Abstract
As fundamental components of cellular membrane, metabolic and energy storage units, and signaling molecules, lipids play key roles in many pathophysiological processes. The systematic study of lipid molecular species and their interactions in biological samples, or the so-called lipidomics study, can assist with the understanding of the general condition of the whole biological system. Qualitative and quantitative determination of the subtle alterations of lipids in biological systems caused by genetics, diet, environment, and therapeutic interventions will not only facilitate in uncovering the pathophysiological role of lipids in disease onset and development but will also help to identify novel biomarkers for early detection, diagnosis, and prognosis of disease. Currently, the tremendous innovations in mass spectrometry and chromatographic techniques have largely driven the development of lipidomics. It has become an emerging approach extensively applied in biomarker research.
Access provided by Autonomous University of Puebla. Download reference work entry PDF
Similar content being viewed by others
Keywords
Key Facts of Nontargeted Lipidomics
-
Nontargeted lipidomics is intended for a comprehensive profiling of lipidome to cover as multiple classes/species as possible.
-
Nontargeted lipidomics is often used in the first stage of biomarker discovery for screening aim.
-
Among various protocols for total lipid extraction, classic Folch method, modified Bligh/Dyer method, and MTBE/MeOH/H2O method are most commonly used.
-
Both Folch method and Bligh/Dyer method were proposed in the 1950s and employ chloroform for extraction.
-
Lipid extraction by MTBE/MeOH/H2O system, proposed in 2008, gains advantage in more convenience, less interference, and toxicity.
-
Methyl tert-butyl ether (MTBE) is used as a substitute for chloroform in lipid extraction.
-
Polar head groups and fatty acyl substitutes in lipid glycerol backbone determine characteristic MS/MS outcome, which highly facilitates identification of lipids, compared with other metabolites.
-
Lipids follow specific retention rules based on polar head group or length/unsaturation of fatty acyls when different types of LC are applied for separation.
-
Multivariate analysis approaches commonly employed in nontargeted lipidomics include principal component analysis (PCA), partial least squares discriminant analysis (PLS-DA), and so on.
Definitions of Words and Terms
Systems Biology
Systems biology is a biology-driven field that aims a comprehensive study of biological interactions or networks on the level of entire biological system, typically ranging from gene to protein and further to downstream metabolites. Therefore, systems biology comprises multi-omics disciplines, e.g., genomics, transcriptomics, proteomics, and metabolomics.
Metabolome
Metabolome is defined as the collectivity of all small molecular metabolites within a biological body with molecular weight typically less than 2,000. Metabolome includes a large spectrum of metabolite classes/molecules with distinct physiochemical properties and dynamic concentration ranges, such as amino acids, nucleic acids, carnitines, bile acids, amines, sugars, hormones, lipids, and so on.
Metabolomics
Metabolomics is an approach for capturing comprehensive and systematic metabolic snapshots or fingerprints of given body fluids/tissues/cells, by qualitative and quantitative measuring of a large set of metabolite molecules, to identify metabolic phenotype under given pathophysiological conditions or metabolic response to stimuli. Two dominating techniques in metabolomics are nuclear magnetic resonance (NMR) and mass spectrometry (MS).
Lipidome
Lipidome is defined as the complete set of all lipids within a biological system such as cells or tissues. It serves as an important part of metabolome. Lipidome comprises diverse lipid molecules including fatty acyls, glycerophospholipids, glycerolipids, sphingolipids, sterol lipids, prenol lipids, saccharolipids, and polyketides.
Lipidomics
Lipidomics is defined as the large-scaled characterization of lipid molecules and their dynamics within a lipidome, as well as the interactions of lipids with other moieties, e.g., proteins. It aims at revealing lipid metabolic perturbations or lipid functions or identifying lipidomics phenotypes or potential lipid biomarkers under studied conditions, e.g., genetics, disease, environmental alterations, and therapeutic intervention. The dominating analytical methods in lipidomics are based on MS technique. It is an integral part of metabolomics and systems biology.
Biomarker
Biomarker (short for biological marker) is broadly defined as any measurable parameter or molecule that is indicative of biological or pathological state or different stages in disease development or response to therapy/treatment. For example, body mass index (BMI) and blood pressure are commonly used biomarkers in clinical practice. In modern omics studies, biomarker specifically refers to a molecular biomarker, including gene, protein, metabolite, and lipid. Biomarkers in disease are applied for screening, disease subtyping, early diagnosis, prognosis, personalized treatment, and drug development. It can be used either alone or in the pattern of combination.
Shotgun Lipidomics
Shotgun lipidomics is one of the two major methods in lipidomics analysis, which features direct infusion into MS without pre-separation and high-throughput analysis by ESI-MS(/MS). Two core techniques of this approach are intrasource separation and multidimensional MS. It was first proposed by Richard W. Gross and Xianlin Han.
Internal Standard
Internal standard used in metabolomics or lipidomics refers to non-endogenous synthetic standard that is spiked into the studied complex biological matrix, to reduce potential analytical bias. In lipidomics, commonly used internal standards are stable-isotope-labeled lipid standards or odd-chain lipid standards.
QC
QC is an abbreviation for quality control samples, which are prepared by pooling equal amount of all studied biological samples, followed by dividing into aliquots and thus can be regarded as being representative of all samples. QC samples are measured by inserting into the whole analytical sequence. It is an effective way to monitor analytical repeatability and to correct possible variations using QC samples.
MRM
MRM is an abbreviation for multiple reaction monitoring, a commonly used scanning mode of tandem MS triple quadrupole (QqQ). By fixing precursor-product ion pairs, higher selectivity and signal-to-noise ratio (S/N) could be obtained in this mode. It is often performed in targeted lipidomics based on the feature of “building blocks” of lipids.
Multivariate Analysis
Multivariate analysis refers to chemometrics tools that enable extracting effective information or identifying important metabolites/lipids in dealing with dataset representing multiple dimensional spaces generated by omics studies, such as metabolomics or lipidomics. It is a useful approach in identifying a potential biomarker. Principal components analysis (PCA), partial least squares discriminant analysis (PLS-DA), orthogonal projections to latent structures (OPLS), etc. are commonly used.
Introduction
Lipids consist of a large and diverse family of structurally distinct biomolecules that are hydrophobic or amphiphilic. In nature, the number of distinct lipid molecular species is conservatively estimated to be around 200,000. The lipid family consists of eight categories as proposed by LIPID MAPS, i.e., fatty acyls, glycerophospholipids (PLs), glycerolipids (GLs), sphingolipids (SLs), sterol lipids, prenol lipids, saccharolipids, and polyketides (Fahy et al. 2005), spanning from polar via neutral to nonpolar. Their structural diversity and complexity are shown in Fig. 1 for several selected lipid categories.
