Abstract
The marine environment is a major source of biodiversity, food, energy and therapeutics. The vastness and significance of marine could be measured by the fact that about 90% of the marine environment is comprised of microbes and covers about 70% of the surface of planet Earth. Conditions like the wide range of temperature, pH, pressure and salinity further facilitate the diversification of marine organisms. Since the marine environment carries a wide range of organisms, therefore, this chapter is limited to marine bacteria only. We explained the therapeutic applications of different bacteria. Furthermore, to discover and harness the potential of marine microorganisms, we have proposed several methods and sophisticated tools that need to be developed for isolation, culturing and identification.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
11.1 Introduction
The marine environment is one of the extremely valuable ecosystems of planet Earth that holds an immense wealth of supplies for the sustenance of this world and its population. Marine environments are comprised of a multitude of organisms ranging from whales to microorganisms. It is estimated that about 90% of marine biomass is microbial. Marine microorganisms are characterized by their habitation. They cover about 70% of Earth’s surface and range from the saltwater of a sea or ocean or the brackish water of a coastal estuary (Jørgensen and Boetius 2007). These microorganisms have an abundance of 105 cells per millilitre, and it is calculated that the oceans altogether contain 3.6 × 1029 microbial cells. Most marine microorganisms are composed of archaea, bacteria, protists and unicellular fungi (Baharum et al. 2010). Marine microorganisms are well-known for maintaining and regulating the biological and chemical cycles in the marine environment. They are fixing carbon and nitrogen and playing a key role in the re-mineralization of organic matter. These microbes are also fundamental to the ocean food webs (Azam and Malfatti 2007). Marine environments are promising ecosystems for biodiscovery. The most noticeable marine microorganisms include archaea, algae, bacteria, fungi, protozoa and viruses (Fig. 11.1) (Brock et al. 2003).
The marine microorganism is so diverse and holds so many industrial applications that a book chapter is not enough to grasp such immense information. For this reason, we limited this chapter to marine bacteria only. This chapter describes the diversity and composition of marine bacteria, as well as tools for isolating living marine bacteria, innovative tools for bacterial identification and their application in various industries.
Marine bacteria, unicellular organisms, are structured like small orbits, rods and spirals. These organisms are about 1/100th the width of a human hair and can perform all sorts of chemical activities in the sea and ocean. For instance, Trichodesimium (a genus of Cyanobacteria) and Crocosphaera fix nitrogen through the nitrogenase enzyme from nitrogen gas into biologically useful nitrogen compounds. Since nitrogenase is sensitive to the presence of oxygen, therefore, these photosynthetic bacteria have adopted a special mechanism through which they can separate photosynthesis and nitrogen fixation separately (a) either spatially by using diverse types of cells for each procedure or (b) temporally, by carrying out photosynthesis in the daytime and nitrogen-fixation at night. In addition, some marine bacteria are involved in ammonium oxidation and convert ammonium to nitrite and nitrate. Betaproteobacteria and gamma proteobacteria are well-known classes of bacteria for nitrification. Bacteria related to these classes encode the ammonia monooxygenase enzyme (amoA). There are other bacteria such as Nitrobacter, Nitrospina and Nitrospira that can subsequently convert nitrite into nitrate—the second step in the nitrification process (Pajares and Ramos 2019). Bacterial-converted nitrates are produced by other bacteria as a source of nutrients, e.g., Prochlorococcus and Synechococcus. Although Prochlorococcus are photosynthetic, they require nitrogen as a nutrient (Partensky et al. 1999).
Marine environments present a diverse and extreme environment for living organisms as it provides temperature that ranges from −2 °C to more than 100 °C. Hydrothermal vents (<200 m in depth) temperature varies with a fluid temperature range of 10 to 119 °C (Tarasov et al. 2005). Such conditions are hostile for most organisms; however, specialized microbes could thrive in these conditions despite the tough environments (Jannasch and Taylor 1984). Bacteria have built an exceptional approach to live in extreme ecological environments through the production of an extracellular polymeric matrix, giving protection to the cells in the hostile microenvironment. The major structural elements of this composite network constitute high-molecular weight hydrophilic macromolecules, known as exopolysaccharides. The deep-sea hydrothermal vent bacteria’s exopolysaccharides are enriched with hexosamines and uronic acids. Alteromonas, Pseuodoalteromonas and Vibrio are a few of the exopolysaccharide-secreting bacteria that dwell in hydrothermal vents and have been cultured in lab conditions (Zykwinska et al. 2019). Contrary to bacteria adapted to hot temperatures, the psychrophile bacteria can thrive and reproduce at low temperatures varying from 10 °C to −20 °C. The psychrophilic enzymes are distinctive in catalytic action and have low activation energy that yields high efficiency at lower temperatures (Podar and Reysenbach 2006).
Besides temperature variation, marine environments are also characterized by different spatial-temporal and other properties—such as salinity, pH, variation in nutrients, etc.—so different microbes inhabit differently according to their nutrient and growth condition requirements. Even, the water depth is favouring the growth of different bacteria. For instance, a study compared bacterial richness and diversity between the sea surface and subseafloor and found marked variation in the bacterial operational taxonomic units (OTUs). Abundant OTUs were spotted in the subseafloor sediments (Walsh et al. 2016).
Nonetheless, the bacterial diversity changes with changes in season. For instance, the South China Sea holds higher bacterial diversity during the summer compared to other seasons. In particular, Alpha proteobacteria is the most abundant taxon in summer and spring samples followed by Cyanobacteria and Gamma proteobacteria. On the contrary, during the winter season, the South China Sea is enriched with cyanobacteria. Even the coastal and offshore waters hold distinct bacterial communities (Du et al. 2013). Most of the bacteria that inhabit the South China Sea are related to phyla Firmicutes, Planctomycetes, Actinobacteria and Chloroflexi Gamma proteobacteria (Zhu et al. 2013).
