied metabolic prerequisites and showed disparate efficiency in the bioremoval process, which can also differ contingent upon the nature of contaminated sites, concentration of mercury dumping off in the field and also seasonal changes. Albeit efficacy, more and deep investigations entail in the following aspects: (1) A key challenge is appropriate screening and selection of novel species that exhibit characteristic metabolic traits and advantageous physiological properties in accelerating mercury removal at both lab and commercial scales. Interestingly, the extremophilic dwellers (e.g., Archaea) are the promising and potent category recommended, owing to their metabolic versatility, adaptability, and tolerance, for xenobiotics remediation. (2) It is highly desired to employ novel OMICs tools (proteomics, metabolomics, genomics, transcriptomics, and fluxomics) combined with bioinformatics (e.g., in silico) and computational platforms. Such integrative ways of these new techniques could predict and optimize mechanism-based models to uplift the removal performance. (3) It is noteworthy to develop monitoring approaches to trace the stability of the remediated phase and residues of Hg in the contaminated field. (4) In this context, it is crucial also to monitor the performance of remediating microbes either GMO or native, especially Hg don’t coexist in the environment solitary but among multiple pollutants either organic or inorganic, which actually influence adaptive behavior, removal rate, survival time of remediating microbes. (5) More comprehensive studies in combined remediation technology necessitate new porous crystalline nanobiosorbent materials such as nanobiosurfactants, metal–organic frameworks (MOFs), and covalent organic frameworks (COFs), which act as carriers or immobilization matrices. Functionalization and chemical modifications of such innovative biosorbents with various functional groups will instigate their chemical stability and adsorptive behavior. (6) The recovery, reusability, and stability of microbe-MOFs/COFs composites should be conducted more through and the impact of harsh environmental conditions like temperature, pH, and salinity should also be operated. (7) Translating the obtained results accurately to full-scale operation and perceiving the whole image, namely, industrial applications and field scale with a precise assessment of expenditure through collaborative groups of researchers, technologists, health specialists, governmental institutions encourage remediation companies to apply long-term sustainable approaches efficiently.
Abbreviations
- ATSDR:
-
Agency for Toxic Substances and Disease Registry
- EPA:
-
US Environmental Protection Agency
- GMOs:
-
Genetically modified organisms
- EPS:
-
Extracellular polymeric substances
- FTIR:
-
Fourier transform infrared analysis
- TEM:
-
Transmission electron microscope
- SEM:
-
Scanning electron microscope
- EDX:
-
Energy Dispersive X-ray
- SRB:
-
Sulfate-reducing bacteria
- MICP:
-
Microbial-induced carbonate precipitation
- AMF:
-
Mycorrhizal fungi
- PGP:
-
Plant growth promoting
- OM:
-
Organic matter
- DNA:
-
Deoxyribonucleic acid
- NMR:
-
Nuclear magnetic resonance spectroscopy
- CFA:
-
Coal fly ash
- MOFs:
-
Metal–organic frameworks
- COFs:
-
Covalent organic frameworks
References
Abo-Alkasem M, Nemat H, Hassan M, Mostafa A (2023) Microbial bioremediation as a tool for the removal of heavy metals. Bull Natl Res Cent 47:31
Agarwal M, Rathore RS, Jagoe C, Chauhan A (2019) Multiple lines of evidences reveal mechanisms underpinning mercury resistance and volatilization by Stenotrophomonas sp. MA5 isolated from the Savannah River Site (SRS), USA. Cells 8:1–14
Aiking H, Govers H, Van’t Riet J (1985) Detoxification of mercury, cadmium, and lead in Klebsiella aerogenes NCTC 418 growing in continuous culture. Appl Environ Microbiol 50(5):1262–1267
Akhtar N, Khan S, Rehman S, Rehman Z, Khatoon A, Rha E, Jamil M (2021) Synergistic effects of zinc oxide nanoparticles and bacteria reduce heavy metals toxicity in rice (Oryza sativa L.) plant. Toxics 9(5):113
