Abstract
In the present work, three-class breast tissue density classification has been carried out using SVM-based hierarchical classifier. The performance of Laws’ texture descriptors of various resolutions have been investigated for differentiating between fatty and dense tissues as well as for differentiation between fatty-glandular and dense-glandular tissues. The overall classification accuracy of 88.2% has been achieved using the proposed SVM-based hierarchical classifier.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The most commonly diagnosed disease among women nowadays is breast cancer [1]. It has been shown that high breast tissue density is associated with high risk of developing breast cancer [2,3,4,5,6,7,8,9,10]. Mortality rate for breast cancer can be increased if detection is made at an early stage. Breast tissue is broadly classified into fatty, fatty-glandular, or dense-glandular based on its density.
Various computer-aided diagnostic (CAD) systems have been developed by researchers in the past to discriminate between different density patterns, thus providing the radiologists with a system that can act as a second opinion tool to validate their diagnosis. Few studies have been carried out on Mammographic Image Analysis Society (MIAS) dataset for classification of breast tissue density patterns into fatty, fatty-glandular, and dense-glandular tissue types [3,4,5,6,7,8,9,10]. Among these, mostly the studies have been carried on the segmented breast tissue (SBT) and rarely on fixed-size ROIs [3,4,5,6,7,8,9,10]. Out of these studies, Subashini et al. [6] report a maximum accuracy of 95.4% using the SBT approach, and Mustra et al. [9] report a maximum accuracy of 82.0% using the ROI extraction approach.
The experienced participating radiologist (one of the authors of this paper) graded the fatty, fatty-glandular, and dense-glandular images as belonging to typical or atypical categories. The sample images of typical and atypical cases depicting different density patterns are shown in Fig. 1.
In the present work, a hierarchical classifier with two stages for binary classification has been proposed. This classifier is designed using support vector machine (SVM) classifier in each stage to differentiate between fatty and dense breast tissues and then between fatty-glandular and dense-glandular breast tissues using Laws’ texture features.
2 Methodology
2.1 Description of Dataset
The MIAS database consists of total 322 mammographic images out of which 106 are fatty, 104 are fatty-glandular, and 112 are dense-glandular [11]. From each image, a fixed-size ROI has been extracted for further processing.
2.2 Selecting Regions of Interest
After conducting repeated experiments, it has been asserted that for classification of breast density, the center area of the tissue is the optimal choice [12]. Accordingly, fixed-size ROIs of size 200 × 200 pixels have been extracted from each mammogram as depicted in Fig. 2.
2.3 Proposed Method
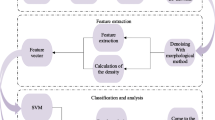
Computer-aided diagnostic systems involve analysis of mammograms through computers which can be used by the radiologists as a second opinion tool for validating their diagnosis as these systems tend to improve the diagnostic accuracy by detecting any lesions that might be missed during subjective analysis [1, 3,4,5,6,7,8,9,10, 13, 14]. The block diagram is shown in Fig. 3.
Feature Extraction Module The texture descriptor vectors (TDVs) derived from Laws’ texture analysis using Laws’ masks of resolutions 3, 5, 7, and 9 have been used in the present work for design of SVM-based hierarchical classifier.
Feature Classification Module Support vector machine classifier has been extensively used for classification of texture patterns in medical images [1, 14,15,16,17,18,19]. In the present work, two binary SVM classifiers arranged in a hierarchical framework have been used for three-class breast tissue density classification. The SVM classifier is implemented using LibSVM library [20].
3 Results
Various experiments were conducted to obtain the classification performance of Laws’ texture features using hierarchical classifier built using two stages of binary SVM classifiers.
3.1 Classification Performance of Laws’ Texture Features Using Hierarchical Classifier
In this work, the performance of TDVs derived using Laws’ masks of length 3, 5, 7, and 9 is evaluated using SVM-based hierarchical classifier. The results obtained are shown in Table 1.
From Table 1, it can be observed that OCA of 91.3, 93.2, 91.9, and 92.5% is achieved for TDV1, TDV2, TDV3, and TDV4, respectively, using SVM-1 sub-classifier, and OCA of 92.5, 84.2, 87.0, and 90.7% is obtained for TDV1, TDV2, TDV3, and TDV4, respectively, using SVM-2 sub-classifier.
The results from Table 1 show that for differentiating between the fatty and dense breast tissues, SVM-1 sub-classifier gives best performance for features extracted using Laws’ mask of length 5 (TDV2), and for further classification of dense tissues into fatty-glandular and dense-glandular classes, SVM-2 sub-classifier gives best performance using features derived from Laws’ mask of length 3 (TDV1). This analysis of the hierarchical classifier is shown in Table 2. The OCA for hierarchical classifier is calculated by adding the misclassified cases at each classification stage.
4 Conclusion
From the exhaustive experiments carried out in the present work, it can be concluded that Laws’ masks of length 5 yield the maximum classification accuracy of 93.2% for differential diagnosis between fatty and dense classes and Laws’ masks of length 3 yield the maximum classification accuracy 92.5% for differential diagnosis between fatty-glandular and dense-glandular classes. Further, for the three-class problem, a single multi-class SVM classifier would construct three different binary SVM sub-classifiers where each binary sub-classifier is trained to separate a pair of classes and decision is made by using majority voting technique. In case of hierarchical framework, the classification can be done using only two binary SVM sub-classifiers.
