Abstract
This paper presents a machine learning classifier, namely, Random Forest to detect abnormalities in retina arising from Diabetic Retinopathy. This is an effort to obtain a computer-aided diagnosis procedure to substitute manual detection. Fundus images from public datasets are used for this purpose. A set of statistical and geometric features were extracted from images in the database which contains the different physical manifestations of the disease. Classification through machine learning can help a physician by giving an indication of the level of the disease. The experimental results show 99.275% of accuracy in prediction of the disease, which is promising.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Rapid advances in health sciences have resulted in the spooling of a huge amount of data, clinical information, and generation of electronic health records [1]. Machine learning and data mining methods are being used to perform intelligent transformation of data into useful knowledge [1]. Application of machine learning in medical imaging has become one of the most interesting challenges of the researchers. In the medical domain, description of a set of diseases in terms of features is supplied to the machine learning classifiers as knowledge base. Depending on the supplied training set, the classifier has to identify the disease for a test set. Performance measures specify the correctness of the classifier in identification of disease from an unknown situation. In this research work, signs of the diseases at different stages of Diabetic Retinopathy (DR) are considered as classes. The first stage of DR is silent in nature as no such clear symptoms are noticeable. The starting of DR includes deformation of retinal capillary resulting in very small spots known as microaneurysms (MA). The next stage contains hard exudates (HE) which are lipid formations from the fragile blood vessels. As DR advances, cotton wool spots (CWS) are formed which are micro-infarcts caused by obstructed blood vessel. Hemorrhages can also occur with further progression of the disease when blood vessels leak blood into retina causing hemorrhages (HAM). Due to poor oxygen supply, new vessels are formed in this state, which challenges the patients eyesight. These stages of DR which comprise the different classes of a machine learning classifier are referred to as Non-proliferative Diabetic Retinopathy leading to Proliferative Diabetic Retinopathy. This is a supervised learning problem owing to the fact that the number of objects to be classified are finite and predetermined [2]. The research aims to find the significance and acceptability of Random Forest classifier to the semiautomated detection of different cases of Diabetic Retinopathy. The performance of the classifier is calculated in terms of number of correct and incorrect classification, and the average accuracy is 99.275%.
2 Related Work
Machine learning can be classified as supervised and unsupervised. In supervised machine learning, every input field is associated with its corresponding target value. On the contrary, unsupervised machine learning deals with only input fields. We use Random Forest, a supervised machine learning technique, to classify the different signs (i.e., lesions) of DR. Random Forest (RF) also known as random decision forests are an ensemble learning method used for classification. This ensemble classifier is a combination of tree predictors. Each tree depends on the values of a random vector which is independently sampled with the same distribution for all the trees of the forest [3]. A number of such decision tree classifiers are applied on subsamples (with same size of original input sample size) of the dataset and averaged in order to improve the predictive accuracy as well as controlling overfitting. A scheme of fusing ill-focused images using Random Forest classifier is proposed in the research work of Kausar et al. [4]. The well-organized images are useful for image enhancement and segmentation. This work aims to generate all-in-focus images by Random Forest classifier. Visibility, spatial features, edge, and discrete wavelet transform are considered as features for machine learning. This scheme outperforms previous approaches like principal component analysis and wavelet transform. Saiprasad et al. [5] used Random Forest classifier to classify pixel-wise an image of abdomen and pelvis into three classes: right adrenal, left adrenal, and background. For this purpose, a training set is formed with a dataset of adrenalin gland images. Manual examination and labeling of a radiologist are used as ground truth. The classification phase is combined with the histogram analysis phase for more accurate result. A Random Forest-based approach for classifying lymphatic disease diagnosis is developed in the report of Almayyan [6]. Segmentation and classification of very high-resolution Remote Sensing data with Random Forests are mentioned in the work of Csillik [7]. Random Forest has been used successfully in the classification of Diabetic Retinopathy Diseases in one of our earlier papers [8], which concludes Random Forest as better than Naive Bayes classifier and support vector machine.
3 Proposed Methodology
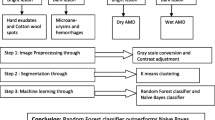
A database of 69 images containing different stages of DR is considered as input dataset. The dataset is formed collecting the images from DIARETDB0 [9] and DIARETDB1 [10]. These images contain bright lesions as hard exudates (HE) and cotton wool spots (CWS) as well as dark lesions like hemorrhages (HAM) and microaneurysms (MA). The retina image database contains 25 images of HE, 15 images of CWS, 15 images of HAM, and 14 MA images. In Fig. 1, the pictorial representation of the system is given.