Alcántara C, Coll-Marqués JM, Jadán-Piedra C, Vélez D, Devesa V, Zúñiga M, Monedero V (2018) Polyphosphate in Lactobacillus and its link to stress tolerance and probiotic properties. Front Microbiol 9:1–10
Al-Mailem DM, Al-Awadhi H, Sorkhoh NA, Eliyas M, Radwan SS (2011) Mercury resistance and volatilization by oil utilizing haloarchaea under hypersaline conditions. Extremophiles 15:39–44
Alviz-Gazitua P, Fuentes-Alburquenque S, Rojas LA, Turner RJ, Guiliani N, Seeger M (2019) The response of Cupriavidus metallidurans CH34 to cadmium involves inhibition of the initiation of biofilm formation, decrease in intracellular c-di-GMP levels, and a novel metal regulated phosphodiesterase. Front Microbiol 10:1499
Amin A, Latif Z (2011) Isolation and characterization of H2S producing yeast to detoxify mercury containing compounds. Int J Microbiol 2:517–525
Amin A, Naveed M, Sarwar A, Rasheed S, Saleem HG, Latif Z, Bechthold A (2022) In vitro and in silico studies reveal Bacillus cereus AA-18 as a potential candidate for bioremediation of mercury-contaminated wastewater. Front Microbiol 13:847806
Anthony E (2014) Bioremediation of mercury by biofilm forming mercury resistant marine bacteria. Doctoral dissertation. National Institute of Technology, Rourkela, India
Anuar SN, Othman F, Chay TC, Nasir NA (2020) Biosorption of mercury ion (Hg2+) using live and dead cells of Rhizopus oryzae and Aspergillus niger: characterization, kinetic and isotherm studies. J Pure Appl Microbiol 14(3):1749–1760
Auernik KS, Maezato Y, Blum PH, Kelly RM (2008) The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl Environ Microbiol 74(3):682–692
Azoddein AAM (2013) Mercury removal from petroleum based industries wastewater by Pseudomonas Putida ATCC 49128 in membrane bioreactor. Doctoral dissertation, UMP
Balan BM, Shini S, Krishnan KP, Mohan M (2018) Mercury tolerance and biosorption in bacteria isolated from Ny-Ålesund, Svalbard. Arctic J Basic Microbiol 58(4):286–295
Barkay T, Miller SM, Summers AO (2003) Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev 27:355–384
Beckers F, Awad YM, Beiyuan J, Abrigata J, Mothes S, Tsang DC, Ok YS, Rinklebe J (2019) Impact of biochar on mobilization, methylation, and ethylation of mercury under dynamic redox conditions in a contaminated floodplain soil. Environ Int 127:276–290
Bouffard A, Amyot M (2009) Importance of elemental mercury in lake sediments. Chemosphere 74(8):1098–1103
Bravo G, Vega-Celedón P, Gentina JC, Seeger M (2020a) Effects of mercury II on Cupriavidus metallidurans strain MSR33 during mercury bioremediation under aerobic and anaerobic conditions. Process 8(8):893
Bravo G, Vega-Celedón P, Gentina JC, Seeger M (2020b) Bioremediation by Cupriavidus metallidurans strain MSR33 of mercury-polluted agricultural soil in a rotary drum bioreactor and its effects on nitrogen cycle microorganisms. Microorganisms 8(12):1952
Cardona GI, Escobar MC, Acosta-González A, Marín P, Marqués S (2022) Highly mercury-resistant strains from different Colombian Amazon ecosystems affected by artisanal gold mining activities. Appl Microbiol Biotechnol 106:2775–2793
Chan CS, Fakra SC, Edwards DC, Emerson D, Banfield JF (2009) Iron oxyhydroxide mineralization on microbial extracellular polysaccharides. Geochim Cosmochim Acta 73(13):3807–3818
Chang J, Yang Q, Dong J, Ji B, Si G, He F, Li B, Chen J (2019) Reduction in Hg phytoavailability in soil using Hg-volatilizing bacteria and biochar and the response of the native bacterial community. Microbiol Biotechnol 12(5):1014–1023
Chang J, Shi Y, Si G, Yang Q, Dong J, Chen J (2020) The bioremediation potentials and mercury(II)-resistant mechanisms of a novel fungus Penicillium spp. DC-F11 isolated from contaminated soil. J Hazard Mater 396:122638
Chen SC, Lin WH, Chien CC, Tsang DC, Kao CM (2018) Development of a two-stage biotransformation system for mercury-contaminated soil remediation. Chemosphere 200:266–273
Choudhury PR, Bhattacharya P, Ghosh S, Majumdar S, Saha S, Sahoo GC (2017) Removal of Cr (VI) by synthesized titania embedded dead yeast nanocomposite: optimization and modeling by response surface methodology. J Environ Chem Eng 5(1):214–221
Christakis CA, Barkay T, Boyd ES (2021) Expanded diversity and phylogeny of mer genes broadens mercury resistance paradigms and reveals an origin for MerA among thermophilic Archaea. Front Microbiol 12:1–20