Since the marine environment is diversified, these environments inhabit different bacteria. Therefore, the following literature is categorized to describe environmental-bacteria associations and their potential industrial application.
11.2 Co-inhabiting Niches for Marine Bacteria
11.2.1 Mangroves
Mangrove forests are a hitherto undiscovered source of microbial diversity that are found at the meeting point of marine and terrestrial ecosystems (Nedwell et al. 1994; Andreote et al. 2012). More than a hundred nations are home to mangroves, which encompass an area of around 150,000 km2. The American continent has the most mangroves, followed by Brazil, which accounts for 8.5% of the world’s mangroves (Colares and Melo 2013). Mangrove jungles are well-known for their great quantities of organic matter (OM) and nutrients, making them very productive habitats (Nedwell et al. 1994). Fish may reproduce and develop in the sediments of mangroves, and they can also find food and safety there. They serve as a transitional area between the terrestrial and marine habitats and are crucial for preserving sea levels (Duke et al. 2007). Additionally, mangrove forests serve as a terminal sink for local waste and nutrient-rich sediments.
Complex mangrove ecosystems offer a dynamic microbial environment. Because of elevated relative moisture and recurring tidal floods, the microbial colonies are exceptionally diverse and adaptable. The key determinants of the development of mangrove ecosystems are environmental elements like nutrient availability, light, salinity and temperature (Holguin et al. 2006). The general bacteria of the mangroves are N2-fixing (Klebsiella sp., Azospirillum, Rhizobium, Azotobacter, Clostridium, etc.), sulphate-reducing (Desulfosarcina, Desulfovibrio, Desulfococcus sp., Desulfotomaculum, etc.), phosphate-solubilizing (Vibrio proteolyticus, Enterobacter, Kluyvera, Bacillus, Paenibacillus, Chryseomonas, Pseudomonas sp., etc.), methanogenic (Methanoccoides methylutens sp., etc.) and photosynthetic anoxygenic (Beggiatoa, Chloronema, Thiopedia, Leucothiobacteria sp., etc.) bacteria (Kathiresan and Bingham 2001). Despite the extensive studies conducted on mangrove bacteria—it is estimated that more than 95% of the species remain uncharacterized. In a study carried out in the pristine and anthropogenically influenced mangrove ecosystems on the Red Sea, a research group recorded 32 bacterial phyla in the mangrove rhizospheres which was dominated by bacteria form phylum Proteobacteria (Ullah et al. 2017).
Mangrove-associated bacteria participate in nitrogen fixation (combining N2 to ammonia or organic nitrogen), phosphate-solubilization, sulphur-oxidization and cellulose degradation. Mangrove environments dwelling bacteria are a valuable source of industrially valuable agents. These bacteria produce a wide range of vital therapeutic compounds that include insecticides, enzymes, anti-tumour agents, immunosuppressants, vitamins and immune modulators. These bacteria encode industrially useful enzymes that are used in the production of biosurfactants, bioplastics and natural bioproducts (Hong et al. 2009).
11.2.2 Marine Sponges: A Niche for Marine Bacteria
In certain species of sponges, up to 60% of the tissue volume is comprised of microbes—that said—a sponge tissue inhabitant more than 109 microbial cells per mL (Webster and Hill 2001). Sponges inhabit mutualistic microbes and develop mutualistic relationships. Sponges provide shelter and food to these microbes and in turn, microbial by products protect sponges against various infectious diseases (Taylor et al. 2007).
So far, at least 10 bacterial phyla that are associated with sponges have been reported, including Bacteroidetes, Actinobacteria, Proteobacteria and Verrucomicrobia (Schloss and Handelsman 2003). Pseudoalteromonas piscicida (a bacterium) is associated with Hymeniacidon perleve (a sponge) to produce β-carboline alkaloids that possess anti-microbial activities, particularly against Staphylococcus aureus, Bacillus subtilis and Agrobacterium tumefaciens (Zheng et al. 2005). Similarly, metabolites extracted from a strain of Suberites domuncula have shown cytotoxic and anti-microbial activities. These metabolites have limited the growth of Staphylococcus aureus and Staphylococcus epidermidis (Thakur et al. 2005). In addition to their anti-microbial action, most of these compounds have also shown anti-cancer properties (Aoyagi et al. 1992). These mutualistic microbes are diverse and produce Sponge-dwelling microbes including bacteria producing alkaloids, peptides, polyketides, fatty acids and terpenes that have exhibited a wide array of biological functions of biotechnological and medical significance such as anti-fungal, immunosuppressive, anti-tumour and anti-protozoal (Imhoff et al. 2011).
Moreover, sponges are in direct contact with pollutants. The hydrolases-producing microbes that inhabit sponges help in the breakdown of organic pollutants into nutrients. For instance, Cytophaga bacteria dwell on Halichondria panacea (a sponge) and encode the agarase enzyme. Other sponge-associated bacteria, such as Arthrobacter, produce another important enzyme called acetylcholinesterase (Imhoff and Stöhr 2003).
11.2.3 Algae–Bacteria Association
(a) Macroalgae (Seaweeds)
Macroalgae, also known as seaweeds, refer to thousands of species of macroscopic and multicellular marine algae. Several studies have been conducted on the interaction of macroalgae-bacteria and bacterial role in macroalgae physiology. The interlinked evolution of macroalgae and bacteria permitted the development of a wide range of relationships defined by symbiotic assistance with growth factors, nutrient interchange and quorum sensing facilitation. Bacteria could inhabit the seaweeds and chemical boundary layer of the seaweed in the microenvironment (Mieszkin et al. 2013; Wichard 2015). Some of the known seaweeds-associated bacteria include Leucobacter sp., Micrococcus and Kocuria palustri (Villarreal-Gómez et al. 2010; Zhuang et al. 2003).
