Abstract
In the present scenario, solid waste management has emerged as a huge problem for the environment and mankind. Most of the wastes are generated from the plant source that is made up of cellulose. Cellulose can serve as the source of glucose by various methods of degradation that can be chemical or enzymatic. The cellulase enzyme of the bacterial system can also be used for cellulose degradation. In the present study, cellulose-degrading bacteria have been isolated from soil obtained from wood habitat. To indicate the cellulase activity of the organisms, the diameter of clear zone around the colony and hydrolytic value on cellulose Congo Red agar media were measured. The strain with maximum cellulolytic potential was selected on the basis of DNS assay. The selected strain was further characterized at the molecular level using 16S RNA sequencing. The characterized strain was later used for the production of glucose by degrading the tissue paper. The glucose such produced was used for the production of bioethanol using Saccharomyces cerevisiae. The production of ethanol was positively tested after 6 days by iodoform test and amount of alcohol produced was obtained by distillation of the fermented mash.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Human activities create wastes and these wastes are handled, stored, collected, and disposed of so that they do not become a risk for the health and environment. With the rapid increase in urbanization and population, the nature of solid waste has become complexed. Municipal solid waste (MSW) is generally categorized as degradable and nondegradable (Mehta et al. 2018). Wastes like paper, textile, fiber, food all those composed of plant components are degradable in nature and other wastes like plastic, metals, glass, e-wastes are termed as nondegradable. Characterization of MSW indicated that the waste consists of 30–45% organic matter, 6–10% recyclables, and the rest as inert matter (Kumar et al. 2009). The nature of waste suggests that the main content of waste is cellulose, which is the component of plant cell wall.
Cellulose is a complex carbohydrate or polysaccharide consisting of several glucose units linked together by β-1,4 glycosidic bond (Klemm et al. 2005). Abundant availability and accessibility of cellulose make it a long-term raw material for producing many industrially important commercial products that will be cost-effective and eco-friendly. But in the present juncture, much of the cellulose are disposed of as waste all over the globe. Cellulose is mainly obtained from plants hence if we are concerned about the environment and global warming, we must find ways to utilize the cellulosic waste generated. This can be done by using cellulolytic system, by which cellulose can be converted into glucose, which is a multiutility product and has industrial value. Cellulolysis is a biological process for the degradation of cellulose that is effectively done by the cellulase enzyme system (Kumakura 1997).
The microbial system is the best source for the extraction of cellulose-degrading enzyme. There is a large range of microorganisms that are responsible for cellulase production including aerobic and anaerobic bacteria, white rot and soft rot fungi, and anaerobic fungi (Lynd et al. 2002). The cellulase enzymes isolated from microbes can be used to degrade cellulose to produce glucose that can be further utilized for the production of several industrially important products such as bioethanol. Bioethanol is a striking alternative fuel as it is a renewable bio-based resource, and it is oxygenated and hence provides the opportunity to reduce particulate emissions in compression–ignition engines (Balat 2007). Therefore, in the present investigation, focus has been made to isolate cellulose-degrading bacteria and utilizes its cellulolytic potential for degradation of paper waste for the production of bioethanol. Several studies showed the production of bioethanol from paper waste using hydrolysis and fermentation (Wang et al. 2013; Maceiras et al. 2016).
2 Materials and Methods
2.1 Sample Collection
For isolation of cellulose-degrading bacteria (CDB), the soil samples were collected from location of wood habitat manufacturing industry of Bawana, New Delhi, India (28.7884° N, 77.0301° E). After collection, the soil samples were stored at 4 °C in sterile containers.
2.2 Screening of Cellulose-Degrading Bacteria
For screening of cellulose-degrading bacteria (CDB), around 2 g of soil sample was homogenized in 0.9% saline solution and serially diluted ranging from 10−1 to 10−3 under sterile condition. Around 100 µl of each serially diluted sample was spread plate on carboxymethyl cellulose (CMC) agar media composed of KH2PO4, 0.5 g; MgSO4, 0.25 g; cellulose, 2 g; agar, 15 g; gelatin, 2 g; distilled water, 1 L; pH, 6.8–7.2. The plates were incubated at 37 °C for 24 h. Colonies obtained were maintained on CMC agar plate and preserved at 4 °C.