3.1 Feature Selection
Several consultations with retina specialists were carried out to identify the process of distinguishing a variety of DR. According to those suggestions, nine features among which six texture-based features and the remaining statistical measure-based features are considered for constructing the training set for Random Forest classifier. The texture-based features are as follows: color, frequency, shape, size, edge, and visibility. Color feature is useful for differentiating dark lesions from bright lesions. Among bright lesions also, for HE, it is generally bright yellow and for CWS, it is generally whitish yellow. In dark lesion cases, MA is normally light red in contrast to the dark red color of HAM. The frequency of occurrence is also important as HE comes in large numbers compared to CWS. Shape and size of MA vary from HAM in a significant manner. The sharpness of edge is also significant as edge of CWS is blurred whereas the edge of HE is sharp. Visibility in terms of human eye is considered as another feature. HE and HAM have a high visibility compared to CWS and MA. Figure 2 shows some variety of DR images. The texture-based features are manually extracted from the original input image. No preprocessing is done on the original input image before feature extraction as the texture-based features are directly accessible from the input image without losing their originality. Table 1 describes the features along with their values for different diseases. Three statistical measure-based features are as follows: mean, standard deviation, and entropy. Mean is the average brightness of an image. Standard deviation describes the distance between ith pixel from mean. Another statistical measure is entropy which is the randomness of an image to realize the texture of the image. These features are defined in equations [1,2,3].
In Eqs. (1) and (2), N is the total number of pixels in the input image, and \( x_i\) is the gray value of ith pixel. In Eq. (3), p contains the histogram counts of the image and operator.* indicates element-by-element multiplication. In Eqs. (1) and (2), N is the total number of pixels in the input image, and \( x_i \) is the gray value of ith pixel. In Eq. (3), p contains the histogram counts of the image and operator.* indicates element-by-element multiplication.
The feature values of each lesion of an input image are evaluated along with the disease type and this contributes a row for the training set. This training set is supplied to the RF classifier. RF classifier is not used for selection of features. Feature selection is totally based on the working principle of retina specialists and literature survey.
3.2 Random Forest Classifier
In Random Forest classification technique, several trees are grown together. For classifying a test data, the new data is added down each of the trees. Each tree generates a class for the test data which is called voting for the class. The most voted class is selected by the classifier as final class of the test data. Random Forest classification is the most popular ensemble model classifier as it runs on large database in a time-saving approach. This model is not suitable to the cases dealing with completely new data.
Random Forest classifier with percentage split (66%) is used for machine learning. Two-third data of the total dataset is selected as training dataset which helps the growing of tree. Cases are selected at random with replacement, that is, the case considered for a tree can be reassigned to another tree. Generally, the square root of total number of feature variables is selected at random from all the prescribed feature values. This value remains fixed during the growth of the forest. The best split on these selected feature variables is used to split a node. The remaining 1/3 data is considered as test dataset. This dataset is called out-of-bag (OOB) data. For each test data, each tree generates a class that is counted as the vote for that class of the test data. The class with maximum votes is assigned to the test data. The term Random is associated in two ways with this classifier: random selection of sample data and random selection of feature variables. It is the characteristic of Random Forest classifier that it needs no separate test dataset. The OOB data is used for calculating the error internally at the time of construction. When each tree of the forest is grown, then OOB cases are put down the tree and the number of votes for the correct class is calculated.
3.3 Algorithm of Random Forest Classifier
Step 1: Initialize total number of classes to L and total number of feature variables to M.
Step 2: Let m be the number of selected feature variable at a node (generally \( m=\sqrt{M}\) ).
Step 3: For each decision tree, randomly select with replacement a subset of dataset containing L different classes.
Step 4: For each node of a decision tree, randomly select m feature variables to calculate the best split and decision at this node.
4 Results and Analysis
The performance analysis of Random Forest classifier is carried out with the basis of confusion matrix. In Weka 3.7 [11] machine learning classifier, Random Forest model is selected for analyzing the input dataset. The database contains 69 images. As Random Forest classifier automatically selects 2/3 data as training set and the rest as test set, 46 data are treated as training dataset, and 23 data are considered as test set. After analyzing the dataset, a confusion matrix is generated by the classifier itself.