Structures of selected lipid categories. (a) Fatty acyls, exemplarily shown are FA 18:2 (9Z, 11Z); (b) glycerolipids; (c) glycerophospholipids, including six subcategories with distinct polar head group; (d) sphingolipids, exemplarily shown are common subcategories derived from sphingoid base d18:1, i.e., sphingosine
As fundamental components of cellular membrane, metabolic and energy storage units, and signaling molecules as well as vital players in membrane transport and anchoring, lipids widely participate in many pathophysiological processes in living cells. Notably, the role of lipids in cellular membrane is known to be profound. Many groups of lipids serve as dynamic entities involved in cellular membrane structure and scaffolding for membrane proteins (Brown and Murphy 2009). Specifically, PLs including phosphatidylcholines (PCs), phosphatidylethanolamines (PEs), phosphatidylserine (PSs), phosphatidylinositols (PIs), phosphatidic acids (PAs), sterols, and sphingolipids (SLs) including sphingomyelins (SMs) and glycosphingolipids (GSLs) together with proteins reside in cell membranes, where they form the lipid bilayer (Santos and Schulze 2012). This complex array of lipids in cell membrane play crucial roles in the activities of the life process, e.g., controlling protein traffic, separating cells and subcellular organelles from each other, and leading to the generation of thousands of biomolecules that carry information both within and across the cells (Brown and Murphy 2009; Piomelli et al. 2007). Therefore, metabolic homeostasis of lipidome (an integral part of metabolome) is fundamental to healthy maintenance.
Specific lipid profiles or lipid compositions can reflect the biological outcome of a series of biochemical events taking place in cell, body fluids, organ, or whole organism in response to stimuli, genetics, environment, diets, nutrition, drugs, disease, etc. Consequently, qualitative and quantitative study of lipidome on a systematic scale will assist with the understanding of the general condition of the biological system (Hu 2012).
The emerging lipidomics tool greatly facilitated lipid-related studies in life science. It is defined as the comprehensive analysis of lipid molecules and their interactions that are context-dependent and vary through time in complex biological systems influenced by biological stimuli, genetics, environment, or their combinations. Driven by advanced mass spectrometric technology, current lipidomics analytical platforms gain unparallel sensitivity, specificity, and accuracy for comprehensive and high-throughput lipid analysis, making it a valuable complementarity of genomics, proteomics, and metabolomics.
The implication of lipids in various diseases determines the wide applications of lipidomics in the area of disease study. Comparative analysis of lipid profiles from different pathophysiological conditions will give indication of the onset and development of ailments or certain diseases that are associated with the disturbance of lipid metabolic enzymes and pathways (Jelonek et al. 2013; Wenk 2005, 2006). Lipidomics applications in this field focus on biomarker development for screening, disease subtyping, early diagnosis , prognosis, personalized treatment, and drug development.
In this chapter, we will firstly summarize current analytical technologies in mass spectrometry (MS)-based lipidomics and then move on to its applications in disease area, with specific focus on biomarker research; finally, challenges of current lipidomics analysis are discussed.
Analytical Methods in Lipidomics
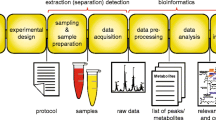
Over the years, many efforts have been made to push the lipidomics field forward. Classic lipidomics methods usually employ chromatographic techniques including thin-layer chromatograph (TLC), gas chromatography (GC), and high-performance liquid chromatography (HPLC) together with complementary procedures such as hydrolysis, chemical derivatization, and enzymatic cleavage. However, these approaches were arduous, time consuming,and low sensitive. In the last decade, lipidomics approaches have been greatly driven by state-of-the-art mass spectrometric technologies with advances in soft ionization technologies and mass analyzers. Frequently used soft ionization technologies in lipidomics include electrospray ionization (ESI), atmospheric pressure chemical ionization (APCI), as well as matrix-assisted laser desorption/ionization (MALDI). Newly emerging technologies, including desorption electrospray ionization (DESI), matrix-free laser desorption ionization (LDI), and laser ablation electrospray ionization (LAESI), are also used. Frequently used mass analyzers include ion trap (IT), quadrupole (Q), time-of-flight (TOF), Fourier transform ion cyclotron resonance (FT-ICR), Orbitrap, and hybrid MS (e.g., triple quadrupole (QqQ), IT-TOF, QTOF, and IT-FT-ICR). MS-based lipidomics approaches can generally be classified into several branches based on various purposes or strategies, as summarized in Fig. 2.
An overview of analytical approaches in lipidomics. Biological samples may be subjected to lipid extraction followed by subsequent analysis or direct analysis by imaging mass spectrometry (IMS). Analytical methods in lipidomics can be classified into several branches based on various purposes or strategies
-
(I)
Nontargeted and targeted lipidomics. Based on experimental purposes, lipidomics approaches can be defined as “nontargeted” and “targeted” lipidomics. Nontargeted approaches aim to cover as broad lipid classes/species as possible, whereas target approaches are intended in specific classes or pathways. As presented in “discovery phase” (Fig. 3) is a general workflow of nontargeted lipidomics approach (note that this nontargeted pipeline is also applicable to other fields, not limited to biomarker discovery). Generally, (i) biological samples, e.g., serum/plasma, tissue, and cells, are collected and subjected to lipid extraction by protocols such as classic Folch method (Folch et al. 1957), modified Bligh/Dyer method (Bligh and Dyer 1959), or methyl tert-butyl ether (MTBE)/MeOH/H2O method (Matyash et al. 2008). Of note, spiking a set of internal standards (ISs) prior to extraction and preparation of quality control (QC) samples is compulsory. (ii) Obtained lipid extract is then subjected to nontargeted data acquisition in shotgun mode or LC-MS mode, which will be elaborated below. (iii) Qualitative and quantitative characterizations are then performed. Lipid database, exact mass, tandem mass spectrometry (MSn), and retention time (TR) are utilized for provisional assignments. This process could be greatly facilitated by lipidomics software tools. For an unambiguous identification, validation with authentic standards is required. Relative quantitative results could be generated by using data processing software tools or by high-resolution extracted ion chromatography (EIC), followed by normalization using ISs. (iv) Statistics using univariate or multivariate analysis are followed to identify potential lipids of interest from huge amount of raw data. (v) The final step involves generation of biomarker candidates, or biological interpretation, which usually requires assistance of bioinformatics. Exemplarily for nontargeted analytical approaches, Taguchi et al. developed a global phospholipid profiling method, using high-mass accuracy and data-dependent MSn of LTQ-Orbitrap, reversed-phase LC (RPLC) separation, and search tool “lipid search.” In total 290 and 248 phospholipids were identified from mouse liver and brain, respectively (Taguchi and Ishikawa 2010). As an example of targeted approaches, Masoodi et al. presented a method for comprehensive profiling of more than 100 bioactive lipids within arachidonic acid pathway cascade. The procedure was composed of multiple optimized steps, from extraction and chromatographic separation to tentative identification based on chromatographic retention, isotopic pattern, and high-resolution tandem MS (Masoodi et al. 2010).
