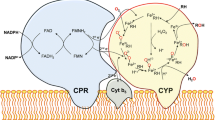
Abstract
Many fungal species exhibit unique and superior metabolic functions, including secondary metabolite production and detoxification of environmental pollutants, which are associated with cytochrome P450-dependent reactions. In the last decade, fungal genome projects have uncovered the astonishing molecular diversity of P450s in the fungal kingdom. The tremendous variation among the P450s implies that fungi have vigorously diversified P450 functions to meet novel metabolic needs. Fungal P450s discovered from genome projects are often categorized into novel families and subfamilies, suggesting that fungal P450s possess greater divergence than the animal, plant, or bacterial P450s. It is a challenging task to exploit the catalytic functions of the numerous P450s to better understand the biology of fungal metabolic systems. Comprehensive information about the functions of the P450s will also give hints about how to use their catalytic potentials in the biotechnology sector; however, experimental screening remains essential to reveal the catalytic potentials of individual P450s. In this chapter, the fungal metabolic systems in which P450s play a role are described, and the molecular and functional diversity and potential uses of fungal P450s are discussed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Alternative electron-transfer pathway
- Ascomycetous fungi
- Basidiomycetous fungi
- Functionomic survey
- Metabolic diversity
- Molecular diversity
1 Metabolic Systems Associated with Cytochromes P450 in Filamentous Fungi
A fungus is a eukaryotic microorganism in a group that includes yeasts, molds, and basidiomycetes. The fungal kingdom encompasses an enormous diversity of taxa, and it has been estimated that there may be from 1.5 to 5.1 million extant fungal species (Hawksworth 2001; Blackwell 2011). Fungi inhabit a broad range of environments, and some play fundamental roles in nutrient cycling by decomposing organic matter on the Earth. As well as the ability to adapt to various nutrient sources, fungi possess unique defense systems to survive exposure to various environmental agents. Consequently, many filamentous fungi rely on P450-dependent reactions to convert environmental pollutants, as do animals (Ichinose et al. 1999; Matsuzaki and Wariishi 2004), and to produce secondary metabolites, as do plants (Ferrer-Sevillano and Fernández-Cañón 2007; Prieto and Woloshuk 1997; Tudzynski et al. 2002; Kimura et al. 2007). Thus, we can assume an evolutionary history in which molecular diversification of fungal P450s conferred survival strategies by meeting novel metabolic requirements.
1.1 Bioconversion of Exogenous Compounds by Basidiomycetous Fungi
Wood-rotting basidiomycetes, often categorized into white-rot and brown-rot basidiomycetes , are common inhabitants of forest litter. White-rot basidiomycetes are capable of degrading all plant cell-wall components, including cellulose, hemicellulose, and lignin. The name “white-rot” originally comes from the observation that these fungi degrade lignin, which then appears as colorless cellulose. Because lignin is the most abundant renewable aromatic polymer, and is known as one of the most recalcitrant biomaterials on earth, the biodegradation of lignin is crucial for the carbon cycle of the biosphere (Kirk and Farrell 1987; Gold et al. 1989; Eriksson et al. 1990). As important enzymes involved in lignin biodegradation, the ligninolytic extracellular enzymes, lignin and manganese peroxidase and laccase, have been identified from a series of white-rot basidiomycetes . These enzymes trigger the decomposition of polymeric lignin via nonspecific one-electron oxidation, resulting in the formation of a variety of aromatic and quinoid fragments that are further metabolized intracellularly. White-rot basidiomycetes therefore possess unique and versatile intracellular metabolic systems to support and assist extracellular ligninolytic activities. In contrast, brown-rot basidiomycetes typically decompose cellulose and hemicellulose components of plant cell walls, but leave lignin. Lignin in brown-rotted wood appears dark brown in color and is almost equal in weight to lignin in sound wood (Kirk and Farrell 1987; Blanchette 1991). However, brown-rot basidiomycetes cause some modification of lignin; for instance, brown-rotted lignins were found to be extensively demethylated and significant quantities of phenolic hydroxyl groups were introduced into brown-rotted lignin (Kirk and Adler 1970; Niemenmaa et al. 2008; Yelle et al. 2011). In general, brown-rot basidiomycetes are thought to have repeatedly evolved from white-rot basidiomycetes. Thus, the cellular mechanisms associated with their secondary metabolism may be considerably more sophisticated. Apart from biological interest in the wood-rotting process, many research studies have focused on using brown-rot basidiomycetes in biotechnological applications; however, relatively little is known about the biology of these fungi (Itoh et al. 1997; Kerem et al. 1998; Fahr et al. 1999; Wetzstein et al. 1999).