2.3 Cellulolytic Ability Assay
Cellulose-degrading ability was analyzed by streaking the isolated bacterial strains on Congo Red agar media composed of CMC agar media supplemented with Congo Red 0.2 g/l. The formation of a clear zone of hydrolysis indicated cellulose degradation ability of the isolated bacterial strain. The colony showing maximum degradation was selected and used for further study.
2.4 Estimation of Carboxymethyl Cellulase (CMCase) Activity
Using the 3, 5-dinitrosalicylic acid (DNS) method, CMCase activity was determined (Miller 1959). Supernatant obtained from overnight-grown culture after centrifugation at 5000 rpm for 10 min at 4 °C was used as crude enzyme source. Assay was carried out by adding around 0.5 ml of crude enzyme to 0.5 ml of 1% CMC (solubilized in 0.05 M phosphate buffer, pH 8) followed by incubation at 50 °C in water bath for 30 min. The reaction was stopped by the addition of 1.5 ml of DNS reagent followed by boiling the reaction mixture for 10 min. Sugars liberated were estimated by measuring absorbance at 540 nm. A unit of activity is defined as the amount of enzyme required to liberate 1 mol of glucose per minute under the assay conditions.
2.5 Molecular Identification and Phylogenetic Analysis
The bacterial strain that showed maximum cellulose activity was used for molecular characterization using 16S rRNA sequencing. The isolated bacterial strain streaked on CMC agar plate was deposited and sequenced at Microbial Type Culture Collection and Gene Bank (MTCC), CSIR-IMTECH, Chandigarh, India. The sequence obtained was compared using BLASTN program and the members of the closely related genera were retrieved from EzTaxon server (Chun et al. 2007) and aligned using the MEGA software version 6.0 (Tamura et al. 2013). Phylogenetic trees were constructed using the neighbor-joining as well as maximum parsimony algorithms. Bootstrap analysis was performed to assess the confidence limits of the branching.
2.6 Production of Bioethanol
The isolated CDB strain was grown in two sets, one in CMC media and another in basal salt media composed of KH2PO4, 2 g; MgSO4, 0.2 g; NaCl, 0.2 g; NaNO3, 2.5 g; CaCl3·6H2O, 0.1 g; Whatman No. 1 filter paper, 2.5 g; distilled water, 1 L; pH 6.8–7.2, for 4 days at 37 °C with constant shaking of 150 rpm. The supernatant obtained from the first set of culture was used as the source of enzymes for saccharification process, where as the broth obtained from the second culture was used for fermentation using Saccharomyces cerevisiae (NCIM Accession No: 3594, NCL, Pune, India). Simultaneous saccharification and fermentation processes were carried out at 27 °C for 6 days as described by Jain et al. 2014. Preparation of yeast growth medium was done using yeast extract 5.0 g, peptone 5.0 g, diluted saccharified slurry to attain an overall sugar percentage of 1%, volume made up to 1000 ml with pH 5.0.
Post fermentation, iodoform test was done to confirm the presence of ethanol. The fermented mash was distilled and alcohol percentage was estimated (Ghosal et al. 2013).
2.7 Iodoform Test
The presence of ethanol in the culture broth was confirmed by iodoform test (Lieben 1870). Around 10 drops of methanol, ethanol, and the culture broth were taken in three separate test tubes. The methanol and ethanol were taken as negative and positive control, respectively. Around 10 drops of diluted NaOH solution were added to each test tube along with 30 drops of iodine solution. The color developed was later analyzed.