-
(II)
Shotgun and LC-MS - based lipidomics. Mass spectrometer is used either as stand-alone instrument, termed shotgun lipidomics (Han and Gross 2005a, b), or hyphenated with liquid chromatography (LC), i.e., LC-MS(/MS)-based lipidomics (de Grauw et al. 2011; Hu et al. 2010). ESI-MS(/MS)-based shotgun lipidomics initially proposed by Han and Gross (2003) was based on techniques of intrasource separation, multidimensional MS, and microfluidics (Han et al. 2008; Yang et al. 2009). Ejsing et al. used automated shotgun lipidomics for quantitative analysis of yeast lipidome (Ejsing et al. 2009). A total of 250 lipid molecules covering 21 major lipid classes were quantitated, covering ~95 % of yeast lipidome. Recent advances in shotgun lipidomics based on multidimensional MS (MDMS) and its novel strategies were summarized in a newly published review (Han et al. 2012). Other representative shotgun lipidomics techniques employed in qualification/quantification of individual lipids or lipid-related biomarkers in biological samples such as HIV (Brugger et al. 2006), mouse myocardium (Yang et al. 2009), Madin-Darby canine kidney cells (Sampaio et al. 2011), and human blood (Zhao et al. 2013) are summarized in Table 1. Despite of ion suppression, shotgun pattern is rapid and high throughput.
Table 1 Representative analytical methods in lipidomics research applying shotgun or LC-MS-based pattern In LC-MS-based lipidomics, three different modes are frequently used including normal phase LC (NPLC)-MS(/MS), reversed-phase LC (RPLC)-MS(/MS), and hydrophilic interaction liquid chromatography (HILIC)-MS(/MS). Lipids are separated into different classes with specific polar head groups by NPLC or HILIC and are eluted in a mixed pattern based on the length/unsaturation of fatty acyl chains in RPLC. Examples of LC-MS-based lipidomics analyses can be referred to a review (Wolf and Quinn 2008). LC separation prior to MS detection not only facilitates the identification of isomeric lipid species with identical m/z and similar fragmentation patterns but also largely decreases the ion suppression, two major problems encountered in front of shotgun lipidomics. In one study on neutral lipids, Hutchins et al. used NPLC-ESI-MS(/MS) to determine cellular lipids including cholesteryl esters (CEs), monoalkylether diacylglycerols, triacylglycerols (TAGs), and diacylglycerols (DAGs) (Hutchins et al. 2008). In the case of RPLC-MS(/MS)-based technology, a RPLC-ESI-Orbitrap system was used for profiling of cardiolipins (CLs) and monolysocardiolipins (MLCLs) in rat liver mitochondria (Bird et al. 2011). By high-resolution full scan and high collision dissociation (HCD) scan, a total of 28 unique CL and 2 MLCL species involved in mitochondrial stress and dysfunction were identified. Other excellent LC-MS-based lipidomics studies are presented in Table 1.
-
(III)
“Top-down” and “bottom-up” lipidomics. Based on strategies for lipid screening, lipidomics approaches can be defined as “top-down” and “bottom-up” lipidomics. “Top-down” approach takes advantage of the unique elemental composition of lipid species of specific classes, which usually lies in high-resolution MS for exact mass measurement, e.g., Orbitrap and FTMS. Schwudke et al. proposed a workflow for high-throughput lipidomics screen, where rapid acquisition of high-resolution survey scan spectra was used as a powerful tool for “top-down” lipidomics based on elemental composition of different lipid classes. As a proof of concept, this method was applied in three populations of C. elegans, showing its capacity in complementing functional genomics (Schwudke et al. 2007). Hybrid linear ion trap-Orbitrap tandem mass spectrometer (LTQ-Orbitrap) was utilized in this work, which enables a high resolution exceeding 100,000 (FWHM) and a mass accuracy better than 2 and 5 ppm, respectively, at internal and external calibration (Makarov et al. 2006).
In contrast, “bottom-up” approach utilizes the “building block” feature of lipids, where tandem MS technologies are applied for screening of lipids with specific product ions or neutral losses indicative of polar heads, fatty acyl chains. QqQ is typical for such strategy by performing product ion scans, precursor ion scans, and neutral loss scans. For detailed work principles of QqQ-MS, interested readers can read one publication (Hou et al. 2008). Rappley et al. utilized ESI-QqQ for profiling of PLs in lipid extracts of mouse brain tissues to investigate the variations between age and gender associated with α-synuclein genotype (Rappley et al. 2009). Schuhmann et al. established a shotgun bottom-up lipidomics method based on higher-energy collision dissociation (HCD) of lipids using LTQ-Orbitrap. High resolution in both MS/MS and survey scan enhanced identification confidence (Schuhmann et al. 2011). This work thus proved the capacity of LTQ-Orbitrap for both “top-down” and “bottom-up” approaches.
Apart from the abovementioned popular ESI-based lipidomics approaches, imaging mass spectrometry (IMS) is a new emerging but versatile tool for lipidomics study. IMS-based lipidomics enables direct visualization of lipid profile of biological tissue, thereby to provide spatial distribution information. No extraction or pre-separation procedures are required before MS analysis. The key of IMS lies in ionization technologies, including MALDI, DESI, LAESI, and second ion MS (SIMS). Readers of interest can refer to reviews (Goto-Inoue et al. 2011; Murphy et al. 2009).
In addition, to acquire not only “snapshot” information of lipid metabolism but also its dynamic process, stable isotope labeled precursors have been introduced into lipidomics (Haynes et al. 2011; McLaren et al. 2011). Li et al. presented a strategy for dynamic analysis of lipid metabolism using stable isotope-assisted lipidomics based on high-resolution UHPLC-MS technology. Figure 4 displays the workflow of the strategy. A UHPLC-ESI-LTQ-Orbitrap system was employed for nontargeted lipidomics of human myotube cells exposed with or without [U-13C]palmitate. Obtained dataset (more than 7,000 ions) was subjected to nontargeted isotopomer filtering, which resulted in 692 isotopomers. These isotopomers were then assembled into 203 labeled lipid species covering 12 lipid subclasses. This strategy possesses the potential to provide in-depth insights into a comprehensive profile of dynamics of lipid metabolisms that helps to understand the pathophysiological mechanisms associated with metabolic perturbations (Li et al. 2013b).