References
Kriti, Virmani, J., Dey, N., Kumar, V.: PCA-PNN and PCA-SVM based CAD systems for breast density classification. In: Hassanien, A.E., et al. (eds.) Applications of Intelligent Optimization in Biology and Medicine, vol. 96, pp. 159–180. Springer (2015)
Wolfe, J.N.: Breast patterns as an index of risk for developing breast cancer. Am. J. Roentgenol. 126(6), 1130–1137 (1976)
Blot, L., Zwiggelaar, R.: Background texture extraction for the classification of mammographic parenchymal patterns. In: Proceedings of Conference on Medical Image Understanding and Analysis, pp. 145–148 (2001)
Bosch, A., Munoz, X., Oliver, A., Marti, J.: Modeling and classifying breast tissue density in mammograms. In: Computer Vision and Pattern Recognition, IEEE Computer Society Conference, 2, pp. 1552–1558. IEEE Press, New York (2006)
Muhimmah, I., Zwiggelaar, R.: Mammographic density classification using multiresolution histogram information. In: Proceedings of 5th International IEEE Special Topic Conference on Information Technology in Biomedicine (ITAB), pp. 1–6. IEEE Press, New York (2006)
Subashini, T.S., Ramalingam, V., Palanivel, S.: Automated assessment of breast tissue density in digital mammograms. Comput. Vis. Image Underst. 114(1), 33–43 (2010)
Tzikopoulos, S.D., Mavroforakis, M.E., Georgiou, H.V., Dimitropoulos, N., Theodoridis, S.: A fully automated scheme for mammographic segmentation and classification based on breast density and asymmetry. Comput. Methods Programs Biomed. 102(1), 47–63 (2011)
Li, J.B.: Mammographic image based breast tissue classification with kernel self-optimized fisher discriminant for breast cancer diagnosis. J. Med. Syst. 36(4), 2235–2244 (2012)
Mustra, M., Grgic, M., Delac, K.: Breast density classification using multiple feature selection. Auotomatika 53(4), 362–372 (2012)
Silva, W.R., Menotti, D.: Classification of mammograms by the breast composition. In: Proceedings of the 2012 International Conference on Image Processing, Computer Vision, and Pattern Recognition, pp. 1–6 (2012)
Suckling, J., Parker, J., Dance, D.R., Astley, S., Hutt, I., Boggis, C.R.M., Ricketts, I., Stamatakis, E., Cerneaz, N., Kok, S.L., Taylor, P., Betal, D., Savage, J.: The mammographic image analysis society digital mammogram database. In: Gale, A.G., et al. (eds.) Digital Mammography. LNCS, vol. 1069, pp. 375–378. Springer, Heidelberg (1994)
Li, H., Giger, M.L., Huo, Z., Olopade, O.I., Lan, L., Weber, B.L., Bonta, I.: Computerized analysis of mammographic parenchymal patterns for assessing breast cancer risk: effect of ROI size and location. Med. Phys. 31(3), 549–555 (2004)
Kumar, I., Virmani, J., Bhadauria, H.S.: A review of breast density classification methods. In: Proceedings of 2nd IEEE International Conference on Computing for Sustainable Global Development (IndiaCom-2015), pp. 1960–1967. IEEE Press, New York (2015)
Virmani, J., Kriti.: Breast tissue density classification using wavelet-based texture descriptors. In: Proceedings of the Second International Conference on Computer and Communication Technologies (IC3T-2015), vol. 3, pp. 539–546 (2015)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: A comparative study of computer-aided classification systems for focal hepatic lesions from B-mode ultrasound. J. Med. Eng. Technol. 37(44), 292–306 (2013)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: SVM-based characterization of liver ultrasound images using wavelet packet texture descriptors. J. Digit. Imaging 26(3), 530–543 (2013)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: Characterization of primary and secondary malignant liver lesions from B-mode ultrasound. J. Digit. Imaging 26(6), 1058–1070 (2013)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: SVM-based characterization of liver cirrhosis by singular value decomposition of GLCM matrix. Int. J. Artif. Intel. Soft Comput. 3(3), 276–296 (2013)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: PCA-SVM based CAD system for focal liver lesions from B-Mode ultrasound. Defence Sci. J. 63(5), 478–486 (2013)
Chang, C.C., Lin, C.J.: LIBSVM, a library of support vector machines. ACM Trans. Intell. Syst. Technol. 2(3), 27–65 (2011)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Virmani, J., Kriti, Thakur, S. (2019). Classification of Breast Tissue Density Patterns Using SVM-Based Hierarchical Classifier. In: Hoda, M., Chauhan, N., Quadri, S., Srivastava, P. (eds) Software Engineering. Advances in Intelligent Systems and Computing, vol 731. Springer, Singapore. https://doi.org/10.1007/978-981-10-8848-3_18
Download citation
DOI: https://doi.org/10.1007/978-981-10-8848-3_18
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-8847-6
Online ISBN: 978-981-10-8848-3
eBook Packages: EngineeringEngineering (R0)