Abstract
The significant issue for image segmentation that performs in the realization and analysis of the tumor and lesion region in multimodal such as CT and MRI images lies in the computational time and accuracy values. Recently, tumor region extraction from medical image that executes with quick response of evolution process will result in the aid of clinical surgical application. The automatic process can be ensured from unsupervised image segmentation algorithm, as it provides the clear identification of the tissue and lesion region in CT and MRI image. In specific, the unsupervised image segmentation process comprises of self-organization map (SOM)-based Modified Fuzzy C-Means Clustering (MFCM) algorithm that results in exact tumor identification and clear segmentation of tumor involved in organ such as liver, lung, brain, and thorax region. The proposed SOM-based Modified Fuzzy C-Means Clustering algorithm is an approach that refers to enhance the image quality measures such as mean squared error (MSE), peak signal-to-noise ratio (PSNR), Jaccard index, and dice overlap index (DOI). Modified Fuzzy K-Means (MFKM) algorithm and the self-organization map (SOM) based fuzzy K-Means (FKM) algorithm are evaluated, and it was finalized that better results are obtained from SOM-based Modified Fuzzy C-Means Clustering algorithm.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In early year, tumor disease is diagnosed through the CT and MRI image, which are mostly helpful for radiotherapy treatment and medical research. The most segmentation method was performed upon graylevel information process, in which similar intensities of several organs make the tumor region complicated to identify. Kohonen [9] introduced an unsupervised approach of SOM neural network with automatic topological mapping process, which provides one- or two-dimensional reduction using competitive learning approach. So far, the image segmentation process offered by SOM neural network tends to be better attractive, significantly due to the topological mapping process. Jiang [7] suggested an automatic classification process of SOM neural network by establishing feature vector (color, shape, texture, etc.) that offers assistance of color image segmentation process. Ong [10] proposed fixed order of two-stage classification techniques that uses SOM neural network in image segmentation process, and it is found that noise removal process for unsupervised approach cannot be offered. Khan [8] employed SOM neural network that instantaneously performs with fuzzy clustering approach resulting in an automatic optimal cluster center. This system also gets assistance from trained network to get offered with dimensionality reduction processes during unsupervised segmentation. The segmentation of SOM with fuzzy clustering approach is compared with efficient graph (EG)-based segmentation approach, in terms of center initialization process. The research group headed by Halder [6] recommended an automatic classification process prepared with SOM neural network, modal analysis, and mutational agglomeration. This segmentation approach relies on mapping process and employed with dimensional reduction, and the quality of classification was quite low. Torbati et al. [14] suggested the 2D discrete wavelet transform that performs with moving average SOM (MASOM) algorithm to achieve efficient segmentation, as it ascertains the least participation of human interaction in the modification of the variables that instigate segmentation process. The multilayer clustering process with a novel hierarchical self-organizing map (HSOM) and vector quantization process achieves efficient classification upon the input image [2]. To solve the time efficiency problem, Gular [5] introduced a novel automatic SOM approach that uses the pixel intensity characteristics of input image for effective segmentation. Hybrid approach utilizing automatic process of image segmentation in tissue and segmentation of tumor region has been proposed by Ortiz. Subsequently, this fully automatic process of image segmentation utilizes hybrid SOM and genetic algorithm that rely on feature vector such as entropy and gradient for clustering MR brain images [11]. Vishnuvarthanan et al. [15] recommended SOM-based FKM algorithm that provides brain tumor identification and segmentation of MR input image. This methodology serves to be an automatic process, and it fuses clustering with self-organizing map to attain effective segmentation of MRI images. The major hindrance of this approach during the segmentation process is the more consumption of computational time.
2 Proposed Method
Combination of SOM and MFCM algorithms can be of potentially important for the automatic unsupervised process in image segmentation. Figure 1 exposes the execution process of SOM-based MFCM algorithm. The initial process for SOM-based MFCM algorithm includes clustering vector followed by the mapping and classification process. This process was extensively executed for the feature extraction in terms of mean and standard deviation of the input image [15]. The significant process of feature extraction is adopted to overcome the over fitting problem. Thus, the mapping process uses \( 8 \times 8 \) local window to support the membership function acquisition from the nearest neighbor. The SOM prototype \( w_{x} \) will be established from the minimum Euclidean distance with nearest neighbor pixel. Let \( X\epsilon J^{r} \), where X represents the input vector and the input data is indicated as \( J^{r} \).