Colombo MJ, Ha J, Reinfelder JR, Barkay T, Yee N (2014) Oxidation of Hg(0) to Hg(II) by diverse anaerobic bacteria. Chem Geol 363:334–340
Deckwer WD, Becker FU, Ledakowicz S, Wagner-Döbler I (2004) Microbial removal of ionic mercury in a three-phase fluidized bed reactor. Environ Sci Technol 38(6):1858–1865
Dekker L, Arsène-Ploetze F, Santini JM (2016) Comparative proteomics of Acidithiobacillus ferrooxidans grown in the presence and absence of uranium. Res Microbiol 167(3):234–239
Deng X, Jia P (2011) Construction and characterization of a photosynthetic bacterium genetically engineered for Hg2+ uptake. Bioresour Technol 102:3083–3088
Eltarahony M, Zaki S, Abd-El-Haleem D (2020) Aerobic and anaerobic removal of lead and mercury via calcium carbonate precipitation mediated by statistically optimized nitrate reductases. Sci Rep 10(1):4029
Eltarahony M, Kamal A, Zaki S, Abd-El-Haleem D (2021) Heavy metals bioremediation and water softening using ureolytic strains Metschnikowia pulcherrima and Raoultella planticola. J Chem Technol Biotechnol 96(11):3152–3165
EPA (2003) EPA protocol for the review of existing national primary drinking water regulations, united states environmental protection agency office of water office of groundwater and drinking water standards and risk management division targeting and Analysis branch1200 pennsylvania avenue, NW (4607M) Washington, DC 20460
Essa AM, Macaskie LE, Brown NL (2002) Mechanisms of mercury bioremediation. Biochem Soc Trans 30:4
Farraji H, Zaman NQ, Tajuddin R, Faraji H (2016) Advantages and disadvantages of phytoremediation: a concise review. Int J Env Tech Sci. 2:69–75
Feng S, Ai Z, Zheng S, Gu B, Li Y (2014) Effects of dryout and inflow water quality on mercury methylation in a constructed wetland. Wat Air Soil Poll 225:1–1
Fleming EJ, Mack EE, Green PG, Nelson DC (2006) Mercury methylation from unexpected sources: molybdate-inhibited freshwater sediments and an iron-reducing bacterium. Appl Environ Microbiol 72(1):457–464
Franco MW, Mendes LA, Windmöller CC, Moura KA, Oliveira LA, Barbosa FA (2018) Mercury methylation capacity and removal of Hg species from aqueous medium by cyanobacteria. Wat Air Soil Poll 229:1–2
François F, Lombard C, Guigner JM, Soreau P, Brian-Jaisson F, Martino G, Vandervennet M, Garcia D, Molinier AL, Pignol D, Peduzzi J (2012) Isolation and characterization of environmental bacteria capable of extracellular biosorption of mercury. Appl Environ Microbiol 78(4):1097–1106
Frey B, Rast BM, Qi W, Stierli B, Brunner I (2022) Long-term mercury contamination does not affect the microbial gene potential for C and N cycling in soils but enhances detoxification gene abundance. Front Microbiol 13:1–18
Gilmour CC, Bullock AL, McBurney A, Podar M, Elias DA (2018) Robust mercury methylation across diverse methanogenic archaea. Mbio 9(2):e02403-e2417
Gionfriddo CM, Soren AB, Wymore AM, Hartnett DS, Podar M, Parks JM, Elias DA, Gilmour CC (2023) Transcriptional control of hgcAB by an ArsR-like regulator in Pseudodesulfovibrio mercurii ND132. Appl Environ Microbiol 89:1–23
Giovanella P, Vieira GA, Otero IVR, Pellizzer EP, de Jesus Fontes B, Sette LD (2020) Metal and organic pollutants bioremediation by extremophile microorganisms. Hazmats 382:121024
Goci MC, Leudjo, Taka A, Martin L, Klink MJ (2023) Chitosan-based polymer nanocomposites for environmental remediation of mercury pollution. Polymer J15(3):482
Gong Y, Zhao D, Wang Q (2018) An overview of field-scale studies on remediation of soil contaminated with heavy metals and metalloids: technical progress over the last decade. Water Res 147:440–460
Goswami M, Chakraborty P, Mukherjee K, Mitra G, Bhattacharyya P, Dey S, Tribedi P (2018) Bioaugmentation and biostimulation: a potential strategy for environmental remediation. J Microbiol Exp 6(5):223–231
Grégoire DS, Poulain AJ (2014) A little bit of light goes a long way: the role of phototrophs on mercury cycling. Metallomics 6(3):396–407
Grégoire DS, Poulain AJ (2016) A physiological role for HgII during phototrophic growth. Nat Geosci 9(2):121–125