3 Results and Discussion
3.1 Isolation of Cellulose-Degrading Bacteria
Cellulose-degrading bacteria were isolated from the soil sample on CMC agar medium using serial dilution. Serial dilution ranging from 10−1 to 10−3 under sterile condition was performed to obtain the isolated colonies (Fig. 1). A total of 37 isolated colonies were selected and named as cellulose-degrading bacteria, CDB 1–37. Isolation of CDB was also reported from the other sources like guts of invertebrates (Gupta et al. 2012), kitchen waste (Kaur 2012), and sheep rumen (Guder and Krishna 2019).
3.2 Cellulolytic Ability Assay
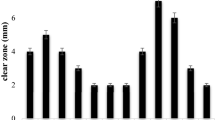
Cellulose degrading ability was analyzed by streaking the isolated bacterial strains on Congo Red agar media. Based on the morphology, six morphologically distinct bacterial isolates that were giving halo zones in CMC agar medium were selected, namely, CDB1, CDB4, CDB29, CDB32, CDB34, and CDB37 (Fig. 2).
3.3 Carboxymethyl Cellulase (CMCase) Activity
Production of cellulase enzyme by six bacterial isolates was analyzed using the 3, 5-dinitrosalicylic acid (DNS) method. The cellulase production ability of the bacterial isolates in the production medium was in the following order, CDB-37 < CDB-32 < CDB-4 < CDB-29 < CDB-34 < CDB-1 (Fig. 3). Among these two bacterial isolates, CDB-1 and CDB-34 showed maximum cellulase production ability. Out of these strains, CDB-1 was chosen for molecular characterization.
3.4 Molecular Identification of Cellulolytic Bacteria
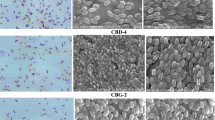
The strain CDB-1 that showed maximum cellulose activity was further subjected to molecular characterization. The 16S rRNA gene sequence obtained was compared using BLASTN program, which showed the strain CDB-1 to be Streptomyces albogriseolus. Multiple sequence alignment analysis with all the closely related genera showed 99–98% similarities at the sequence level. Phylogenetic tree analysis showed that the strain CDB-1 is closely related to other Streptomyces species available in the database (Fig. 4). Several actinomycetes are known to degrade cellulose (Saini et al. 2015).
3.5 Production of Bioethanol from Tissue Paper
The strain CDB-1 was used for its cellulolytic potential to degrade tissue paper to obtain glucose as the product that was, for further, used for fermentation. The strain Sacchromyces cerevisiae was used in the fermentation process for the conversion of glucose to ethanol. The fermentation product was obtained from the broth that was further confirmed by iodoform test (Fig. 5). After distillation of the fermented mash, the percentage of alcohol in distillate was calculated to be 7.8%.
4 Conclusion
Rapid increase in population and urbanization has led to the generation of huge municipal solid waste. The percentage ofv cellulose waste is around 17%, which mainly consists of woods, papers, old furniture, etc. Using biotechnology tools, research has been accelerated to find ways to convert cellulose into industrial important products. Due to its cellulolytic potential, microbes are the excellent system for the degradation of cellulose. Therefore, in the present investigation, we have screened and isolated cellulose-degrading bacteria (CBD) from the soil sample. Out of 37 bacterial strains, 6 showed cellulose-degrading ability. The cellulase production ability of the six was in the following order—CDB-37 < CDB-32 < CDB-4 < CDB-29 < CDB-34 < CDB-1. Molecular characterization of CBD-1 showed that the strain is closely related to Streptomyces species. The isolated CBD-1 bacterial strain was later used for the saccharification along with S. cerevisiae during fermentation. After distillation of the fermented mash, the production of alcohol was confirmed using iodoform test. Thus, the biodiversity of microbes like bacteria, fungi gives us the chance to identify and isolate strains that could assist in the biological conversion of cellulose into glucose, which can later be used for the production of some industrial useful products.