Fig. 4 Strategy for dynamic analysis of lipid metabolism using stable isotope-assisted lipidomics based on UHPLC-Orbitrap-MS technology. MID analysis, mass isotopomer distribution analysis (Reprinted with permission from Li J, Hoene M, Zhao X, et al. Stable isotope-assisted lipidomics combined with nontargeted isotopomer filtering, a tool to unravel the complex dynamics of lipid metabolism. Anal Chem. 2013b;85(9):4651–4657. Copyright (2013) American Chemical Society)
Lipidomics Applications in Biomarker Discovery
Lipid metabolic dysregulations have been widely recognized being relevant in the pathogenesis of many devastating diseases. Lipidomics profiling of cells, body fluids, or tissues enables a global snapshot of lipid concentrations and structural compositions. Meanwhile, novel bioinformatics tools and collaborative works from multidisciplinary fields enable researchers to build a comprehensive picture of lipid metabolic interconnections, to reveal lipid-based regulations and identify potential lipid biomarkers that are indicative of disease. So far, lipidomics approaches to biomarker discovery have been widely applied in studies of many diseases. Figure 3 displays the pipeline for lipid-based biomarker discovery. Body fluids (e.g., blood/serum/plasma, urine, cerebrospinal fluid, etc.) are preferred to be employed in biomarker study due to their relatively easy collection and less invasiveness. A complete and robust biomarker study is recommended to comprise two stages. At first stage (discovery phase), well-designed populations are enrolled in discovery phase. The case-control individuals or time-series individuals should be representative and matched in other factors, e.g., age, sex, and collection conditions. A survey study is often performed in this stage using nontargeted methodology, which enables a comprehensive screening of altered lipids without prior knowledge. Generated biomarker candidates in this stage are then subjected to large-scaled validation (validation phase), where targeted approaches are often performed. Multiple reaction monitoring mode (MRM) is typically used in targeted way. Finally, the remaining lipids after strict validation are identified as potential biomarkers.
Herein the current states of lipid biomarker research are reviewed on several representative subjects including neuropathic disease, epidemic disease, cancer, and drug therapy.
Lipid biomarker discovery in neuropathic disease. Alzheimer’s disease (AD) is the most common dementia among neuropathic diseases. One of the pathological features of AD is brain structural lipid dysregulation in PLs and SLs. A recent shotgun lipidomics study reported alterations in plasma sphingolipidome in early AD. Eight sphingomyelins (SMs) particularly with long aliphatic chains were observed to be significantly decreased while two ceramides (Cers) to be significantly increased in AD patients as compared to normal controls. Notably, the ratios of Cer to SM molecular species with identical fatty acyl chain exhibited more robust power in distinguishing AD versus controls than that by either species alone (Han et al. 2011). Another recent lipidomics study showed that differences in fatty acid profiles between AD, mild cognitive impairment (MCI), and normal controls were most outstanding in free fatty acids (FFAs) and PLs in plasma than that in brain context. In detail, the alterations in plasma metabolism of both DHA (docosahexaenoic acid, a typical n-3 fatty acid) and other fatty acids irrelevant to DHA are associated with AD (Cunnane et al. 2012). Iuliano et al. reported that increased levels of arachidic, erucic, and vaccenic acids and decreased levels of cerotic and linoleic acids were observed in MCI and AD compared with normal controls. Particularly, linoleic acid and mead acid were, respectively, decreased and increased progressively from controls to MCI to AD patients and also correlated inversely in patients with AD and MCI (Iuliano et al. 2013). These findings provide the relevance between lipid biochemistry and neuronal dysfunction in early AD and indicated that peripheral circulating lipidome is promising for novel biomarker discovery. Typical applications of lipidomics in biomarker discovery of other neuropathic diseases (e.g., schizophrenia, neuronal injury, and juvenile bipolar disorder) are summarized in Table 2.
Lipid biomarker discovery in epidemic disease. Epidemic diseases are highly correlative with the lifestyle-related factors which have become leading causes of morbidity and mortality worldwide. Lipidomics for disease biomarker discovery has been successfully used for many epidemic risks/diseases such as hypertension, obesity, diabetes, arteriosclerosis, and coronary heart disease. In one comparative study of circulating and aortic lipidome and plasma metabolome from the hamster using LC-MS/MS lipidomics, potential plasma biomarkers were defined for early formation of atheromatous plaque. It further revealed that lipidomics profile of aorta exhibited more significant changes in response to high-fat diet, compared with that of plasma. Specifically, docosahexaenoic acid and ceramide (d18:1/24:1) showed significant variation in both plasma and aorta. In addition, levels of free fatty acids were altered due to intake of the high-fat diet, suggesting the impact of high-fat diet on free fatty acid metabolism in the plasma (Jove et al. 2013). In another study, a comprehensive lipidomics study was performed in plasma samples from 189 patients with type 2 diabetes and 189 age-matched healthy individuals using LC-MS-based technology. The results revealed that lipid species with shorter acyl chain length and lower double bond were positively correlated with the risk of diabetes, whereas lipid species with longer acyl chain length and higher double bond were negatively correlated with the risk. Besides, levels of TAG species associated with risk for diabetes were found to be correlated with response to insulin action and insulin resistance. Therefore, TAG species could serve as the predicating biomarkers for type 2 diabetes. This study identifies a lipidomics signature of diabetes risk by revealing the relationship between lipid acyl chain/double bond and diabetes risk, demonstrating the potential of lipidomics in assisting clinical assessment of disease risk (Rhee et al. 2011). Other representative applications of lipidomics in biomarker discovery of epidemic diseases are summarized in Table 2 that include hypertension, dilated cardiomyopathy, cardiovascular disease, and steatohepatitis.
Lipid biomarker discovery in cancer. Currently, lipidomics strategy for detection and evaluation of potential biomarkers has been increasingly and extensively utilized in cancer. Herein lipidomics applications for biomarker discovery in two types of cancer, breast cancer and prostate cancer, are exemplarily discussed.
Breast cancer is the most prevalent female cancer worldwide. By using a nano-LC-MS/MS lipidomics approach, PCs and PEs were comparatively analyzed in urine from healthy controls and women with breast cancer before and after surgery. It was found that the urinary levels of PCs and PEs were significantly increased in cancer individuals as compared to healthy controls. Specifically, urinary levels of 6 PC species containing 16:0 or 18:0 chain at sn-1 position and 4 PE species containing 18:2 chain at sn-2 position were significantly changed in patients after breast cancer surgery versus the controls (Kim et al. 2009). In another subsequent study, four groups of PLs (PS, PI, PG, PA) were comparatively analyzed in urine samples of the same study groups using the same lipidomics platform. Significant increase of urinary PSs (18:1/18:1, 18:2/18:0) and PA (18:1/20:1) and significant decrease of PI (18:0/20:4) were found in cancer patients versus healthy controls (Min et al. 2010). The results imply the potential of urinary PLs in serving as diagnostic markers of breast cancer. Another study compared 142 serum samples from breast cancer patients before and after neoadjuvant chemotherapy. Lipid profiling by UPLC-ESI-QTOF using positive ionization mode showed that TAGs containing mainly oleic acid side chain(s) were decreased in patients showing pathologic complete response before chemotherapy. Besides, monounsaturated TAGs especially containing oleic acid were found to be associated with estrogen receptor status in premenopausal patients. These results indicated that oleic acid-containing TAG species are involved in metabolic response to neoadjuvant chemotherapy as revealed in serum lipidome in breast cancer patients (Hilvo et al. 2013).