From Eq. (1), the nearest minimum neighbor pixel was obtained such that xϵX, where \( x\left( z \right) \) refers to the input vector at instants of time ‘n’ and \( w\left( z \right) \) represents the sample vector function, which are nearer to the input vector and associated with the membership function value. In addition, from the nearest neighbor values, input vector and weight function, best matching unit (BMU) of nearest neuron was found. But, the trained network is still initiated by the input vector and it is updated with BMU from the nearest neighbor within time, which relies on exponential decay function. Thus, updating SOM prototype is expressed as:
Here, \( \propto \left( z \right) \) refers to the learning factor of exponential decay at instants of time ‘z’. In order to provide the efficient classification process of input information, the updated SOM prototype is used. The updated SOM prototype provides two-dimensional information for classification process. MFCM algorithm receives inputs from the updated SOM prototype for the entire clustering process. The standard MFCM objective function introduced by Ahmed et al. [1] for clustering process is describes as
Let \( N_{k} \) refers to the nearest neighbor function that corresponds to each other of \( x_{c} \) and \( N_{R} \). The parameter refers to the support for assessment of the nearest neighbor that provides the effective segmentation result. In that process, the parameter \( \propto \) value is realized from \( 0 \) to \( 100 \). The value of parameter \( \propto \) in clustering process must be chosen as high as possible due to its inversely proportional characteristic with the SNR value. Chen and Zhang [4] proposed the advance clustering process for finding the nearest neighbors and also provided the lowest processing time. In order to achieve the objective function, the term \( \left\| {\bar{x}_{c} - v_{i} } \right\|^{2} \) was substituted in Eq. (3) instead of \( \frac{1}{{N_{R} }}\sum\nolimits_{{x_{R} \in N_{k} }} {\left\| {x_{R} - v_{i} } \right\|}^{2} \). Then, the objective function of MFCM algorithm becomes
Thus, the SOM prototype provides the automatic cluster center for clustering process. The final segmentation process is done by MFCM algorithm, when provided the efficient segmentation result.
where ‘\( U = \left( {u_{i,c} } \right)_{k,N} \)’ refers to the membership matrix, ‘\( V = \left( {v_{1,} v_{2,} \ldots v_{c} } \right) \)’ refers to the cluster center vectors. Finally, they update the values of membership function and cluster center, which supports of given equation is
3 Result and Discussion
Table 1 explicates the average values of image quality parameter offered by the proposed SOM-based MFCM algorithm when compared with other traditional image segmentation algorithm, which helps to verify the effectiveness of the proposed SOM-based MFCM algorithm.
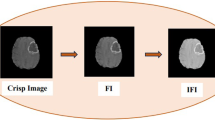
Figure 1 represents the clear understanding of the proposed SOM-based MFCM algorithm. Generally, SOM neural network is trained by the spatial characteristics in the input medical image. An effective classification process of pixels can be derived a \( 8 \times 8 \) map formation, and this map formed by the SOM is offered as the input to MFCM algorithm. Thus, classification derived from map structure becomes a strong motivation for the re-clustered process to obtain the automatic cluster center. Finally, the tumor and lesion regions were explicitly defined in the input and segmented images.
Figure 2 briefly discusses the effective segmentation provided by SOM-based MFCM algorithm. Figure 2a illustrates the larger cyst tumor affecting the right liver lobe region, which is efficiently identified with aid of the proposed SOM-based MFCM algorithm. Figure 2b demonstrates the lesion affecting the right lung lobe which is segmented by the proposed SOM-based MFCM algorithm, which greatly proves the successfulness of lesion region identification.
Figure 3 exhibits the brain tumor of the patients who have been affected by glioma and metastatic bronchogenic carcinoma. Figure 3a, b represents the input image of the proposed SOM-based MFCM algorithm. A concise demarcation between the exact tumor region and the gray matter (GM) and white matter (WM) was obtained using the proposed method.
Table 2 exhibits the average values of evaluation parameter determined for the competitive soft computing algorithms. The proposed SOM-based MFCM algorithm provides the average performance values when compared the other traditional algorithms. Vishnuvarthanan et al. [15] have offered the average estimation values of SOM-based FKM algorithm, which are evaluated for pixel size of input brain image.