Grégoire DS, Lavoie NC, Poulain AJ (2018) Heliobacteria reveal fermentation as a key pathway for mercury reduction in anoxic environments. Environ Sci Technol 52(7):4145–4153
Groudev S, Georgiev P, Spasova I, Nicolova M (2014) Decreasing the contamination and toxicity of a heavily contaminated soil by in situ bioremediation. J Geochem Explor 144:374–379
Gupta A, Joia J, Sood A, Sood R, Sidhu C, Kaur G (2016) Microbes as potential tool for remediation of heavy metals: a review. J Microbol Biochem Technol 8(4):364–372
Håkansson T, Suer P, Mattiasson B, Allard B (2008) Sulphate reducing bacteria to precipitate mercury after electrokinetic soil remediation. Int J Environ Sci Technol 5:267–274
He F, Gao J, Pierce E, Strong PJ, Wang H, Liang L (2015) In situ remediation technologies for mercury-contaminated soil. Environ Sci Poll Res 22:8124–8147
Herath BM, Madushan KW, Lakmali JP, Yapa PN (2021) Arbuscular mycorrhizal fungi as a potential tool for bioremediation of heavy metals in contaminated soil. World J Adv Res Rev 10(3):217–228
Hobson AM, Frederickson J, Dise NB (2005) CH4 and N2O from mechanically turned windrow and vermicomposting systems following in-vessel pre-treatment. Waste Manag Res 25(4):345–352
Huang Q, Chen W, Xu L (2005) Adsorption of copper and cadmium by Cu-and Cd-resistant bacteria and their composites with soil colloids and kaolinite. Geomicrobiol J 22(5):227–236
Hyma M, Dupont RR (2001) Groundwater and soil remediation: process design and cost estimating of proven technologies. American Society of Civil Engineers, Chicago
Jafari SA, Cheraghi S, Mirbakhsh M, Mirza R, Maryamabadi A (2015) Employing response surface methodology for optimization of mercury bioremediation by Vibrio parahaemolyticus PG02 in coastal sediments of Bushehr, Iran. CLEAN–Soil Wat Air Soil Poll 43(1):118–126
Jain AN, Udayashankara TH, Lokesh KS (2012) Review on bioremediation of heavy metals with microbial isolates and amendments on soil residue. Int J Sci Res 6:2319–7064
Jaiswal S, Shukla P (2020) Alternative strategies for microbial remediation of pollutants via synthetic biology. Front Microbiol 19:11
Jaiswal G, Singh R, Porwal S (2019) Bioremediation of mercury through encapsulation of the clone carrying meroperon. J Pure Appl Microbiol 13(1):553–561
Jeyakumar P, Debnath C, Vijayaraghavan R, Muthuraj M (2023) Trends in bioremediation of heavy metal contaminations. Environ Eng Res 28(4)
Jin Y, Luan Y, Ning Y, Wang L (2018) Effects and mechanisms of microbial remediation of heavy metals in soil: a critical review. Appl Sci 8(8):1336
Kim YM, Chung JY, An HS, Park SY, Kim BG, Bae JW, Han M, Cho YJ, Hong YS (2015) Biomonitoring of lead, cadmium, total mercury, and methylmercury levels in maternal blood and in umbilical cord blood at birth in South Korea. Int J Environ Res Public Health 12(10):13482–13493
Kim Y, Abdilla B, Yuan K, De Andrade V, Sturchio NC, Lee SS, Fenter P (2021) Replacement of calcium carbonate polymorphs by cerussite. ACS Earth Space Chem 5(9):2433–2441
Kritee K, Motta LC, Blum JD, Tsui MT, Reinfelder JR (2017) Photomicrobial visible light-induced magnetic mass independent fractionation of mercury in a marine microalga. ACS Earth Space Chem 2(5):432–440
Krout IN, Scrimale T, Vorojeikina D, Boyd ES, Rand MD (2022) Organomercurial lyase (merB)-mediated demethylation decreases bacterial methylmercury resistance in the absence of mercuric reductase (merA). Appl Environ Microbiol 88:1–14
Kulshreshtha A, Agrawal R, Barar M, Saxena S (2014) A review on bioremediation of heavy metals in contaminated water. IOSR J Environ Sci Toxicol Food Technol 8(7):44–50
Li Y, Li D, Song B, Li Y (2022a) The potential of mercury methylation and demethylation by 15 species of marine microalgae. Water Res 215:118266
Li X, Yang Z, Zhang G, Si S, Wu X, Cai L (2022b) Plasmid genomes reveal the distribution, abundance, and organization of mercury-related genes and their co-distribution with antibiotic resistant genes in gamma proteobacteria. Genes 13:1–13
Li XC, Yang ZZ, Zhang C, Wei JJ, Zhang HQ, Li ZH, Ma C, Wang MS, Chen JQ, Hu JW (2019) Effects of different crystalline iron oxides on immobilization and bioavailability of Cd in contaminated sediment. J Chem Eng 373:307–317