Lipidomics-driven biomarker research is also carried out in prostate cancer, which is the most common lethal malignancy in men worldwide. A nano-LC-MS/MS lipidomics analysis of the urine of patients with prostate cancer and age-matched healthy male adults revealed that levels of PC (18:0/20:4), PE (18:2/20:4), 6 PSs (18:0/20:5, 16:0/22:6, 18:1/22:6, 18:1/18:0, 18:1/18:1, 18:0/18:1), and 2 PIs (16:1/20:2, 18:0/18:1) showed prominent differentiations in cancer patients. A 2.5–3.6-fold increase of PC, PE, and PS groups and a more than fourfold decrease of PI, PG, and PA groups were observed in advanced prostate cancer patients with Gleason score higher than 6. This result indicated that PL composition is critically changed upon the progression of prostate cancer (Min et al. 2011). Using ESI-MS/MS, 390 individual lipid species were analyzed in plasma from 105 prostate cancer patients and 36 age-matched healthy males. 35 lipid species showed potential in distinguishing prostate cancer from the controls, and 12 out of them were identified as diagnostic biomarkers with high sensitivity, specificity, and accuracy. Three lipid classes including PE, ether-linked PE, and ether-linked PC could serve as biomarkers for diagnosis of prostate cancer (Zhou et al. 2012). Another recent investigation focused on concentrations and distribution of plasma PLs in prostate cancer. Plasma PLs were analyzed in samples from 57 prostate cancer patients and 43 age-matched controls. The levels of plasma PLs were significantly lower in patients. Among them, lyso-PC (LPC) and PE showed the most significant reduction (Cvetkovic et al. 2012).
MS-based lipid profiles were also performed for cancer tissue samples; the result of one typical study on ovarian cancer is summarized in Table 2 (Liu et al. 2010). In addition, two representative applications of lipidomics in biomarker discovery of colorectal cancer (Li et al. 2013a) and lung cancer (Guo et al. 2012) are also summarized in Table 2.
Lipid biomarker discovery in drug therapy. Lipid-regulating-based therapies are believed to be a key to improvement of global health care due to the fact that pathological features of many diseases involve metabolic disorders of lipids. Therefore, investigations of lipid biomarker research in drug therapies have been largely carried out in recent years. A typical example comes from a study by Laaksonen et al., in which the authors employed UPLC-MS-based lipidomics approach together with bioinformatics of whole genome expression profiling of muscle specimens to investigate the muscular metabolic response to high-dose statin. Plasma samples from patients treated either with high-dose simvastatin, atorvastatin, or placebo were collected for the lipidomics study. The changes in plasma lipidomics profiling caused by simvastatin treatment were observed to be associated with the expression of the arachidonate 5-lipoxygenase-activating proteins, suggesting that the plasma lipid fingerprint may be used as a biomarker for drug treatment in clinic with appropriate drug dosage (Laaksonen et al. 2008). In another more recent well-designed study on early lipid metabolic changes in subjects with schizophrenia, lipidomics was used to evaluate the effects of atypical antipsychotics by comparing lipid metabolic profiles between study groups and within groups before and after treatment with atypical antipsychotics, risperidone, and aripiprazole. As compared with controls, n-3 polyunsaturated fatty acids such as 20:5 n-3, 22:5 n-3, and 22:6 n-3 within the PC and PE lipid classes were significantly reduced in a fist episode of schizophrenia group, indicating that n-3 fatty acid deficits are present early in the course of schizophrenia and can serve as the diagnostic biomarkers for schizophrenia (McEvoy et al. 2013). Besides, studies of lipid biomarker discovery in drug therapy on obesity, brain injury, and transient acute synovitis are summarized in Table 2.
Of note, though a number of lipid biomarker candidates were identified and reported in studies of a wide range of diseases, lipidomics-driven biomarker research is still in its infancy. These reported potential biomarkers at the current state are only the beginning of the long process of defining the biomarkers for a clinical use, or a real diagnostic aid, large cohort validation is highly in need for ultimate verification.
Potential Applications of Lipidomics in Prognosis, Other Diseases or Conditions
Lipidomics tools have been widely used in identifying biomarkers for various purposes including disease subtyping, early diagnosis, prognosis, personalized treatment, drug development, etc. Prognosis is a promising part among them. For instance, a potential prognosis signature of plasma lipidome was identified in cystic fibrosis patients by LC-ESI-MS-based lipidomics (Ollero et al. 2011). In addition to diseases that have been the focal areas for lipidomics application as described above, other disease/health status which may be associated with lipid metabolism is also gaining increasing attraction. An interesting study identified a sex-specific lipid signature characteristic for familial longevity, by using LC-ESI-MS-based lipidomics (Gonzalez-Covarrubias et al. 2013).
Challenges of Current Lipidomics Analysis
For a successful application of lipidomics approach, sensitive, structural-specific, accurate, and reliable lipidomics platforms are highly required and play pivotal roles in outputs. It is especially of significance in the field of this chapter’s focus, lipid biomarker research in disease, because a useful biomarker should be robust in clinical context. In brief, challenges of current lipidomics analysis lie in two aspects, (1) analytical capacity and (2) data handling. From the perspective of analytical capacity, challenges come from the multiplex physiochemical nature of lipidome. Lipids exhibit enormous complexity and diversity in chemical structure and function as described above. In addition, lipids vary in concentration over 5 ~ 6 orders of magnitude in biological systems. From the perspective of lipidomics data handling, challenges come from the multidisciplinary nature of lipidomics field. Consequently, optimized strategies are required for sample preparation, qualitative and quantitative characterization, as well as data mining and interpretation.
-
(I)
Sample preparation. It is an almost impossible task to develop an unbiased sample preparation method that enables to cover the whole lipidome. In practice, compromises are often made based on the sample type and experimental purpose. In some case, as broad a scope of lipids as possible is required to cover (nontargeted), whereas in other cases, only lipids belonging to a specific class or pathway (targeted) are intended during sample preparation. For instance, solid-phase extraction (SPE) or mild alkaline methanolysis was utilized for SL extraction to remove high abundant interfering glycerolipids (GLs). Besides, the instability of some lipids (e.g., acyl-CoA) also adds difficulties in sample preparation.