3.1 Mean Square Error (MSE)
MSE value represents an estimate of error in image quality that extends the deliberation of input and segmented image. It is a theoretical paradigm for an algorithm to provide better segmentation result. MSE value provided by an algorithm must approach nearby zero [3, 15].
Figure 4 briefly explains the average values of MSE values for soft computing algorithm. Thus, the average MSE value presented by the proposed SOM-based MFCM algorithm is lesser than the MFKM and SOM-based FKM algorithms.
3.2 Peak Signal-to-Noise Ratio (PSNR)
PSNR explains the image quality extent of evaluating the ratio of utmost possible pixels present in the input image and the relevant MSE values [15]. PSNR value is described as
3.3 Jaccard (Tanimoto) Index
Jaccard index is significantly used in for the evaluation of identical extent of pixels presence in the input and segmented image [13, 15]. It has been explicated as
3.4 Dice Overlap Index (DOI)
DOI represents the overlap function and is interrelated with the Jaccard index while resolving the segmentation accuracy [12, 15]. The expansion of DOI value is indicated by,
Figure 5 exposes the estimation of average values of PSNR, Jaccard index, and dice overlap index for the MR brain image segmentation algorithm. The clear-cut proposed segmentation process provides higher PSNR value, and it is par above the PSNR values rendered by other segmentation algorithms (40 dB). The proposed SOM-based MFCM algorithm achieves 64.97 dB, and it is higher than the PSNR results of MFKM and SOM-based FKM algorithms. The average Jaccard index of the proposed SOM-based MFCM algorithm is 57.35%, which is higher than the SOM-based FKM algorithm. Efficient segmentation accuracy was achieved by proposed SOM-based MFCM algorithm, and it offers a dice overlap index of 72.85%, which is greater than the SOM-based FKM algorithm, and can be verified in Fig. 5.
3.5 Computational Time
The significant process that achieves the segmentation results must meet the time requirement, which is usually indicated by seconds. Thus, a segmentation algorithm is considered useful, when it provides effective segmentation result satisfying the condition that execution time should be minimal. The proposed algorithm requires the lowest execution time and also provides effective segmentation results.
Figure 6 reveals the average computational time for soft computing algorithms. The computational time used to process a four number of patient’s medical image was used as factor of comparison, in which the average computational time of the proposed SOM-based MFCM algorithm is affordable than the SOM-based FKM and MFKM algorithms.
3.6 Memory Requirement
As the demand for storage is potentially important in future, the segmentation process must be carefully designed such that the resultant image must consume lesser bytes. The proposed segmentation algorithm is in need of minimum memory space for providing the segmentation results.
Figure 7 exhibits the average memory requirement for soft computing algorithms. The proposed SOM-based MFCM algorithm consumes less memory space for executing the input images. Figure 7 clearly exposes that the proposed SOM-based MFCM algorithm requires 2.11E+13 bytes for performing the segmentation upon the four patient’s medical image, which is the smallest among the soft computing algorithms.
4 Conclusion
In this paper, image segmentation using the novel SOM-based MFCM algorithm was discussed. Combination of SOM and MFCM algorithm provides efficacious segmentation result when compared with the MFKM and SOM-based FKM algorithms. The proposed algorithm, for the segmentation of medical image processing, provides better PSNR values among SOM-based FKM and MFKM algorithms. The SOM-based MFCM algorithm enhances the poorly available medical image through segmentation, thus augmenting the image visualization and quality. The proposed SOM-based MFCM algorithm provides segmentation results in lesser processing time when compared with other traditional algorithms. So, the proposed SOM-based MFCM algorithm is recommended as a more accurate process for the identification of tumor regions. Finally, the obtained segmentation results describe that the functioning of the SOM-based MFCM algorithm is better than the SOM-based FKM and MFKM algorithms.