Li X, Zhou M, Shi F, Meng B, Liu J, Mi Y, Dong C, Su H, Liu X, Wang F, Wei Y (2023) Influence of arbuscular mycorrhizal fungi on mercury accumulation in rice (Oryza sativa L.): from enriched isotope tracing perspective. Ecotoxicol Environ Saf 255:114776
Lin H, Hurt RA Jr, Johs A, Parks JM, Morrell-Falvey JL, Liang L, Elias DA, Gu B (2014) Unexpected effects of gene deletion on interactions of mercury with the methylation-deficient mutant Δ hgcAB. Environ Sci Technol Lett 1(5):271–276
Lin H, Ascher DB, Myung Y, Lamborg CH, Hallam SJ, Gionfriddo CM, Holt KE, Moreau JW (2021) Mercury methylation by metabolically versatile and cosmopolitan marine bacteria. ISME J 15:1810–1825
Lin H, Moody ER, Williams TA, Moreau JW (2023) On the origin and evolution of microbial mercury methylation. Genome Biol Evol 15:1–12
Liu D, Yang X, Zhang L, Tang Y, He H, Liang M, Tu Z, Zhu H (2022) Immobilization of biomass materials for removal of refractory organic pollutants from wastewater. Int J Environ Res Public Health 19(21):13830
Lu X, Liu Y, Johs A, Zhao L, Wang T, Yang Z, Lin H, Elias DA, Pierce EM, Liang L, Barkay T (2016) Anaerobic mercury methylation and demethylation by Geobacter bemidjiensis Bem. Environ Sci Technol 50(8):4366–4373
Luo H, Cheng Q, He D, Sun J, Li J, Pan X (2023) Recent advances in microbial mercury methylation: a review on methylation habitat, methylator, mechanism, and influencing factor. Process Safe Environ Prot 170:286–296
Ma F, Peng C, Hou D, Wu B, Zhang Q, Li F, Gu Q (2015) Citric acid facilitated thermal treatment: an innovative method for the remediation of mercury contaminated soil. Hazmats 300:546–552
Mahbub KR, Krishnan K, Naidu R, Andrews S, Megharaj M (2017a) Mercury toxicity to terrestrial biota. Ecol Indic 74:451–462
Mahbub KR, Subashchandrabose SR, Krishnan K, Naidu R, Megharaj M (2017b) Mercury alters the bacterial community structure and diversity in soil even at concentrations lower than the guideline values. Appl Microbiol Biotechnol 101:2163–2175
Mahbub KR, Bahar MM, Labbate M, Krishnan K, Andrews S, Naidu R, Megharaj M (2017c) Bioremediation of mercury: not properly exploited in contaminated soils. Appl Microbiol Biotechnol 101:963–976
Malla MA, Dubey A, Yadav S, Kumar A, Hashem A, Abd_Allah EF (2018) Understanding and designing the strategies for the microbe-mediated remediation of environmental contaminants using omics approaches. Front Microbiol 4:9
Mao X, Han FX, Shao X, Guo K, McComb J, Arslan Z, Zhang Z (2016) Electro-kinetic remediation coupled with phytoremediation to remove lead, arsenic and cesium from contaminated paddy soil. Ecotoxicol Environ Saf 125:16–24
Marques CR (2018) Extremophilic microfactories: applications in metal and radionuclide bioremediation. Front Microbiol 9:1191
McCarthy D, Edwards GC, Gustin MS, Care A, Miller MB, Sunna A (2017) An innovative approach to bioremediation of mercury contaminated soils from industrial mining operations. Chemosphere 184:694–699
Millacura FA, Janssen PJ, Monsieurs P, Janssen A, Provoost A, Van Houdt R, Rojas LA (2018) Unintentional genomic changes endow Cupriavidus metallidurans with an augmented heavy-metal resistance. Genes 9(11):551
Mokone JG, Tutu H, Chimuka L, Cukrowska EM (2018) Optimization and characterization of Cladophora sp. alga immobilized in alginate beads and silica gel for the biosorption of mercury from aqueous solutions. Wat Air Soil Poll 229:1–14
Mukherjee A, Wheaton GH, Blum PH, Kelly RM (2012) Uranium extremophily is an adaptive, rather than intrinsic, feature for extremely thermoacidophilic Metallosphaera species. Proc Natl Acad Sci 109(41):16702–16707
Nakamura K, Hagimine M, Sakai M, Furukawa K (1999) Removal of mercury from mercury-contaminated sediments using a combined method of chemical leaching and volatilization of mercury by bacteria. Biodegradation 10:443–447
Nurfitriani S, Arisoesilaningsih E, Nuraini Y, Handayanto E (2020) Bioaccumulation of mercury by bacteria isolated from small scale gold mining tailings in Lombok, Indonesia. J Ecol Eng 21(6)
O’Connor D, Peng T, Li G, Wang S, Duan L, Mulder J, Cornelissen G, Cheng Z, Yang S, Hou D (2018) Sulfur-modified rice husk biochar: a green method for the remediation of mercury contaminated soil. Sci Total Environ 621:819–826
