Abstract
Exploring the potential efficacy of a drug is a valid approach for drug discovery with shorter development times and lower costs. Recently, several computational drug repositioning methods have been introduced to learn multi-features for potential association prediction. A drug repositioning knowledge graph of drugs, diseases, targets, genes and side effects was introduced in our study to impose an explicit structure to integrate heterogeneous biomedical data. We revealed drug and disease embeddings from the constructed knowledge graph via a two-layer graph convolutional network with an attention mechanism. Finally, KGCN-DDA achieved superior performance in drug-disease association prediction with an AUC value of 0.8818 and an AUPR value of 0.5916, a relative improvement of 31.67% and 16.09%, respectively, over the second-best results of the four existing state-of-the-art prediction methods. Meanwhile, case studies have verified that KGCN-DDA can discover new associations to accelerate drug discovery.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In recent decades, drug discovery techniques and biological systems have been intensively studied by multidisciplinary researchers. However, drug development is still a time-consuming, costly and labor-intensive process. Drug repositioning is a strategy for identifying new uses for approved or investigational drugs that are outside the scope of the original medical indications [1]. It could ease the drug development process, shorten the required time to 6.5 years, reduce costs to $300 million and reduce the risk of failure.
In recent years, computational drug repositioning methods [2] have attracted continuous attentions with explosive growth of large-scale genomic and phenotypic data. The previous computational methods can be roughly divided into three categories: complex network method [3], machine learning method [4], and deep learning method [5]. Besides, the knowledge organization method [6], for example ontologies and knowledge graph, has also been gradually applied to the research of drug disease relationship prediction recently.
With the explosion of the total amount of drug discovery knowledge, the relationships between entities, such as drugs, diseases, targets, symptoms, etc., become progressively more complex. There is a wealth of associations hidden in literature, clinical guidelines, encyclopedias, and structured databases. Semi-structured and unstructured knowledge needs further exploration and exploitation. More hidden drug-disease associations can be found by fully utilizing public databases and literature knowledge related to drug development and disease treatment. This can reduce the risk of failure, shorten the time needed for research and development, and save money, manpower, and material resources. In this study, we first construct a drug repositioning knowledge graph and then propose a novel drug-disease association prediction method called KGCN-DDA based on multiple features in the knowledge graph and graph convolutional neural network. KGCN-DDA has achieved good performance in the prediction of unknown drug disease association. This method can find new indications of drugs, and also provide methodological reference and theoretical basis for drug relocation.
2 Methods and Materials
2.1 Dataset
Data for drug repositioning knowledge graph construction were primarily collected from various data sources including Comparative Toxicology Database (CTD), Drugbank, SIDER, MeSH and PubMed scientific literature from PubMed. Taking as a starting point, 269 drugs, 598 diseases and 18416 drug-disease associations originated from Comparative Toxicology Database (CTD). We extracted drug-target associations from Drugbank and drug-side effect associations from SIDER for drug repositioning knowledge graph construction. Biological semantic relationships between drugs, diseases, targets, genes, and side effects were also discovered from 12056 PubMed scientific literature which titles or abstracts containing drugs or diseases from the CTD dataset. Besides, drug chemical structures (represented by SMILES) from Drugbank, and diseases’ tree numbers from MeSH served as entities attributes to in our study.
2.2 Drug–Disease Association Prediction Based on Knowledge Graph and GCN
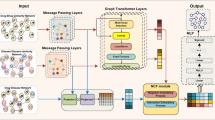
In this study, we presented a comprehensive knowledge graph of drug repositioning with relevant drugs, diseases, targets, genes and side effects. Meanwhile, graph convolutional neural network worked as an efficient way to extract multi-features from the constructed knowledge graph. The workflow of KGCN-DDA was briefly shown in Fig. 1.
Drug Repositioning Knowledge Graph Construction.
Our drug-centric knowledge graph data model comprised five types of entities includes drugs, diseases, and other entities that interact with the two entities, such as targets, side effects and genes. It curates and normalizes data from the four publicly available databases mentioned above, as well as information from PubMed publications based on a pre-training and fine-tuning BERT model. The eight relationship types in drug repositioning knowledge graph include treat (between drugs and diseases), interact (between two drugs), cause (between drugs and side effects), target (between drugs and targets), associate (between drugs and genes), associate (between two genes), biomarker (between diseases and genes), and target (between diseases and targets).
Drug–Disease Feature Representation and Association Prediction.