-
(II)
Qualitative and quantitative characterization. Although a large number of software tools as well as databases are available that greatly facilitate lipid identification and (semi-)quantitation, several aspects hamper an accurate analysis still: (i) one concern is about determining low abundant bioactive lipids. Efforts have been made by researchers to enhance sensitivity, e.g., Wang et al. developed a high-sensitive method for detecting 4-hydroxyalkenal species by derivatization (Wang et al. 2012); (ii) it is not possible to obtain adequate commercially available authentic lipid standards for confirmation and quantification of detected lipid species; (iii) sophisticated technologies are highly required to fulfill a detailed characterization of lipid structure, e.g., the positional determination of fatty acyl chain in glycerol backbone and the localization of double bond; and (iv) thorough and systemic validations of analytical methods in lipidomics study are not drawn enough attention, which influences to a great extent the reliability of the data.
-
(III)
Data mining and interpretation. As an integral branch of systems biology, lipidomics requires the integration of analytical chemistry, instrumental science together with mathematics and bioinformatics, biology, and biochemistry, to convert complex and diverse lipid-related datasets into explainable knowledge (Bou Khalil et al. 2010). Firstly, like other omics disciplines, lipidomics study often generates huge amount of data covering multiple classes and hundreds of species. Secondly, systems biology requires establishing the linkage between lipidomics observations and lipid phenotypes or metabolic/regulation pathways to elucidate their biological relevance, which often involves in-depth studies of interactions on lipid-lipid, lipid-protein and lipid-gene scale. For these purposes, bioinformatics is in need regarding lipidomics data processing, multivariate/univariate statistics, pathway analysis, and lipidome remodeling (Oresic 2011). Notably, it is of high relevance to establish a thorough database containing substrate-product lipid metabolic pathway maps and to integrate lipidomics outputs with genetic, proteomic, and metabolic outputs. Although some resources for lipid databases such as Lipid Library (http://www.lipidlibrary.co.uk/), LIPIDAT (http://www.lipidat.ul.ie/), LMSD (http://www.lipidmaps.org/data/structure/index.html), Cyberlipid Center (http://www.cyberlipid.org/), SphinGOMAP (http://sphingolab.biology.gatech.edu/), KEGG (http://www.genome.jp/kegg/pathway.html), etc., are online available, each encompasses information on a small part of lipid species. For instance, KEGG database is limited to pathway maps containing fatty acid biosynthesis and degradation, sterol metabolism, and PL pathway, and SphinGOMAP focuses on pathway map for SL biosynthesis (Hou et al. 2008). Obviously, continuous worldwide collaboration on establishing open access and multidisciplinary lipid-related databases with constant updates is highly in need for lipidomics researchers. Once achieved, it can provide powerful tools for the search of lipid-related physicochemical, biological, metabolic, and medical information based on the subtle lipid metabolic changes, yielding indicators of certain diseases or pharmacological responses to a therapeutic intervention. Such combined efforts from multidisciplinary scientists will highly facilitate the study on lipid biomarker discovery and pathophysiological mechanisms associated with lipid-related disorders.
Perspectives
Currently lipidomics is very promising in characterizing the constitutes of normal/perturbed lipid metabolism and to a great extent shows its great potential in identifying disease biomarkers for preclinical and clinical trials in early detection diagnosis, prognosis, therapy evaluation, drug development, etc. However, most of lipidomics studies were initially performed with a relatively small group, which may make the outputted model error prone and unstable. To overcome this, the robustness and accuracy of the analytical methods are firstly needed to be guaranteed; secondly, appropriate statistical tools are compulsory to reduce potential bias; thirdly and most importantly, validation in large cohort multicenter clinical studies is required to ensure the reliability of the generated biomarkers. Continued developments in analytical technology and bioinformatics tools will further enhance the effectiveness and robustness of lipidomics approach. It may allow lipidomics to deliver disease biomarkers in daily clinical practice.
Summary Points
-
This chapter reviews current analytical methods and challenges in lipidomics, additionally with focus on potential impact of lipidomics on the research of disease biomarker discovery.
-
Lipids play vital roles in constructing cellular membrane, energy storage, signaling, and membrane transport/anchoring and thereby are highly relevant in many pathophysiological processes in living cells.
-
Driven by advances in modern mass spectrometry (MS), MS-based lipidomics gains enhanced sensitivity, specificity, robustness, accuracy, and dynamic range for qualitative and quantitative characterization of lipids, making it a dominating tool for lipidomics studies.
-
However, lipidomics is still hampered by analytical challenges in dealing with tremendous complexity and diversity of lipidome, effective data mining/interpretation, as well as integration with other omics studies and disciplines.
-
Shifted lipid homeostasis is indicative of pathophysiological state of many diseases. Therefore, comparative lipidomics on body fluids or tissues from healthy and diseased subjects has been a major way to identify altered lipid species that may serve as potential biomarkers.
-
Lipidomics has been broadly applied for biomarker discovery in many devastating diseases including epidemic diseases (e.g., obesity, diabetes, hypertension, cardiovascular disease, steatohepatitis), neuropathic diseases (e.g., Alzheimer’s disease, schizophrenia, neuronal injury, juvenile bipolar disorder), cancer (e.g., breast cancer, prostate cancer), etc.
Abbreviations
- AD:
-
Alzheimer’s Disease
- APCI:
-
Atmospheric Pressure Chemical Ionization
- CE:
-
Cholesteryl Ester
- Cer:
-
Ceramide
- CL:
-
Cardiolipin
- DAG:
-
Diacylglycerol
- DESI:
-
Desorption Electrospray Ionization
- DHA:
-
Docosahexaenoic Acid
- EIC:
-
Extracted Ion Chromatography
- ESI:
-
Electrospray Ionization
- FFA:
-
Free Fatty Acid
- FT-ICR:
-
Fourier Transform Ion Cyclotron Resonance
- FWHM:
-
Full Width at Half Maximum
- GC:
-
Gas Chromatography
- GL:
-
Glycerolipid
- GSL:
-
Glycosphingolipid
- HCD:
-
High Collision Dissociation
- HILIC:
-
Hydrophilic Interaction Liquid Chromatography
- HPLC:
-
High-Performance Liquid Chromatography
- IMS:
-
Imaging Mass Spectrometry
- IS:
-
Internal Standard
- IT:
-
Ion Trap
- LAESI:
-
Laser Ablation Electrospray Ionization
- LC:
-
Liquid Chromatography
- LDI:
-
Matrix-Free Laser Desorption Ionization
- LPC:
-
Lyso-PC
- LTQ-Orbitrap:
-
Hybrid Linear Ion Trap-Orbitrap Tandem Mass Spectrometer
- MALDI:
-
Matrix-Assisted Laser Desorption/Ionization
- MCI:
-
Mild Cognitive Impairment
- MDMS:
-
Multidimensional MS
- MLCL:
-
Monolysocardiolipin
- MRM:
-
Multiple reaction monitoring
- MS:
-
Mass Spectrometry
- MSn :
-
Tandem Mass Spectrometry
- MTBE:
-
Methyl Tert-Butyl Ether
- NPLC:
-
Normal Phase Liquid Chromatography
- PA:
-
Phosphatidic Acid
- PC:
-
Phosphatidylcholine
- PE:
-
Phosphatidylethanolamine
- PI:
-
Phosphatidylinositol
- PL:
-
Phospholipid
- PS:
-
Phosphatidylserine
- Q:
-
Quadrupole
- QC:
-
Quality Control
- QqQ:
-
Triple Quadrupole
- RPLC:
-
Reversed-Phase Liquid Chromatography
- SIMS:
-
Second Ion Ms
- SL:
-
Sphingolipid
- SM:
-
Sphingomyelin
- SPE:
-
Solid-Phase Extraction
- TAG:
-
Triacylglycerol
- TLC:
-
Thin-Layer Chromatography
- TOF:
-
Time-of-Flight
- TR:
-
Retention Time
- UHPLC:
-
Ultrahigh-Performance Liquid Chromatography
References
Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal Chem. 2011;83(3):940–9.
Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–7.
Bou Khalil M, Hou W, Zhou H, et al. Lipidomics era: accomplishments and challenges. Mass Spectrom Rev. 2010;29(6):877–929.
Brown HA, Murphy RC. Working towards an exegesis for lipids in biology. Nat Chem Biol. 2009;5(9):602–6.
Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. The HIV lipidome: a raft with an unusual composition. Proc Natl Acad Sci U S A. 2006;103(8):2641–6.
Clayton EH, Hanstock TL, Hirneth SJ, Kable CJ, Garg ML, Hazell PL. Long-chain omega-3 polyunsaturated fatty acids in the blood of children and adolescents with juvenile bipolar disorder. Lipids. 2008;43(11):1031–8.
Cunnane SC, Schneider JA, Tangney C, et al. Plasma and brain fatty acid profiles in mild cognitive impairment and Alzheimer’s disease. J Alzheimers Dis. 2012;29(3):691–7.
Cvetkovic B, Vucic V, Cvetkovic Z, Popovic T, Glibetic M. Systemic alterations in concentrations and distribution of plasma phospholipids in prostate cancer patients. Med Oncol. 2012;29(2):809–14.
de Grauw JC, van de Lest CH, van Weeren PR. A targeted lipidomics approach to the study of eicosanoid release in synovial joints. Arthritis Res Ther. 2011;13(4):R123.
Dennis EA, Deems RA, Harkewicz R, et al. A mouse macrophage lipidome. J Biol Chem. 2010;285(51):39976–85.
Ejsing CS, Sampaio JL, Surendranath V, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A. 2009;106(7):2136–41.
Fahy E, Subramaniam S, Brown HA, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46(5):839–61.
Folch J, Lees M, Stanley GHS. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226(1):497–509.
Fonteh AN, Chiang J, Cipolla M, et al. Alterations in cerebrospinal fluid glycerophospholipids and phospholipase A2 activity in Alzheimer’s disease. J Lipid Res. 2013;54(10):2884–97.
Gonzalez-Covarrubias V. Lipidomics in longevity and healthy aging. Biogerontology. 2013;14:663–72.
Gonzalez-Covarrubias V, Beekman M, Uh H-W, et al. Lipidomics of familial longevity. Aging Cell. 2013;12(3):426–34.
Goto-Inoue N, Hayasaka T, Zaima N, Setou M. Imaging mass spectrometry for lipidomics. Biochim Biophys Acta. 2011;1811(11):961–9.
Gregory KE, Bird SS, Gross VS, et al. Method development for fecal lipidomics profiling. Anal Chem. 2013;85(2):1114–23.
Guo Y, Wang X, Qiu L, et al. Probing gender-specific lipid metabolites and diagnostic biomarkers for lung cancer using Fourier transform ion cyclotron resonance mass spectrometry. Clin Chim Acta. 2012;414:135–41.
Han XL, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003;44(6):1071–9.
Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005a;24(3):367–412.
Han X, Gross RW. Shotgun lipidomics: multidimensional MS analysis of cellular lipidomes. Expert Rev Proteomics. 2005b;2(2):253–64.
Han X, Yang K, Gross RW. Microfluidics-based electrospray ionization enhances the intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: development of an automated high-throughput platform for shotgun lipidomics. Rapid Commun Mass Spectrom. 2008;22(13):2115–24.
Han X, Rozen S, Boyle SH, et al. Metabolomics in early Alzheimer’s disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS One. 2011;6(7):e21643.
Han X, Yang K, Gross RW. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev. 2012;31(1):134–78.
Haynes CA, Allegood JC, Wang EW, Kelly SL, Sullards MC, Merrill Jr AH. Factors to consider in using [U-C]palmitate for analysis of sphingolipid biosynthesis by tandem mass spectrometry. J Lipid Res. 2011;52(8):1583–94.
Hilvo M, Gade S, Hyotylainen T, et al. Monounsaturated fatty acids in serum triacylglycerols are associated with response to neoadjuvant chemotherapy in breast cancer patients. Int J Cancer. 2013;134:1725–33.
Hou W, Zhou H, Elisma F, Bennett SA, Figeys D. Technological developments in lipidomics. Brief Funct Genomic Proteomic. 2008;7(5):395–409.
Hu C. Systems biology for evaluating system-based medicine. Thesis, Leiden University. ISBN: 978-90-74538-77-0. Printed by Wöhrmann Print Service, Zutphen, The Netherlands; 2012, p. 6–12.
Hu C, Hoene M, Zhao X, et al. Lipidomics analysis reveals efficient storage of hepatic triacylglycerides enriched in unsaturated fatty acids after one bout of exercise in mice. PLoS One. 2010;5(10):e13318.
Hutchins PM, Barkley RM, Murphy RC. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J Lipid Res. 2008;49(4):804–13.
Iuliano L, Pacelli A, Ciacciarelli M, et al. Plasma fatty acid lipidomics in amnestic mild cognitive impairment and Alzheimer’s disease. J Alzheimers Dis. 2013;36(3):545–53.
Jelonek K, Ros M, Pietrowska M, Widlak P. Cancer biomarkers and mass spectrometry-based analyses of phospholipids in body fluids. Clin Lipidol. 2013;8(1):137–50.
Ji J, Kline AE, Amoscato A, et al. Lipidomics identifies cardiolipin oxidation as a mitochondrial target for redox therapy of brain injury. Nat Neurosci. 2012;15(10):1407–13.
Jove M, Ayala V, Ramirez-Nunez O, et al. Lipidomic and metabolomic analyses reveal potential plasma biomarkers of early atheromatous plaque formation in hamsters. Cardiovasc Res. 2013;97(4):642–52.
Kim H, Min HK, Kong G, Moon MH. Quantitative analysis of phosphatidylcholines and phosphatidylethanolamines in urine of patients with breast cancer by nanoflow liquid chromatography/tandem mass spectrometry. Anal Bioanal Chem. 2009;393(6–7):1649–56.