Okoh MP, Olobayetan IW, Machunga-Mambula SS (2018) Bioleaching, a technology for metal extraction and remediation: mitigating health consequences for metal exposure. Int J Develop Sustain 7:2103–2118
O'rear DJ, Cooper RE, Sheu FR, Belue JT (2014) Inventors; Chevron USA Inc, assignee. Process, method, and system for removing mercury from fluids. United States patent US 8,790,427
Ozdemir S, Kilinc E, Celik KS, Okumus V, Soylak M (2017) Simultaneous preconcentrations of CO2+, Cr6+, Hg2+ and Pb2+ ions by Bacillus altitudinis immobilized nanodiamond prior to their determinations in food samples by ICP-OES. Food Chem 215:447–453
Panda J, Sarkar P (2012) Bioremediation of chromium by novel strains Enterobacter aerogenes T2 and Acinetobacter sp. PD 12 S2. Environ Sci Poll Res 19:1809–1817
Pan-Hou HS, Imura N (1981) Role of hydrogen sulfide in mercury resistance determined by plasmid of Clostridium cochlearium T-2. Arch Microbiol 129:49–52
Peng Q, Liu Y, Zeng G, Xu W, Yang C, Zhang J (2010) Biosorption of copper(II) by immobilizing Saccharomyces cerevisiae on the surface of chitosan-coated magnetic nanoparticles from aqueous solution. JHM Lett 177(1–3):676–682
Pepi M, Focardi S, Tarabelli A, Volterrani M, Focardi SE (2013) Bacterial strains resistant to inorganic and organic forms of mercury isolated from polluted sediments of the Orbetello Lagoon, Italy, and their possible use in bioremediation processes. In: E3S web of conferences.1, 31002. EDP Sciences
Poulin BA, Aiken GR, Nagy KL, Manceau A, Krabbenhoft DP, Ryan JN (2016) Mercury transformation and release differs with depth and time in a contaminated riparian soil during simulated flooding. Geochim Cosmochim Acta 176:118–138
Priyadarshanee M, Chatterjee S, Rath S, Dash HR, Das S (2022) Cellular and genetic mechanism of bacterial mercury resistance and their role in biogeochemistry and bioremediation. JHM Lett 423:1–20
Rafique A, Amin A, Latif Z (2015) Screening and characterization of mercury-resistant nitrogen fixing bacteria and their use as biofertilizers and for mercury bioremediation. Pak J Zool 47(5):1271–1277
Ravikumar S, Williams GP, Shanthy S, Gracelin NAA, Babu S, Parimala PS (2007) Effect of heavy metals (Hg and Zn) on the growth and phosphate solubilising activity in halophilic phosphobacteria isolated from Manakudi mangrove. J Environ Biol 28(1):109–114
Ravindran A, Sajayan A, Priyadharshini GB, Selvin J, Kiran GS (2020) Revealing the efficacy of thermostable biosurfactant in heavy metal bioremediation and surface treatment in vegetables. Front Microbiol 10(11):222
Riseh RS, Vazvani MG, Hajabdollahi N, Thakur VK (2022) Bioremediation of heavy metals by Rhizobacteria. Appl Biochem Biotechnol 1–23
Rojas LA, Yáñez C, González M, Lobos S, Smalla K, Seeger M (2011) Characterization of the metabolically modified heavy metal-resistant Cupriavidus metallidurans strain MSR33 generated for mercury bioremediation. PLoS ONE 6(3):17555
Rothschild LJ, Mancinelli RL (2001) Life in extreme environments. Nat
Ruiz ON, Alvarez D, Gonzalez-Ruiz G, Torres C (2011) Characterization of mercury bioremediation by transgenic bacteria expressing metallothionein and polyphosphate kinase. BMC Biotechnol 11:1–8
Saranya K, Sundaramanickam A, Shekhar S, Swaminathan S, Balasubramanian T (2017) Bioremediation of mercury by Vibrio fluvialis screened from industrial effluents. Bio Med Res Int
Sarao LK, Kaur S (2021) Chapter-3 microbial bioremediation. New Vistas Microbial Sci 47
Sargın İ, Arslan G, Kaya M (2016) Efficiency of chitosan–algal biomass composite microbeads at heavy metal removal. React Funct Polym 98:38–47
Schaefer JK, Kronberg RM, Morel FMM, Skyllberg U (2014) Detection of a key Hg methylation gene hgcA, in wetland soils. Environ Microbiol Rep 6:441–447
Schelert J, Rudrappa D, Johnson T, Blum P (2013) Role of MerH in mercury resistance in the archaeon Sulfolobus solfataricus. Microbiol 159:1198
Shahpiri A, Mohammadzadeh A (2018) Mercury removal by engineered Escherichia coli cells expressing different rice metallothionein isoforms. Ann Microbiol 68:145–152
Sharma KR, Naruka A, Raja M, Sharma RK (2022) White rot fungus mediated removal of mercury from wastewater. Water Environ Res 94(7):10769