Seaweed-associating bacteria secrete certain molecules, known as algal development and morphogenesis-promoting factors, that are important for the seaweed’s development and facilitate macroalgal adaptation to environmental changes (Wichard 2015). For instance, the transition of Ectocarpus (macroalgae) from high-saline water to lower-saline water is dependent on intrinsic microbial communities. In a study Dittami et al. (2016) treated Ectocarpus with antibiotics to remove the associated bacteria and noted that Ectocarpus could not adapt to a new environment without associated bacteria. Similarly, macroalgae-bacteria share several strategies for metal detoxification. For example, bacteria (such as Bacillus spp. and Pseudomonas aeruginosa) eliminate cadmium and lead metals through their setup in extracellular polymeric substances (De et al. 2008). Seaweed-associated bacteria produce secondary metabolites that have the potential to inhibit colorectal cancer cells. For instance, Bacillus sp. which is associated with seaweed produces metabolites that have inhibited the development of HCT-116 cancer cells (Villarreal-Gómez et al. 2010).
Algal–bacterial interaction is historically well-known and has been explored for wastewater treatment as early as the 1950s (Oswald and Gotaas 1957). This interaction holds great promise in the metal bioremediation and degradation of organic pollutants (Boivin et al. 2007; Tang et al. 2010).
(b) Microalgae (Diatoms)
Microalgae are photosynthetic and unicellular microorganisms—that regularly live in aqueous surroundings such as marine systems, blackish and freshwater. The most important types of microalgae include Bacillariophyta (diatoms), golden algae (chrysophyta), green algae (chlorophyta) and blue–green algae (cyanophyta) (Chen et al. 2010).
Diatoms contribute to one-fifth of the photosynthesis on Earth, whereas bacteria re-mineralize a big part of the fixed carbon in the water. Diatoms and bacteria have lived in common habitats for millions of years and have fostered connections with world-wide biogeochemical significances. The diatoms-bacterial interaction occurs in the microenvironment surrounding diatoms, known as the phycosphere (Amin et al. 2012). Bacteria inhabit the diatoms’ phycosphere either through random encounters, chemotaxis or vertical transmission (Seymour et al. 2017). Several organic nutrients that are essential for the growth of microalgae are provided by bacteria in marine environments. For instance, microalgae, including diatoms, need ammonia but these organisms cannot obtain ammonia directly by fixing N2. Instead, nitrogen-fixing bacteria and cyanobacteria provide ammonia to algae in the marine setting (Foster et al. 2011).
Some of the most commonly diatom-associated bacteria include Gammaproteobacteria (Marinobacter), Alphaproteobacteria (Sulfitobacter), Bacteroidetes (Croceibacter) and Betaproteobacteria (Limnobacter) (Schäfer et al. 2002). However, different diatoms harbour different bacterial species. A study that investigated the bacterial communities of six diatoms genera (namely Ditylum, Asterionella, Thalassiosira, Chaetoceros, Coscinodiscus and Leptocylindrus) found distinctive bacteria associated with each genus (Grossart et al. 2005; Sapp et al. 2007a, b; Schäfer et al. 2002). Moreover, the evaluation of diatom-bacteria interaction is important to decipher nutrients and biochemical rounds in the marine setting. Their mutualistic existence cycles nutrients between reduced and oxidized states that facilitate the bioavailability of nutrients to higher trophic levels.
11.2.4 Fish
Fish-linked symbiotic gut bacteria contribute to nutritional provisioning, metabolic equilibrium and immunological defence, just like humans and other mammals (Sullam et al. 2012). Fish gut microbe research has been going on since the first part of the twentieth century, but lately, as the aquaculture sector has grown, interest in this area has accelerated. Some early studies that investigated bacteria related to fish eggs suggest that the dominating species at this point are related to the bacterial phylum Cytophaga, and the genera Flavobacterium and Pseudomonas (Austin 1982). The first colonizing bacteria are now recognized as being species-specific, with distinctions being governed by variations in binding glycoproteins on the egg exterior (Larsen 2014). Fascinatingly, as fish grow, so does the diversity of bacteria, just like in humans. In the various fish GIT parts, there are variations in microbiota density, composition and function (Clements et al. 2014).
The gut microbiota (GM), gut-dwelling microbes, has been assessed in a few model fish species, such as rainbow trout, guppy and zebrafish. In addition, the gut microbes of a few other valuable marine animals, such as Atlantic salmon, Atlantic cod and sturgeon, have also been catalogued (Kim et al. 2021). It is observed that the GM of fish is strongly influenced by the host habitat.
Much focus has been given to the nutritional exploitation and alteration of GM to meet the demands of fish farming whereas maintaining the quality and welfare of fish. Various prebiotic and probiotic methodologies are being developed to replace the use of chemicals and antibiotics in fish farming (Abelli et al. 2009). Some of the commonly examined probiotics include lactic acid producing Lactococcus, Shewanella and Aeromonas (Merrifield and Carnevali 2014).
11.3 Tools for the Identification of Marine Bacteria
The discovery and extraction of bioactive compounds from the marine environment demands the identification and isolation of marine bacteria in in-vitro conditions. Since the microbial world is so diverse, their growth conditions are difficult to replicate in the lab conditions. Even though many new methods have brought previously uncultured bacteria into laboratory culture, there are still numerous most wanted bacteria that need to be cultured from marine environments. In addition, the screening of marine bacteria is also needed as they could transfer antibiotic resistance genes from marine resources to humans (Khan et al. 2019). Following, we will discuss the innovative tools that are in best practice to identify and isolate bacterial colonies from the marine environment.