Laaksonen R, Janis MT, Oresic M. Lipidomics-based safety biomarkers for lipid-lowering treatments. Angiology. 2008;59(2 Suppl):65S–8.
Lagarde M, Bernoud-Hubac N, Calzada C, Vericel E, Guichardant M. Lipidomics of essential fatty acids and oxygenated metabolites. Mol Nutr Food Res. 2013;57(8):1347–58.
Li F, Qin X, Chen H, et al. Lipid profiling for early diagnosis and progression of colorectal cancer using direct-infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 2013a;27(1):24–34.
Li J, Hoene M, Zhao X, et al. Stable isotope-assisted lipidomics combined with nontargeted isotopomer filtering, a tool to unravel the complex dynamics of lipid metabolism. Anal Chem. 2013b;85(9):4651–7.
Liu Y, Chen Y, Momin A, et al. Elevation of sulfatides in ovarian cancer: an integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol Cancer. 2010;9:186.
Llorente A, Skotland T, Sylvanne T, et al. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta. 2013;1831(7):1302–9.
Makarov A, Denisov E, Kholomeev A, et al. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem. 2006;78(7):2113–20.
Masoodi M, Eiden M, Koulman A, Spaner D, Volmer DA. Comprehensive lipidomics analysis of bioactive lipids in complex regulatory networks. Anal Chem. 2010;82(19):8176–85.
Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49(5):1137–46.
McEvoy J, Baillie RA, Zhu H, et al. Lipidomics reveals early metabolic changes in subjects with schizophrenia: effects of atypical antipsychotics. PLoS One. 2013;8(7):e68717.
McLaren DG, He T, Wang SP, et al. The use of stable-isotopically labeled oleic acid to interrogate lipid assembly in vivo: assessing pharmacological effects in preclinical species. J Lipid Res. 2011;52(6):1150–61.
Min HK, Kong G, Moon MH. Quantitative analysis of urinary phospholipids found in patients with breast cancer by nanoflow liquid chromatography-tandem mass spectrometry: II. Negative ion mode analysis of four phospholipid classes. Anal Bioanal Chem. 2010;396(3):1273–80.
Min HK, Lim S, Chung BC, Moon MH. Shotgun lipidomics for candidate biomarkers of urinary phospholipids in prostate cancer. Anal Bioanal Chem. 2011;399(2):823–30.
Murphy RC, Hankin JA, Barkley RM. Imaging of lipid species by MALDI mass spectrometry. J Lipid Res. 2009;50(Suppl):S317–22.
Ollero M, Astarita G, Guerrera IC, et al. Plasma lipidomics reveals potential prognostic signatures within a cohort of cystic fibrosis patients. J Lipid Res. 2011;52(5):1011–22.
Oresic M. Informatics and computational strategies for the study of lipids. Biochim Biophys Acta. 2011;1811(11):991–9.
Patterson AD, Maurhofer O, Beyoglu D, et al. Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer Res. 2011;71(21):6590–600.
Piomelli D, Astarita G, Rapaka R. A neuroscientist’s guide to lipidomics. Nat Rev Neurosci. 2007;8(10):743–54.
Rappley I, Myers DS, Milne SB, et al. Lipidomic profiling in mouse brain reveals differences between ages and genders, with smaller changes associated with alpha-synuclein genotype. J Neurochem. 2009;111(1):15–25.
Rhee EP, Cheng S, Larson MG, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121(4):1402–11.
Sampaio JL, Gerl MJ, Klose C, et al. Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci U S A. 2011;108(5):1903–7.
Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279(15):2610–23.
Sato Y, Bernier F, Suzuki I, Kotani S, Nakagawa M, Oda Y. Comparative lipidomics of mouse brain exposed to enriched environment. J Lipid Res. 2013;54(10):2687–96.
Schuhmann K, Herzog R, Schwudke D, Metelmann-Strupat W, Bornstein SR, Shevchenko A. Bottom-up shotgun lipidomics by higher energy collisional dissociation on LTQ Orbitrap mass spectrometers. Anal Chem. 2011;83(14):5480–7.
Schwudke D, Hannich JT, Surendranath V, et al. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal Chem. 2007;79(11):4083–93.
Smilowitz JT, Zivkovic AM, Wan YJ, et al. Nutritional lipidomics: molecular metabolism, analytics, and diagnostics. Mol Nutr Food Res. 2013;57(8):1319–35.
Song J, Liu X, Wu J, et al. A highly efficient, high-throughput lipidomics platform for the quantitative detection of eicosanoids in human whole blood. Anal Biochem. 2013;433(2):181–8.
Taguchi R, Ishikawa M. Precise and global identification of phospholipid molecular species by an Orbitrap mass spectrometer and automated search engine Lipid Search. J Chromatogr A. 2010;1217(25):4229–39.
Vilella F, Ramirez LB, Simon C. Lipidomics as an emerging tool to predict endometrial receptivity. Fertil Steril. 2013;99(4):1100–6.
Wang M, Fang H, Han X. Shotgun lipidomics analysis of 4-hydroxyalkenal species directly from lipid extracts after one-step in situ derivatization. Anal Chem. 2012;84(10):4580–6.
Wei H, Hu C, Wang M, et al. Lipidomics reveals multiple pathway effects of a multi-components preparation on lipid biochemistry in ApoE*3Leiden.CETP mice. PLoS One. 2012;7(1):e30332.
Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4(7):594–610.
Wenk MR. Lipidomics in drug and biomarker development. Expert Opin Drug Discov. 2006;1(7):723–36.
Wolf C, Quinn PJ. Lipidomics: practical aspects and applications. Prog Lipid Res. 2008;47(1):15–36.
Yang K, Cheng H, Gross RW, Han X. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal Chem. 2009;81(11):4356–68.
Yang K, Dilthey BG, Gross RW. Identification and quantitation of fatty acid double bond positional isomers: a shotgun lipidomics approach using charge-switch derivatization. Anal Chem. 2013;85(20):9742–50.
Zhao C, Mao J, Ai J, et al. Integrated lipidomics and transcriptomic analysis of peripheral blood reveals significantly enriched pathways in type 2 diabetes mellitus. BMC Med Genomics. 2013;6 Suppl 1:S12.
Zhou X, Mao J, Ai J, et al. Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS One. 2012;7(11):e48889.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media Dordrecht
About this entry
Cite this entry
Hu, C., Li, J., Xu, G. (2015). Mass Spectrometry-Based Lipidomics for Biomarker Research. In: Preedy, V., Patel, V. (eds) General Methods in Biomarker Research and their Applications. Biomarkers in Disease: Methods, Discoveries and Applications. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-7696-8_36
Download citation
DOI: https://doi.org/10.1007/978-94-007-7696-8_36
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-7695-1
Online ISBN: 978-94-007-7696-8
eBook Packages: Biomedical and Life SciencesReference Module Biomedical and Life Sciences