References
Balat M (2007) Global bio-fuel processing and production trends. Energ Explor Exploit 25(3):195–218. https://doi.org/10.1260/014459807782009204
Chun J, Lee JH, Jung Y et al (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57(10):2259–2261. https://doi.org/10.1099/ijs.0.64915-0
Ghosal A, Banerjee S, Chatterjee S (2013) Biofuel precursor from potato waste. Int J Res Eng Technol 2(3):213–219
Guder DG, Krishna MSR (2019) Isolation and characterization of potential cellulose degrading bacteria from sheep rumen. J Pure Appl Microbiol 13(3):1831–1839. https://doi.org/10.22207/jpam.13.3.60
Gupta P, Samant K, Sahu A (2012) Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int J Microbiol 2012:1–5. https://doi.org/10.1155/2012/578925
Jain M, Gupta AK, Chatterjee S (2014) To optimize the process of alcohol production from banana peel. Recent Adv Bioenergy Res 3:208–217
Kaur M (2012) Isolation and Screening of cellulose degrading bacteria in kitchen waste and detecting their degrading potential. IOSR J Mech Civil Eng 1(2):33–35. https://doi.org/10.9790/1684-0123335
Klemm D, Heublein B, Fink HP et al (2005) Cellulose: fascinating biopolymer and sustainable raw material. Angew Chem Int Edit 44:3358–3393. https://doi.org/10.1002/anie.200460587
Kumakura M (1997) Preparation of immobilized cellulase beads and their application to hydrolysis of cellulosic materials. Process Biochem 32(7):555–559. https://doi.org/10.1016/s0032-9592(97)00011-3
Kumar S, Bhattacharyya JK, Vaidya AN et al (2009) Assessment of the status of municipal solid waste management in metro cities, state capitals, class I cities, and class II towns in India: An insight. Waste Manag 29(2):883–895. https://doi.org/10.1016/j.wasman.2008.04.011
Lieben A (1870) The formation of iodoform and the application of this reaction in the chemical analysis. Liebigs Ann Chem Suppl 7:218. https://doi.org/10.1002/9780470114735.hawley09809
Lynd LR, Weimer PJ, Van Zyl WH et al (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66(3):506–577. https://doi.org/10.1128/mmbr.66.3.506-577.2002
Maceiras R, Alvonsin V, Poole JE (2016) Bioethanol production from waste office paper. J Environ Sci 6(7)
Mehta YD, Shastri Y, Joseph B (2018) Economic analysis and life cycle impact assessment of municipal solid waste (MSW) disposal: a case study of Mumbai, India. Waste Manag Res 36(12):1177–1189
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428
Saini A, Aggarwal NK, Sharma A et al (2015) Actinomycetes: a source of lignocellulolytic enzymes. Enzym Res 2015:1–15. https://doi.org/10.1155/2015/279381
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. MolBiolE 30(12):2725–2729 https://doi.org/10.1093/molbev/mst197
Wang L, Sharifzadeh M, Templer R, Murphy RJ (2013) Bioethanol production from various waste papers: economic feasibility and sensitivity analysis. App Energy 111:1172–1182. https://doi.org/10.1016/j.apenergy.2012.08.048A
Acknowledgements
SC, KT, and RSP like to thank GGS Indraprastha University, New Delhi for all the laboratory space and financial support provided.
Conflict of Interest
The authors declare that they have no conflict of interest.
Declaration by authors
Appropriate permissions were obtained from responsible authorities for collecting soil samples for the study from manufacturing industry of Bawana, New Delhi.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Chatterjee, S., Tripathi, K., Purty, R.S. (2021). Isolation of Cellulose-Degrading Bacteria and to Use Their Cellulolytic Potential for Production of Bioethanol from Paper Waste. In: Ramkrishna, D., Sengupta, S., Dey Bandyopadhyay, S., Ghosh, A. (eds) Advances in Bioprocess Engineering and Technology . Lecture Notes in Bioengineering. Springer, Singapore. https://doi.org/10.1007/978-981-15-7409-2_1
Download citation
DOI: https://doi.org/10.1007/978-981-15-7409-2_1
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-7408-5
Online ISBN: 978-981-15-7409-2
eBook Packages: EngineeringEngineering (R0)