11.3.1 Culturomics
Culturomics is a culturing approach that uses multiple culture conditions and identification technologies (such as MALDI-TOF mass spectrometry and 16S rRNA sequencing) to bring the bacterial species into the lab conditions. A cultivation-based method is a gold standard for isolating and characterizing bacteria. This method is essential for being able to isolate pure colonies. The bacterial community belong to widespread groups, isolation and characterization of bacteria offer insight into their role and functions in natural ecosystems. In addition, culture-based techniques enable us to understand metabolic interactions between microorganisms and their surroundings (Kaeberlein et al. 2002). Culturomics is an indispensable technique used to explore cultivable bacterial diversity.
There are several reasons for our inability to cultivate marine microorganisms in the laboratory, including a lack of adequate growth conditions, low growth rates, poor development of colonies, requirements for metabolites generated by other microbes and the presence of dormant cells (Gutleben et al. 2018). Numerous attempts have been made to unveil the hidden secrets of bacteria through culture-based techniques. Cultivation-based and molecular-based techniques have been used for decades to investigate untapped environments.
Many functions and applications of marine bacteria are unknown as growing these microbes in a laboratory is challenging. Therefore, the field of Culturomics came into being to design and formulate growth media and conditions that could support maximum bacterial diversity. Besides, the rigorous cultivation methods, the number of bacteria gained through Culturomics is lesser than that seen via sequencing.
(a) Identification Through Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS)
This is the fastest technique for purified bacterial identification. Samples are obtained from the marine setup and then pass-through homogenization and serial dilution. The dilution is then evenly spread on the growth medium to obtain the master culture. After the predetermined culture time, the master plate is sub-plated for isolation of the pure colonies. These pure colonies are then processed through the preparation phase for identification through the MALDI-TOF MS. Briefly, each bacterial isolate is spread on a MALDI-TOF target plate and then poured with a 1 μL matrix solution. Matrix solution is then obtained by adding 475 μL HPLC grade water in 500 μL acetonitrile and 25 μL trifluoroacetic acid—in the end, 5 mg of α-cyano-4-hydroxycinnamic acid is dissolved. The solution is vortexed until it becomes clear. Each spot on the plate is targeted with a laser and spectra are obtained through flex control software and analysed by MALDI-Biotyper software. If the spectra aren’t clear, then the colony is speared again until the spectrum is measured with a score ≥ 1.9). If a colony remained unidentified, then 16S RNA sequencing is employed for the amplification and sequencing of the 16S rRNA gene. This technique is graphically illustrated in Fig. 11.2.
(b) Colony PCR and Sanger Sequencer-Based Identification
This is the second fastest method of bacterial identification. Purified bacterial colonies on the media plates are selected and processed for DNA isolation. First, a 5% Chelex is prepared in sterile distilled water (1 g of chelex/20 mL). A volume of 150 μL of 5% Chelex is pipetted into 1.5 mL Eppendorf tubes. Approximately, five to six pure colonies of sample bacteria are carefully mixed with this 150 μL CHELEX and the contents were thoroughly mixed. This step is conducted inside a laminar flow hood setup to restrict contamination. All the remaining samples are processed in the same manner. The samples are warmed in a water bath set at 95 °C for 25 to 30 min. At this temperature, bacterial cells lyse. The main function of Chelex is to protect DNA from DNases. After heating, the sample tubes are centrifuged at 5000 rpm for 1 min. This centrifugation pellets out the Chelex and cell debris and DNA being obtained in the supernatant. Finally, the supernatant is collected and transferred to new, sterile and properly labelled Eppendorf tubes for further processing. The obtained DNA is stored at 4 °C until used for PCR.
Generally, PCR is performed with the universal set of primers 27F 5′-AGAGTTTGATCMTGGCTCAG-3′ and 1492R 5′-GGTTACCTTGTTACGACTT-3′ to amplify the highly variable region of the 16S rRNA gene. The master mix is prepared by mixing 12.5 μL of master mix with 1 μL of each reverse and forward primer and 9 μL of de-ionized water. The mixture is slightly vortexed for 5 s and then 1.5 μL of the template DNA is added and again vortexed. The tubes are then centrifuged for 5 s. After the PCR step, the amplified region of the 16S rRNA gene is then targeted for sequencing using Sanger sequencer by following the manufacturer protocol. This technique is graphically illustrated in Fig. 11.3.
11.3.2 Culture-Independent Method (Metagenomics)
Metagenomics is a beautiful combination of biological sciences and bioinformatics. Arguably, the invention of metagenomics is one of the greatest events in the field of microbiology in the past two decades. Metagenomics describes the genetic analysis of the genomes of an environment. This field originally began with the cloning of environmental DNA, followed by functional expression screening. As has been discussed below, metagenomics can be classified into two categories:
(a) 16S Amplicon Sequencing
The study of the interaction between organisms and their habitat is fundamental for the understanding of ecology and evolution. This mutuality led to a novel view of complex lifeforms, known as metaorganisms. A metaorganism is a grouping of all the organisms (mostly microorganisms) that live in a given habitat. But it is extremely tough and time-taking to catalogue the metaorganism of a habitat timely and efficiently. Currently, metagenomics is the gold standard to identify and characterize the metaorganism. Metagenomic was first utilized to describe the diversity and composition of dense microbial communities (Venter et al. 2004). Metagenomics has been broadly applied to recognize microorganisms that are related to an ecological and physiological state.