We calculated drug-drug similarities and disease-disease similarities based on multi features based on the drug repositioning knowledge graph, including: (1) drug-side effect associations, drug-target associations, drug-gene associations, drug molecular fingerprints, (2) disease-target associations, disease-gene target associations, disease MeSH tree-numbers. We then proposed this multi-feature fusion similarities and drug-disease associations in the knowledge graph to compute an association feature matrix. Finally, two GCN layers were applied to learn drug and disease embeddings of with an attention mechanism. An inner product decoder was used to discover unknown drug-disease associations.
3 Results and Discussion
3.1 Performances and Comparison with State-of-the-Art Methods
In this study, we constructed a drug repositioning knowledge graph based on structured knowledge and semantic information from biomedical literature. Specifically, a knowledge graph of drugs, diseases, targets, genes and side effects was constructed. There are in total of 8374 entities (269 drugs, 598 diseases, 266 targets, 3793 side effects, and 2938 genes) and 67350 triples (18416 drug-disease, 43508 drug-side effect, 722 drug-target, 4081 disease-gene, and 623 disease-target) in knowledge graph. For feature fusion and similarity computation, an adjusted weight for each measurement was applied to achieve optimal performance by a step of 0.01. Finally, the AUC and AUPR of our predictive model reached 0.8801 and 0.5961 optimality. Compared with four existing state-of-the-art prediction methods [7,8,9,10], KGCN-DDA achieved superior performance in drug-disease association prediction, shown in Table 1, which were 33.89% and 16.09% relative improvements than the second-best result.
3.2 Case Study
To demonstrate KGCN-DDA's ability to discover new indications and new therapies, we conducted three case studies with validation from clinical indications already in use, Clinical Trials, CTD and public literature from PubMed: (1) Top 10 drug–disease associations, (2) Top 10 associated diseases for given drugs (Doxorubicin).
We listed the top 10 drug-disease associations predicted by KGCN-DDA in Table 2, and seven out of them can be demonstrated by the verification methods mentioned above. For example, we found olanzapine and fluoxetine together are more effective than duloxetine alone for treating severe depression in terms of improving physical and sleep quality [11]. Researchers examined how rosiglitazone inhibits hepatocellular carcinoma and showed that the medication can cause liver cancer cells to undergo apoptosis [12]. According to study from Johns Hopkins University in the United States, taking a certain amount of caffeine might enhance the body's memory function temporarily [13]. Cimetidine is a medication that can be used clinically to treat arrhythmia and chronic hepatitis B hepatitis. This therapeutic approach aligns with the expected management of inflammation and cardiac disease. Besides, several predictions have been confirmed effective by ClinicalTrials and CTD records.
The top 10 combinations in drug-disease prediction were examined from the viewpoint of a single medication, using doxorubicin as an example (Table 3). Doxorubicin is an anti-tumor medication that mostly inhibits DNA synthesis, but it can also limit RNA synthesis as well. It has a broad anti-tumor range and is mostly used in clinical practice to treat individuals with acute leukemia, including acute lymphocytic leukemia and acute myeloid leukemia. Combinations 1, 2, 3, 6, 8 [14,15,16,17,18] have been clinically treated and validated by literature, including doxorubicin, which has a certain ameliorative impact on non-small cell lung cancer, acute myeloid leukemia, trigeminal neuralgia, glioma, and osteosarcoma. Meanwhile, the remaining three combinations have not received much attention but have been predicted by the KGCN-DDA model. To some extent, this might give researchers fresh ideas for drug repositioning. As a result, it is feasible to predict drug-disease association by KGCN-DDA.
4 Conclusions
In this study, we proposed a method called KGCN-DDA for drug-disease association prediction. Due to the huge amount of information contained in biomedical public databases and scientific literature, we constructed a drug repositioning knowledge graph and compute drug-drug and disease-disease similarities by knowledge graph multi-feature fusion. Two GCN layers were utilized to capture structural embeddings from association feature matrix. The proposed method achieved superior performance compared to four state-of-the-art methods, and we demonstrated its potential for identifying novel drug-disease associations in clinical practice.
However, there are still some limitations in our work that require an in-depth investigation. First, more association features should be further considered in our work. We can collect more prior biological knowledge from literature or datasets, such as drug-protein, drug-gene, disease-gene and drug-pathway from DisGeNET, Gene Ontology (GO) and so on, to improve similarity accuracy. Second, the two-layer GCN is a basic model for learning on graph-structured data, while some other graph neural network models are worth investigating in the future.
Above all, KGCN-DDA is able to learn scattered multidimensional information from heterogeneous networks and identify latent drug-disease associations. It gives researchers, pharmacologists, and pharmaceutical companies a tremendous opportunity to study and validate predictive associations that are more likely to exist. We expect KGCN-DDA to be an efficient approach that can improve drug repositioning in the future and shorten its cost and time.