Singh A, Singh AK (2017) Haloarchaea: worth exploring for their biotechnological potential. Biotechnol Lett 39:1793–1800
Singh A, Kumari R, Yadav AN (2021) Fungal secondary metabolites for bioremediation of hazardous heavy metals. Recent Trends Mycol Res Environ Ind Perspect 2:65–98
Singh M, Jayant K, Bhutani S, Mehra A, Kaur T, Kour D, Suyal DC, Singh S, Rai AK, Yadav AN (2022) Bioremediation—sustainable tool for diverse contaminants management: current scenario and future aspects. J Appl Biol Biotechnol 10(2):48–63
Sinha A, Khare SK (2012) Mercury bioremediation by mercury accumulating Enterobacter sp. cells and its alginate immobilized application. Biodegradation 23:25–34
Sorkhoh NA, Ali N, Al-Awadhi H, Dashti N, Al-Mailem DM, Eliyas M, Radwan SS (2010) Phytoremediation of mercury in pristine and crude oil contaminated soils: contributions of rhizobacteria and their host plants to mercury removal. Ecotoxicol Environ Saf 73(8):1998–2003
Tada Y, Marumoto K, Iwamoto Y, Takeda K, Sakugawa H (2023) Distribution and phylogeny of mercury methylation, demethylation, and reduction genes in the Seto Inland Sea of Japan. Mar Pollut Bull 186:1–13
Taha A et al (2023) Bioremediation of heavy metals in wastewaters: a concise review. Egypt J Aquat Biol Fish 27(1):143–166
Takeuchi F, Iwahori K, Kamimura K, Negishi A, Maeda T, Sugio T (2001) Volatilization of mercury under acidic conditions from mercury-polluted soil by a mercury-resistant Acidithiobacillus ferrooxidans SUG 2–2. Biosci Biotechnol Biochem 65(9):1981–1986
Tarangini K (2009) Biosorption of heavy metals using individual and mixed cultures of Pseudomonas aeruginosa and Bacillus subtilis. Doctoral dissertation
Tarekegn M, Zewdu Salilih F, Ishetu AI (2020) Microbes used as a tool for bioremediation of heavy metal from the environment. Cogent Food Agric 6(1):1783174
Tarfeen N, Nisa KU, Hamid B, Bashir Z, Yatoo AM, Dar MA, Mohiddin FA, Amin Z, Ahmad RA, Sayyed RZ (2022) Microbial remediation: A promising tool for reclamation of contaminated sites with special emphasis on heavy metal and pesticide pollution: a review. Process 10(7):1358
Tay PK, Nguyen PQ, Joshi NS (2017) A synthetic circuit for mercury bioremediation using self-assembling functional amyloids. ACS Synth Biol 6(10):1841–1850
Tazaki K, Asada R (2007) Transmission electron microscopic observation of mercury-bearing bacterial clay minerals in a small-scale gold mine in Tanzania. Geomicrobiol J 24(6):477–489
Tiodar ED, Văcar CL, Podar D (2021) Phytoremediation and microorganisms-assisted phytoremediation of mercury-contaminated soils: challenges and perspectives. Int J Environ Res Public Health 18:1–37
Tripathi SM, Ram S (2018) Bioremediation of groundwater: an overview. Int J Appl Eng Res 13(24):16825–16832
Văcar CL, Covaci E, Chakraborty S, Li B, Weindorf DC, Frențiu T, Pârvu M, Podar D (2021) Heavy metal-resistant filamentous fungi as potential mercury bioremediators. J Fungus 7(5):386
Vela-García N, Guamán-Burneo MC, González-Romero NP (2019) Efficient bioremediation of metallurgical effluents through the use of microalgae isolated from the amazonic and highlands of ecuador. Rev Int Contam Ambient 4:917–929
Velkova Z, Kirova G, Stoytcheva M, Kostadinova S, Todorova K, Gochev V (2018) Immobilized microbial biosorbents for heavy metals removal. Eng Life Sci 18(12):871–881
Verma A (2021) Bioremediation techniques for soil pollution: an introduction. IntechOpen, London, UK
Verma A (2022) Bioremediation techniques for soil pollution: an introduction. Biodegrad Technol Organ Inorgan Poll
Vidali M (2001) Bioremediation. An overview. Pure Appl Chem 73(7):1163–1172
Vitor G, Palma TC, Vieira B, Lourenço JP, Barros RJ, Costa MC (2015) Start-up, adjustment and long-term performance of a two-stage bioremediation process, treating real acid mine drainage, coupled with biosynthesis of ZnS nanoparticles and ZnS/TiO2 nanocomposites. Miner Eng 89:18–85
Volarić A, Svirčev Z, Tamindžija D, Radnović D (2021) Microbial bioremediation of heavy metals. Hem Ind 75(2):103–115
Wagner-Döbler I (2003) Pilot plant for bioremediation of mercury-containing industrial wastewater. Appl Microbiol Biotechnol 62:124–133