Unlike bacterial Culturomics, metagenomics suffers to determine the ratio of dead or alive microorganisms in a sample. Metagenomics sequences a part of the total DNA pool. At the start, metagenomics was struggling with bacterial identification at the species level due to the shorter read length, which hindered the wider application and precision of metagenomics in various applied fields (Scher et al. 2013). However, recent advancements in sequencing have supported bacterial detection beyond the strain level (Truong et al. 2017). The big development in bioinformatics has aided the investigation of complete sequence data sets by utilizing homogeneity algorithms or clustering tools without taxonomic annotation (Truong et al. 2017). Before the metagenomic experiment, the isolated DNA must be quantified using nanodrop equipment and subsequently visualize through gel electrophoresis to ensure that the DNA is not denatured. To this day, several sequencing platforms are available for 16S rRNA gene sequencing such as (Illumina, PacBio and Oxford Nanopore Sequencing). And these sequencing platforms have adopted different chemistry. Designing and enriching cultivation media and conditions based on information obtained from metagenomic datasets and growth condition is a promising strategy in development for cataloguing the most wanted marine bacteria.
(b) Metagenomics Shotgun Sequencing
Beyond taxonomic identification that relies on the sequencing of the 16S rRNA gene, shotgun sequencing empowers the functional assignment of genes and provides the details about the functional potential of a community (Glass and Meyer 2011). In other words, shotgun sequencing enables scientists to comprehensively catalogue all genes in the meta-organism present in each complex environment.
Shotgun sequencing holds the lead in species and strain-level classification of bacteria. In addition to that, it enables researchers to assess the functional associations between hosts/habitats and metaorganisms (bacteria, viruses, fungi) by identifying the functional content. Beyond that, shotgun sequencing enables scientists to explore the unknown microbial life that remains unclassifiable. However, its high cost is limiting the shotgun sequencing application from a wider perspective (Jovel et al. 2016). Hopefully, the cost of shotgun metagenomics will go down in the same manner as 16S rRNA gene sequencing methods. I expect that this technique will become a standard approach for researchers around the world to comprehensively analyse and understand the function of microbial colonies and their association with the host and surrounding. A workflow of the metagenomic experiment is shown in Fig. 11.4.
11.4 Application of Marine Bacteria in the Medicinal Industry
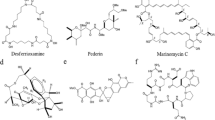
Despite the massive interest in isolating and storing marine bacteria, metabolites obtained from these organisms are increasingly captivating to researchers due to their wide-ranging application, especially those with unique colour pigments. Great interests are being developed to explore the application of marine bacteria in various industries such as food, medicine, animal feed and colouring agents. Particularly, marine bacteria have been extensively investigated for medicinal purposes and several studies have isolated secondary metabolites from marine bacteria that possess anti-cancer, anti-microbial, anti-parasitic and immunosuppressive activities (Ramesh et al. 2019). Some of the studied metabolites are discussed below and their structures are shown in Fig. 11.5.
-
1.
Phenazine (C6H4)2 N2, is a small nitrogen-containing organic compound that is produced by several marine bacteria such as Actinomycetes, Pelagibacter, Vibrio and Pseudomonas. Phenazine holds a broad spectrum of anti-microbial, insecticidal, anti-protozoal and anti-tumour activities (Pierson and Pierson 2010; Soliev et al. 2011).
-
2.
Carotene, C40 Polyunsaturated hydrocarbon, is a fat-soluble precursor of vitamin A that is produced plants, fungi, yeast and bacteria. The carotenes include β-carotene, α-carotene and γ-carotene. Marine bacteria that produce carotene include Arthrobacter sp., Rhodotorula rubra and Rhodotorula brasiliensis. They are well-known for their strong antioxidant activities. In addition, carotene is an important product for cosmetics and a good source of natural food colourants (Asker 2017; Ligia et al. 2017).
-
3.
Marine bacteria can also produce melanin pigment, which has exhibited anti-viral, anti-cancer and anti-bacterial activities. Most melanin-producing bacteria are sponge symbionts. Bacteria that produce melanin include Vibrio sp., Bacillus sp., Providencia sp., S. algae, S. roseus, S. sciuri, P. maritimus and G. creatinolyticus (Vijayan et al. 2017). In addition, the bacterial produce melanin also possesses heavy metal treatment and preventing fouling (Manirethan et al. 2018, 2020).
-
4.
Prodigiosin is a red-coloured secondary metabolite compound that is produced by bacteria such as Serratia marcescens, Zooshikella sp. S2.1 and Streptomyces sp. Prodigiosin has shown solid anti-microbial, anti-fungal, cytotoxic and immune suppressive activities (Huryn and Wipf 2008; Ramesh et al. 2020). This compound has also shown anti-bacterial activity against S. aureus (Danevčič et al. 2016).
-
5.
Violacein is another compound which is water-insoluble in nature and purple-coloured produced by Janthinobacterium lividum, Pseudoalteromonas tunicata D2, Chromobacterium violaceum and Janthinobacterium lividum (Masuelli et al. 2016). The formation of violacein originates from the oxidation of tryptophan. This compound has been comprehensively investigated and reported to be an effective anti-bacterial, anti-viral, anti-tumour, anti-protozoal and anti-parasitic agent (Masuelli et al. 2016). This compound exhibits anti-cancer properties by enhancing mitochondrial membrane hyperpolarization (Masuelli et al. 2016).
-
6.
Quinones are abundant biological pigments that have a yellow crystalline appearance and a distinctive irritating odour like that of chlorine. Quinones are produced by bacteria, fungi, plants and a few animals. Quinone displays anti-viral, anti-cancer, anti-microbial and insecticidal activities where the colour ranges from yellow to red (Soliev et al. 2011). Streptomyces sp. is a well-known bacterium for quinone production (Liang et al. 2016). This compound is usually prepared by the oxidation of aromatic amines, polyhydric phenols and polynuclear hydrocarbons.