\(TP_ i \) (True Positive): number of members of class \(X_ i\) correctly classified as class \(X_ i\),
\(FN_ i \) (False Negative): number of members of class \(X_ i\) incorrectly classified as not in class \( X_ i\),
\(FP_ i \) (False Positive): number of members which are not of class \(X_ i \) but incorrectly classified as class \( X_ i\),
\(TN_ i \) (True Negative): number of members which are not of class \( X_ i\) and correctly classified as not of class \( X_ i\),
Accuracy (Acc): the ability of the classifier to correctly classify a member
Sensitivity (Sn): the ability of the classifier to detect the positive class
Specificity (Sp): the ability of the classifier to detect the negative class
F-measure: statistical test of prediction accuracy of the classifier in terms of Precision and Recall
where \( Precision_i=\frac{TP_i}{TP_i+FP_i} \) and \(Recall_i=\frac{TP_i}{TP_i+FN_i} .\)
Mathew Correlation Coefficients (MCC): estimate of over prediction and under prediction
MCC generates three different types of values: \(-1\), 0, and 1. The value \(-1\) means the classifier’s prediction is incorrect, 0 means random prediction, and 1 means fully correct prediction. In Table 2, TP, TN, FP, and FN are calculated using the values of confusion matrix. Table 3 represents different performance measures for each class. For each class, receiver operating characteristic (ROC) is plotted. An ROC curve is plotted with false positive rate along X-axis against true positive rate along Y-axis. Figure 3 shows ROC for class CWS.
5 Conclusion
In this research work, Random Forest classifier is used for determining different stages of retinal abnormalities due to DR using machine learning techniques. Being an ensemble classifier, Random Forest constructs several decision trees at training time and generates the classification for each tree. In this research work, a dataset containing several retinal images having abnormalities is formed. The images are collected from various sources like DIARETDB0 [9] and DIARETDB1 [10]. A set of nine features including three statistical features and six texture-based features are selected for the machine learning. For each input image, the feature values are calculated, and thus, a dataset is formed for 69 images. In Weka 3.7 [11], the dataset is supplied as input to the Random Forest classifier. Performance measures like accuracy, sensitivity, and specificity are calculated depending on the classification result. The accuracy of HAM and MA classes is 100% each as the feature size distinctly separates these two classes. The size of HAM is considered to be medium to large while MA is very small. In future, a collection of large database with more features can be added to the feature set, and the number of images can be increased to get more complex training set for the classifier. The average accuracy is 99.275% which is promising.
References
Kavakiotis I, Tsave O, Salifoglou A, Maglaveras N, Vlahavas I, Chouvarda I (2017) Machine learning and data mining methods in diabetes research. Comput Struct Biotechnol J 15:104–116
Jingar R, Verma M (2012) Semantic segmentation of an image using random forest and single histogram class model. Technical report, Indian Institute of Technology Kanpur
Kausar N, Majid A (2016) Random forest based scheme using feature and decision levels information for multi-focus image fusion. Pattern Anal Appl 19:221–236
Saiprasad G, Chang C, Safdar N, Saenz N, Seigel E (2013) Adrenal gland abnormality detection using random forest classification. J Digit Imaging 26:891–897
Almayyan W (2016) Lymph disease prediction using random forest and particle swarm optimization. J Intell Learn Syst and Appl 8:51–62
Csillik O (2017) Fast segmentation and classification of very high resolution remote sensing data using SLIC superpixels. Remote Sens 9(3):243
Roychowdhury A, Banerjee S, Machine learning in the classification of diabetic retinopathy lesions from abnormal retina images (submitted)
Kauppi T, Kalesnykiene V, Kamarainen J-K, Lensu L, Sorri I, Uusitalo H, Klviinen H, Pietil J, DIARETDB0: evaluation database and methodology for diabetic retinopathy algorithms, Technical report
Kauppi T, Kalesnykiene V, Kamarainen J-K, Lensu L, Sorri I, Raninen A, Voutilainen R, Uusitalo H, Klviinen H, Pietil J, DIARETDB1: diabetic retinopathy database and evaluation protocol, Technical report
Frank E, Hall MA, Witten IH (2016) The Weka Workbench, Online Appendix for Data Mining : Practical Machine Learning Tools and Techniques, 4th edn. Morgan Kaufman
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Roychowdhury, A., Banerjee, S. (2018). Random Forests in the Classification of Diabetic Retinopathy Retinal Images. In: Bhattacharyya, S., Gandhi, T., Sharma, K., Dutta, P. (eds) Advanced Computational and Communication Paradigms. Lecture Notes in Electrical Engineering, vol 475. Springer, Singapore. https://doi.org/10.1007/978-981-10-8240-5_19
Download citation
DOI: https://doi.org/10.1007/978-981-10-8240-5_19
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-8239-9
Online ISBN: 978-981-10-8240-5
eBook Packages: EngineeringEngineering (R0)