References
Pushpakom, S., et al.: Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 18(1), 41–58 (2019)
Deng, J., Yang, Z., Ojima, I., Samaras, D., Wang, F.: Artificial intelligence in drug discovery: applications and techniques. Brief Bioinform. 23(1), bbab430 (2022)
Wang, W., Yang, S., Zhang, X., Li, J.: Drug repositioning by integrating target information through a heterogeneous network model. Bioinformatics 30(20), 2923–2930 (2014)
Napolitano, F., et al.: Drug repositioning: a machine-learning approach through data integration. J. Cheminform. 5(1), 30 (2013)
Fatehifar, M., Karshenas, H.: Drug-Drug interaction extraction using a position and similarity fusion-based attention mechanism. J. Biomed. Inform. 115(3), 103707 (2021)
Karim, M.R., Cochez, M., Jares, J., Uddin, M., Beyan, O., Decker, S.: Drug-drug interaction prediction based on knowledge graph embeddings and convolutional-LSTM network. ACM (2019). arXiv:1908.01288
Gao, C.Q., Zhou, Y.K., Xin, X.H., Min, H., Du, P.F.: DDA-SKF: predicting drug-disease associations using similarity Kernel fusion. Front. Pharmacol. 12, 784171–784186 (2022)
Cai, L., et al.: Drug repositioning based on the heterogeneous information fusion graph convolutional network. Brief Bioinform. 22(6), bbab319 (2021)
Yu, Z., Huang, F., Zhao, X., Xiao, W., Zhang, W.: Predicting drug-disease associations through layer attention graph convolutional network. Brief Bioinform. 22(4), bbaa243 (2021)
Meng, Y., Lu, C., Jin, M., Xu, J., Zeng, X., Yang, J.: A weighted bilinear neural collaborative filtering approach for drug repositioning. Brief Bioinform. 23(2), bbab581 (2022)
Qu, W., Gu, S., Luo, H., et al.: Effects of olanzapine-fluoxetine combination treatment of major depressive disorders on the quality of life during acute treatment period. Cell Biochem. Biophys. 70(3), 1799–1802 (2014)
Bo, Q., Sun, X., Jin, L., et al.: Antitumor action of the peroxisome proliferator-activated receptor-γ agonist rosiglitazone in hepatocellular carcinoma. Oncol. Lett. 10(4), 1979–1984 (2015)
Borota, D., Murray, E., Keceli, G., et al.: Post-study caffeine administration enhances memory consolidation in humans. Nature Neurosci. 17(2), 201–212 (2014)
Ghosh, S., Lalani, R., Maiti, K., et al.: Synergistic co-loading of vincristine improved chemotherapeutic potential of pegylated liposomal doxorubicin against triple negative breast cancer and non-small cell lung cancer. Nanomedicine 31(2), e102320 (2021)
Perry, J.M., Tao, F., Roy, A., et al.: Overcoming Wnt-β-catenin dependent anticancer therapy resistance in leukaemia stem cells. Nat. Cell Biol. 22(6), 689–700 (2020)
Zheng, B., Song, L., Liu, H.: Gasserian ganglion injected with Adriamycin successfully relieves intractable trigeminal nerve postherpetic neuralgia for an elderly patient: a case report. Medicine (Baltimore) 97(38), e12388 (2018)
Niu, W., Xiao, Q., Wang, X., et al.: A biomimetic drug delivery system by integrating grapefruit extracellular vesicles and doxorubicin-loaded heparin-based nanoparticles for glioma therapy. Nano Lett. 21(3), 1484–1492 (2021)
Wei, H., Chen, J., Wang, S., et al.: A nanodrug consisting of doxorubicin and exosome derived from mesenchymal stem cells for osteosarcoma treatment in vitro. Int. J. Nanomed. 14(1), 8603–8610 (2019)
Funding
This work was supported by The National Social Science Fund of China (22CTQ024), Innovation Project of Chinese Academy of Medical Sciences (2021-I2M-1-001), The National Key Research and Development Program of China (2022YFB2702801).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Kang, H., Hou, L., Li, J., Li, Q. (2024). KGCN-DDA: A Knowledge Graph Based GCN Method for Drug-Disease Association Prediction. In: Cruz, C., Zhang, Y., Gao, W. (eds) Intelligent Computers, Algorithms, and Applications. IC 2023. Communications in Computer and Information Science, vol 2036. Springer, Singapore. https://doi.org/10.1007/978-981-97-0065-3_12
Download citation
DOI: https://doi.org/10.1007/978-981-97-0065-3_12
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-97-0064-6
Online ISBN: 978-981-97-0065-3
eBook Packages: Computer ScienceComputer Science (R0)