References
Ahmed MN, Yamany SM, Mohamed N, Farag AA, Moriarty T (2002) A modified fuzzy C-means algorithm for bias field estimation and segmentation of MRI data. IEEE Trans Med Imaging 21:193–199
Bhandarkar SM, Koh J, Suk M (1997) Multiscale image segmentation using a hierarchical self-organizing map. Neurocomputing 14:241–272
Chang CY, Lei YF, Tseng CH, Shih SR (2010) Thyroid segmentation and volume estimation in ultrasound images. IEEE Trans Biomed Eng 57:1348–1357. https://doi.org/10.1109/TBME.2010.2041003
Chen S, Zhang D (2004) Robust image segmentation using FCM with spatial constraints based on new kernel-induced distance measure. IEEE Trans Syst Man Cybern 34:1907–1916. https://doi.org/10.1109/TSMCB.2004.831165
Guler I, Demirhan A, Karakis R (2009) Interpretation of MR images using self-organizing maps and knowledge-based expert systems. Digit Signal Process 19:668–677. https://doi.org/10.1016/j.dsp.2008.08.002
Halder A, Dalmiya S, Sadhu T (2014) Color image segmentation using semi-supervised self-organization feature map. In: Advances in intelligent systems and computing, vol 264. Springer, pp 591–598
Jiang Y, Chen KJ, Zhou ZH (2003) SOM based image segmentation. In: Wang G, Liu Q, Yao Y, Skowron A (eds) Rough sets, fuzzy sets, data mining, and granular computing. RSFDGrC 2003. Lecture Notes in Computer Science, vol 2639, Springer, Berlin, Heidelberg, pp 640–643. https://doi.org/10.1007/3-540-39205-x_107
Khan A, Jaffar MA, Choi TS (2013) SOM and fuzzy based color image segmentation. Multi Med Tools Appl 64:331–344. https://doi.org/10.1007/s11042-012-1003-6
Kohonen T (1982) Self-organized formation of topologically correct feature maps. Biol Cybern 43:59–69
Ong SH, Yeo NC, Lee KH, Venkatesh YV, Cao DM (2002) Segmentation of color images using a two-stage self-organizing network. Image Vis Comput 20:279–289
Ortiz A, Gorriz JM, Ramirez J, Salas-Gonzalez D (2014) Improving MR brain image segmentation using self-organising maps and entropy-gradient clustering. Inf Sci 262:117–136. https://doi.org/10.1016/j.ins.2013.10.002
Song J, Yang C, Fan L, Wang K, Yang F, Liu S, Tian J (2016) Lung lesion extraction using a Toboggan based growing automatic segmentation approach. IEEE Trans Med Imaging 35:337–353. https://doi.org/10.1109/TMI.2015.2474119
Sun S, Guo Y, Guan Y, Ren H, Fan L, Kang Y (2014) Juxta—Vascular nodule segmentation based on flow entropy and geodesic distance. IEEE J Biomed Health Inf 18:1355–1362
Torbati N, Ayatollahi A, Kermani A (2014) An efficient neural network based method for medical images segmentation. Comput Biol Med 44:76–87
Vishnuvarthanan G, Rajasekaran MP, Subbaraj P, Vishnuvarthanan A (2015) An unsupervised learning method with a clustering approach for tumor identification and tissue segmentation in magnetic resonance brain images. Appl Soft Comput 38:190–212. https://doi.org/10.1016/j.asoc.2015.09.016
Acknowledgements
The authors thank Dr. K.G. Srinivasan, MD, RD, Consultant Radiologist and Dr. K.P. Usha Nandhini, DNB, KGS Advanced MR and CT Scan—Madurai, Tamil Nadu, India, for supporting the research with the patient information. Also, the authors thank the Department of Electronics and Communication Engineering of Kalasalingam University (Kalasalingam Academy of Research and Education), Tamil Nadu, India, for permitting to use the computational facilities available in Center for Research in Signal Processing and VLSI Design which was set up with the support of the Department of Science and Technology (DST), New Delhi, under FIST Program in 2013 (Reference No: SR/FST/ETI-336/2013 dated November 2013).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Vigneshwaran, S., Vishnuvarthanan, G., Pallikonda Rajasekaran, M., Arun Prasath, T. (2019). Segmentation of Tumor Region in Multimodal Images Using a Novel Self-organizing Map-Based Modified Fuzzy C-Means Clustering Algorithm. In: Kulkarni, A., Satapathy, S., Kang, T., Kashan, A. (eds) Proceedings of the 2nd International Conference on Data Engineering and Communication Technology. Advances in Intelligent Systems and Computing, vol 828. Springer, Singapore. https://doi.org/10.1007/978-981-13-1610-4_71
Download citation
DOI: https://doi.org/10.1007/978-981-13-1610-4_71
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1609-8
Online ISBN: 978-981-13-1610-4
eBook Packages: EngineeringEngineering (R0)