References
Abelli L, Randelli E, Carnevali O, Picchietti S (2009) Stimulation of gut immune system by early administration of probiotic strains in Dicentrarchus labrax and Sparus aurata. Ann N Y Acad Sci 1163(1):340–342
Amin SA, Parker MS, Armbrust EV (2012) Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 76(3):667–684
Andreote FD, Jiménez DJ, Chaves D, Dias ACF, Luvizotto DM, Dini-Andreote F, Fasanella CC, Lopez MV, Baena S, Taketani RG, de Melo IS (2012) The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS One 7(6):e38600. https://doi.org/10.1371/journal.pone.0038600
Aoyagi T, Hatsu M, Kojima F, Hayashi C, Hamada M, Takeuchi T (1992) Benarthin: a new inhibitor of pyroglutamyl peptidase. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot 45(7):1079–1083. https://doi.org/10.7164/antibiotics.45.1079
Asker D (2017) Isolation and characterization of a novel, highly selective Astaxanthin-producing marine bacterium. J Agric Food Chem 65(41):9101–9109. https://doi.org/10.1021/acs.jafc.7b03556
Austin B (1982) Taxonomy of bacteria isolated from a coastal, marine fish-rearing unit. J Appl Bacteriol 53(2):253–268
Azam F, Malfatti F (2007) Microbial structuring of marine ecosystems. Nat Rev Microbiol 5(10):782–791. https://doi.org/10.1038/nrmicro1747
Baharum SN, Beng EK, Mokhtar MAA (2010) Marine microorganisms: potential application and challenges
Boivin MEY, Greve GD, Garcia-Meza JV, Massieux B, Sprenger W, Kraak MHS, Breure AM, Rutgers M, Admiraal W (2007) Algal–bacterial interactions in metal contaminated floodplain sediments. Environ Pollut 145(3):884–894
Brock TD, Madigan MT, Martinko JM, Parker J (2003) Brock biology of microorganisms. Prentice-Hall, Upper Saddle River (NJ), p 2003
Chen P, Min M, Chen Y, Wang L, Li Y, Chen Q, Wang C, Wan Y, Wang X, Cheng Y (2010) Review of biological and engineering aspects of algae to fuels approach. Int J Agric Biol Eng 2(4):1–30
Clements KD, Angert ER, Montgomery WL, Choat JH (2014) Intestinal microbiota in fishes: what’s known and what’s not. Wiley Online Library
Colares GB, Melo VMM (2013) Relating microbial community structure and environmental variables in mangrove sediments inside Rhizophora mangle L. habitats. Appl Soil Ecol 64:171–177
Danevčič T, Borić Vezjak M, Tabor M, Zorec M, Stopar D (2016) Prodigiosin induces autolysins in actively grown Bacillus subtilis cells. Front Microbiol 7:27
De J, Ramaiah N, Vardanyan L (2008) Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury. Mar Biotechnol (New York, NY) 10(4):471–477. https://doi.org/10.1007/s10126-008-9083-z
Dittami SM, Duboscq-Bidot L, Perennou M, Gobet A, Corre E, Boyen C, Tonon T (2016) Host-microbe interactions as a driver of acclimation to salinity gradients in brown algal cultures. ISME J 10(1):51–63. https://doi.org/10.1038/ismej.2015.104
Du J, Xiao K, Li L, Ding X, Liu H, Lu Y, Zhou S (2013) Temporal and spatial diversity of bacterial communities in coastal waters of the South China Sea. PLoS One 8(6):e66968. https://doi.org/10.1371/journal.pone.0066968
Duke NC, Meynecke J-O, Dittmann S, Ellison AM, Anger K, Berger U, Cannicci S, Diele K, Ewel KC, Field CD, Koedam N, Lee SY, Marchand C, Nordhaus I, Dahdouh-Guebas F (2007) A world without mangroves? Science 317(5834):41–42. https://doi.org/10.1126/science.317.5834.41b
Foster RA, Kuypers MMM, Vagner T, Paerl RW, Musat N, Zehr JP (2011) Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. ISME J 5(9):1484–1493. https://doi.org/10.1038/ismej.2011.26
Glass EM, Meyer F (2011) The metagenomics RAST server: a public resource for the automatic phylogenetic and functional analysis of metagenomes. In: Handbook of molecular microbial ecology I: metagenomics and complementary approaches. https://doi.org/10.1002/9781118010518.ch37
Grossart H-P, Levold F, Allgaier M, Simon M, Brinkhoff T (2005) Marine diatom species harbour distinct bacterial communities. Environ Microbiol 7(6):860–873. https://doi.org/10.1111/j.1462-2920.2005.00759.x
Gutleben J, Chaib De Mares M, van Elsas JD, Smidt H, Overmann J, Sipkema D (2018) The multi-omics promise in context: from sequence to microbial isolate. Crit Rev Microbiol 44(2):212–229. https://doi.org/10.1080/1040841X.2017.1332003
Holguin G, Gonzalez-Zamorano P, de Bashan LE, Mendoza R, Amador E, Bashan Y (2006) Mangrove health in an arid environment encroached by urban development--a case study. Sci Total Environ 363(1–3):260–274. https://doi.org/10.1016/j.scitotenv.2005.05.026
Hong K, Gao A-H, Xie Q-Y, Gao H, Zhuang L, Lin H-P, Yu H-P, Li J, Yao X-S, Goodfellow M, Ruan J-S (2009) Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar Drugs 7(1):24–44. https://doi.org/10.3390/md7010024
Huryn DM, Wipf P (2008) Natural product chemistry and anticancer drug discovery. Cancer Drug Design Discov:91–120
Imhoff JF, Stöhr R (2003) In: Müller WEG (ed) Sponge-associated bacteria: general overview and special aspects of bacteria associated with Halichondria panicea BT - sponges (Porifera). Springer, Berlin, pp 35–57. https://doi.org/10.1007/978-3-642-55519-0_2
Imhoff JF, Labes A, Wiese J (2011) Bio-mining the microbial treasures of the ocean: new natural products. Biotechnol Adv 29(5):468–482. https://doi.org/10.1016/j.biotechadv.2011.03.001