Wang L, Hou D, Cao Y, Ok YS, Tack FM, Rinklebe J, O’Connor D (2020) Remediation of mercury contaminated soil, water, and air: a review of emerging materials and innovative technologies. Environ Int 134:105281
Wang L, Cheng WC, Xue ZF, Rahman MM, Xie YX, Hu W (2023) Immobilizing lead and copper in aqueous solution using microbial-and enzyme-induced carbonate precipitation. Front Bioeng Biotechnol 11:1146858
Winardi W, Haryono E, Sudrajat S, Soetarto ES (2020) In situ bioremediation strategies for the recovery of mercury-contaminated land in abandoned traditional gold mines in Indonesia. Biosaintifika: J Biol Educ 12(3):469–477
Wu C, Tang D, Dai J, Tang X, Bao Y, Ning J, Zhen Q, Song H, St. Leger RJ, Fang W (2022) Bioremediation of mercury-polluted soil and water by the plant symbiotic fungus Metarhizium robertsii. Proc Natl Acad Sci 119(47):e2214513119
Xie F, Dong K, Wang W, Asselin E (2020) Leaching of mercury from contaminated solid waste: a mini-review. Miner Process Extr Metall Rev 41(3):187–197
Xue Y, Du P, Shendi AA, Yu B (2022a) Mercury bioremediation in aquatic environment by genetically modified bacteria with self-controlled biosecurity circuit. J Clean Prod 337:130524
Xue Y, Qiu T, Sun Z, Liu F, Yu B (2022b) Mercury bioremediation by engineered Pseudomonas putida KT2440 with adaptationally optimized biosecurity circuit. Environ Microbiol 7:3022–3036
Yadav V, Manjhi A, Vadakedath N (2023) Mercury remediation potential of mercury-resistant strain Rheinheimera metallidurans sp. nov. isolated from a municipal waste dumping site. Ecotoxicol Environ Saf 257:1–12
Yan J, Ye W, Jian Z, Xie J, Zhong K, Wang S, Hu H, Chen Z, Wen H, Zhang H (2018) Enhanced sulfate and metal removal by reduced graphene oxide self-assembled Enterococcus avium sulfate-reducing bacteria particles. Bioresour Technol 266:447–453
Yan C, Qu Z, Wang J, Cao L, Han Q (2022) Microalgal bioremediation of heavy metal pollution in water: recent advances, challenges, and prospects. Chemosphere 286:131870
Yao H, Wang H, Ji J, Tan A, Song Y, Chen Z (2023) Isolation and identification of mercury-tolerant bacteria LBA119 from molybdenum-lead mining soils and their removal of Hg2+. Toxics 11(3):261
Yu RQ, Barkay T (2022) Microbial mercury transformations: molecules, functions and organisms. In: Gadd GM, Sariaslani S (eds) Advances in applied microbiology. Elsevier, Amsterdam, Netherlands, pp 31–90
Zhai X, Li Z, Huang B, Luo N, Huang M, Zhang Q, Zeng G (2018) Remediation of multiple heavy metal-contaminated soil through the combination of soil washing and in situ immobilization. Sci Total Environ 635:92–99
Zhang M, Wang H (2016) Preparation of immobilized sulfate reducing bacteria (SRB) granules for effective bioremediation of acid mine drainage and bacterial community analysis. Miner Eng 92:63–71
Zhang W, Chen L, Liu D (2012) Characterization of a marine-isolated mercury-resistant Pseudomonas putida strain SP1 and its potential application in marine mercury reduction. Appl Microbial Biotechnol 93:1305–1314
Zhang X, Liu X, Liang Y, Fan F, Zhang X, Yin H (2016) Metabolic diversity and adaptive mechanisms of iron-and/or sulfur-oxidizing autotrophic acidophiles in extremely acidic environments. Environ Microbiol Rep 8(5):738–751
Zhang J, Zeng Y, Liu B, Deng X (2020) MerP/MerT-mediated mechanism: a different approach to mercury resistance and bioaccumulation by marine bacteria. JHM Lett 388:1–10
Zheng R, Wu S, Ma N, Sun C (2018) Genetic and physiological adaptations of marine bacterium Pseudomonas stutzeri 273 to mercury stress. Front Microbiol 9:1–14
Acknowledgements
The graphical illustrations were performed via biorender.com and without permission from biorender.com.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Eltarahony, M., Ibrahim, E., Hegazy, G., Sabry, A. (2023). Microbial Remediation of Mercury: An Overview. In: Kumar, N. (eds) Mercury Toxicity. Environmental Science and Engineering. Springer, Singapore. https://doi.org/10.1007/978-981-99-7719-2_8
Download citation
DOI: https://doi.org/10.1007/978-981-99-7719-2_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-7718-5
Online ISBN: 978-981-99-7719-2
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)