Jannasch HW, Taylor CD (1984) Deep-sea microbiology. Annu Rev Microbiol 38:487–514. https://doi.org/10.1146/annurev.mi.38.100184.002415
Jørgensen BB, Boetius A (2007) Feast and famine—microbial life in the deep-sea bed. Nat Rev Microbiol 5(10):770–781. https://doi.org/10.1038/nrmicro1745
Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL, Wong GK-S (2016) Characterization of the gut microbiome using 16S or shotgun metagenomics. Front Microbiol 7. https://doi.org/10.3389/fmicb.2016.00459
Kaeberlein T, Lewis K, Epstein SS (2002) Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science (New York, NY) 296(5570):1127–1129. https://doi.org/10.1126/science.1070633
Kathiresan K, Bingham BL (2001) Biology of mangroves and mangrove ecosystems
Khan I, Yasir M, Farman M, Kumosani T, Albasri SF, Bajouh OS, Azhar EI (2019) Evaluation of gut bacterial community composition and antimicrobial resistome in pregnant and non-pregnant women from Saudi population. Infection Drug Resistance 12:1749–1761. https://doi.org/10.2147/IDR.S200213
Kim PS, Shin N-R, Lee J-B, Kim M-S, Whon TW, Hyun D-W, Yun J-H, Jung M-J, Kim JY, Bae J-W (2021) Host habitat is the major determinant of the gut microbiome of fish. Microbiome 9(1):166. https://doi.org/10.1186/s40168-021-01113-x
Larsen A (2014) Studies on the microbiota of fishes and the factors influencing their composition
Liang Y, Xie X, Chen L, Yan S, Ye X, Anjum K, Huang H, Lian X, Zhang Z (2016) Bioactive polycyclic Quinones from marine Streptomyces sp. 182SMLY. Mar Drugs 14(1):10. https://doi.org/10.3390/md14010010
Ligia A d CC, Karen YFK, Susan GK (2017) Microbial production of carotenoids A review. Afr J Biotechnol 16(4):139–146
Manirethan V, Raval K, Rajan R, Thaira H, Balakrishnan RM (2018) Data on the removal of heavy metals from aqueous solution by adsorption using melanin nanopigment obtained from marine source: Pseudomonas stutzeri. Data Brief 20:178–189. https://doi.org/10.1016/j.dib.2018.07.065
Manirethan V, Raval K, Balakrishnan RM (2020) Adsorptive removal of trivalent and pentavalent arsenic from aqueous solutions using iron and copper impregnated melanin extracted from the marine bacterium Pseudomonas stutzeri. Environ Pollut (Barking, Essex: 1987) 257:113576. https://doi.org/10.1016/j.envpol.2019.113576
Masuelli L, Pantanella F, La Regina G, Benvenuto M, Fantini M, Mattera R, Di Stefano E, Mattei M, Silvestri R, Schippa S, Manzari V, Modesti A, Bei R (2016) Violacein, an indole-derived purple-colored natural pigment produced by Janthinobacterium lividum, inhibits the growth of head and neck carcinoma cell lines both in vitro and in vivo. Tumour Biol 37(3):3705–3717. https://doi.org/10.1007/s13277-015-4207-3
Merrifield DL, Carnevali O (2014) Probiotic modulation of the gut microbiota of fish. Aquaculture nutrition: gut health, probiotics and prebiotics, 185–222
Mieszkin S, Callow ME, Callow JA (2013) Interactions between microbial biofilms and marine fouling algae: a mini review. Biofouling 29(9):1097–1113. https://doi.org/10.1080/08927014.2013.828712
Nedwell D, Blackburn T, Wiebe W (1994) Dynamic nature of the turnover of organic carbon, nitrogen and Sulphur in the sediments of a Jamaican mangrove forest. Mar Ecol Prog Ser 110:223–231
Oswald WJ, Gotaas HB (1957) Photosynthesis in sewage treatment. Trans Am Soc Civ Eng 122(1):73–105
Pajares S, Ramos R (2019) Processes and microorganisms involved in the marine nitrogen cycle: knowledge and gaps. Front Mar Sci 6. https://doi.org/10.3389/fmars.2019.00739
Partensky F, Hess WR, Vaulot D (1999) Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev 63(1):106–127. https://doi.org/10.1128/MMBR.63.1.106-127.1999
Pierson LS, Pierson EA (2010) Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol 86(6):1659–1670. https://doi.org/10.1007/s00253-010-2509-3
Podar M, Reysenbach A-L (2006) New opportunities revealed by biotechnological explorations of extremophiles. Curr Opin Biotechnol 17(3):250–255. https://doi.org/10.1016/j.copbio.2006.05.002
Ramesh CH, Vinithkumar NV, Kirubagaran R (2019) Marine pigmented bacteria: a prospective source of antibacterial compounds. J Nat Sci Biol Med 10:104
Ramesh C, Vinithkumar NV, Kirubagaran R, Venil CK, Dufossé L (2020) Applications of prodigiosin extracted from marine red pigmented bacteria Zooshikella sp. and actinomycete streptomyces sp. Microorganisms 8(4). https://doi.org/10.3390/microorganisms8040556
Sapp M, Schwaderer AS, Wiltshire KH, Hoppe H-G, Gerdts G, Wichels A (2007a) Species-specific bacterial communities in the phycosphere of microalgae? Microb Ecol 53(4):683–699. https://doi.org/10.1007/s00248-006-9162-5
Sapp M, Wichels A, Wiltshire KH, Gerdts G (2007b) Bacterial community dynamics during the winter-spring transition in the North Sea. FEMS Microbiol Ecol 59(3):622–637. https://doi.org/10.1111/j.1574-6941.2006.00238.x
Schäfer H, Abbas B, Witte H, Muyzer G (2002) Genetic diversity of “satellite” bacteria present in cultures of marine diatoms. FEMS Microbiol Ecol 42(1):25–35. https://doi.org/10.1111/j.1574-6941.2002.tb00992.x
Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, Huttenhower C, Littman DR (2013) Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. elife 2:e01202. https://doi.org/10.7554/eLife.01202
Schloss PD, Handelsman J (2003) Biotechnological prospects from metagenomics. Curr Opin Biotechnol 14(3):303–310. https://doi.org/10.1016/s0958-1669(03)00067-3
Seymour JR, Amin SA, Raina J-B, Stocker R (2017) Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat Microbiol 2:17065. https://doi.org/10.1038/nmicrobiol.2017.65
Soliev AB, Hosokawa K, Enomoto K (2011) Bioactive pigments from marine bacteria: applications and physiological roles. Evid Based Complement Alternat Med 2011:670349. https://doi.org/10.1155/2011/670349
Sullam KE, Essinger SD, Lozupone CA, O’Connor MP, Rosen GL, Knight R, Kilham SS, Russell JA (2012) Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol 21(13):3363–3378. https://doi.org/10.1111/j.1365-294X.2012.05552.x
Tang X, He LY, Tao XQ, Dang Z, Guo CL, Lu GN, Yi XY (2010) Construction of an artificial microalgal-bacterial consortium that efficiently degrades crude oil. J Hazard Mater 181(1–3):1158–1162
Tarasov VG, Gebruk AV, Mironov AN, Moskalev LI (2005) Deep-sea and shallow-water hydrothermal vent communities: two different phenomena? Chem Geol 224(1–3):5–39
Taylor MW, Radax R, Steger D, Wagner M (2007) Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71(2):295–347. https://doi.org/10.1128/MMBR.00040-06
Thakur AN, Thakur NL, Indap MM, Pandit RA, Datar VV, Müller WEG (2005) Antiangiogenic, antimicrobial, and cytotoxic potential of sponge-associated bacteria. Marine Biotechnol (New York, NY) 7(3):245–252. https://doi.org/10.1007/s10126-004-4085-y
Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N (2017) Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27(4):626–638. https://doi.org/10.1101/gr.216242.116
Ullah R, Yasir M, Khan I, Bibi F, Sohrab SS, Al-Ansari A, Al-Abbasi F, Al-Sofyani AA, Daur I, Lee S-W, Azhar EI (2017) Comparative bacterial community analysis in relatively pristine and anthropogenically influenced mangrove ecosystems on the Red Sea. Can J Microbiol 63(8):649. https://doi.org/10.1139/cjm-2016-0587
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Smith HO et al (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science (New York, NY) 304(5667):66–74. https://doi.org/10.1126/science.1093857
Vijayan V, Jasmin C, Anas A, Parakkaparambil Kuttan S, Vinothkumar S, Perunninakulath Subrayan P, Nair S (2017) Sponge-associated bacteria produce non-cytotoxic melanin which protects animal cells from photo-toxicity. Appl Biochem Biotechnol 183(1):396–411. https://doi.org/10.1007/s12010-017-2453-0
Villarreal-Gómez LJ, Soria-Mercado IE, Guerra-Rivas G, Ayala-Sánchez NE (2010) Antibacterial and anticancer activity of seaweeds and bacteria associated with their surface. Rev Biol Mar Oceanogr 45(2):267–275
Walsh EA, Kirkpatrick JB, Rutherford SD, Smith DC, Sogin M, D’Hondt S (2016) Bacterial diversity and community composition from seasurface to subseafloor. ISME J 10(4):979–989. https://doi.org/10.1038/ismej.2015.175
Webster NS, Hill RT (2001) The culturable microbial community of the great barrier reef sponge Rhopaloeides odorabile is dominated by an α-Proteobacterium. Marine Biol 138(4):843–851. https://doi.org/10.1007/S002270000503
Wichard T (2015) Exploring bacteria-induced growth and morphogenesis in the green macroalga order Ulvales (Chlorophyta). Front Plant Sci 6:86
Zheng L, Chen H, Han X, Lin W, Yan X (2005) Antimicrobial screening and active compound isolation from marine bacterium NJ6-3-1 associated with the sponge Hymeniacidon perleve. World J Microbiol Biotechnol 21(2):201–206. https://doi.org/10.1007/s11274-004-3318-6
Zhu D, Tanabe S-H, Yang C, Zhang W, Sun J (2013) Bacterial community composition of South China Sea sediments through pyrosequencing-based analysis of 16S rRNA genes. PLoS One 8(10):e78501. https://doi.org/10.1371/journal.pone.0078501
Zhuang W-Q, Tay J-H, Maszenan AM, Krumholz LR, Tay ST-L (2003) Importance of gram-positive naphthalene-degrading bacteria in oil-contaminated tropical marine sediments. Lett Appl Microbiol 36(4):251–257. https://doi.org/10.1046/j.1472-765x.2003.01297.x
Zykwinska A, Marchand L, Bonnetot S, Sinquin C, Colliec-Jouault S, Delbarre-Ladrat C (2019) Deep-sea hydrothermal vent bacteria as a source of glycosaminoglycan-mimetic exopolysaccharides. Molecules (Basel, Switzerland) 24(9):1703. https://doi.org/10.3390/molecules24091703
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Khan, I., Ullah, R., Ali, S., Shah, M.D. (2023). Bacterial Diversity in the Marine Environment and the Cutting-Edge Tools for Isolation, Identification and Characterization of the Marine Microbiome. In: Shah, M.D., Ransangan, J., Venmathi Maran, B.A. (eds) Marine Biotechnology: Applications in Food, Drugs and Energy. Springer, Singapore. https://doi.org/10.1007/978-981-99-0624-6_11
Download citation
DOI: https://doi.org/10.1007/978-981-99-0624-6_11
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-0623-9
Online ISBN: 978-981-99-0624-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)