During the past two decades, a number of studies have highlighted the metabolic diversity of basidiomycetes that is associated with unique P450s. The possible involvement of a cytochrome P450-like enzyme in the metabolism of polycyclic aromatic hydrocarbon (PAH ) by the white-rot basidiomycete Phanerochaete chrysosporium was first proposed by Sutherland and coworkers (1991). The reaction of benzo[a]pyrene hydroxylation was then demonstrated in vitro using the microsomal and cytosolic fractions of P. chrysosporium and Pleurotus pulmonarius (Masaphy et al. 1995, 1996). The metabolic capability of P. chrysosporium was investigated widely using a series of compounds that were known to be P450 substrates in other organisms, and the bioconversions of benzoic acid, camphor, 1,8-cineol, cinnamic acid, p-coumaric acid, coumarin, cumene, 1,12-dodecanediol, 1-dodecanol, 4-ethoxybenzoic acid, and 7-ethoxycoumarin were demonstrated (Matsuzaki and Wariishi 2004). Using these 11 molecules as substrates, 23 hydroxylation reactions and two de-ethylation reactions were determined and six novel products were observed, suggesting that P. chrysosporium possesses P450s with unique and novel functions. In addition to product analysis, inhibitory experiments have been conducted widely using the P450 inhibitors piperonyl butoxide (PB) and 1-aminobenzotriazole (ABT). For example, the S-oxidation and hydroxylation steps involved in the bioconversion of 4-methyldibenzothiophene by the white-rot basidiomycete Coriolus versicolor were significantly inhibited in the presence of PB (Ichinose et al. 1999). Currently, a number of reports have demonstrated that PB and ABT effectively inhibit a wide range of bioconversion steps in many white-rot basidiomycetes including Coriolus (Trametes) spp. (Marco-Urrea et al. 2009; Prieto et al. 2011), Phanerochaete spp. (Teramoto et al. 2004a, b; Subramanian and Yadav 2009; Wang et al. 2012), Phlebia spp. (Xiao et al. 2011a, b), and Pleurotus spp. (Golan-Rozen et al. 2011). In addition, inhibitor experiments have shown that the brown-rot basidiomycete Tyromyces palustris metabolizes aromatic compounds via P450-dependent hydroxylation steps (Kamada et al. 2002). However, the biochemical characterization of basidiomycetous P450s has been limited because the poor level of protein expression in fungal cells and the paucity of available gene sequences have hindered subsequent investigations.
1.2 Biosynthesis of Secondary Metabolites in Ascomycetous Fungi
Ascomycetes are the largest known group of fungi and important ecologically, nutritionally, and medically. Many researchers have characterized some of the biosynthetic pathways of secondary metabolites in ascomycetous fungi ; for example, the metabolic pathways of mycotoxin s such as the aflatoxins and trichothecenes have been well established. Mycotoxins have been implicated as causative agents of adverse health effects in humans and animals who have consumed fungus-infected agricultural products. Consequently, fungi that produce mycotoxins are potential problems from both public health and economic perspectives. Because the different enzymes involved in secondary metabolite biosynthesis pathways are often encoded by genes in gene clusters, some of the P450s found in gene clusters were investigated intensively even in the pre-genomic era.
Aflatoxins , a group of polyketide-derived furanocoumarins, are toxic and carcinogenic compounds produced by Aspergillus spp. (Kensler et al. 2011). The aflatoxins were first discovered in the late 1950s and, soon after, it was revealed that toxicity was associated with the presence of Aspergillus flavus. Thus, the name “aflatoxin” was assigned to the toxic agents from A. flavus. Among the 14 different types of aflatoxin known in nature, aflatoxin B1 is considered to be the most toxic of the compounds produced by both A. flavus and Aspergillus parasiticus. In these fungi, 25 genes, including six P450s, are clustered for aflatoxin production (Yu et al. 1995, 2004). The P450s involved in the aflatoxin biosynthesis pathway have been categorized into family/subfamily, namely, CYP58B (aflU; epoxidase), CYP59 (aflN; versicolorin A oxidase), CYP60A (aflG; averantin hydroxylase), CYP60B (aflL; versicolorin B desatulase), CYP62 (aflV; function unknown), and CYP64 (aflQ; O-methylsterigmatocystin hydroxylase) (Ehrlich et al. 2004; Yu et al. 2004).
As well as the aflatoxin mycotoxin s , the trichothecene mycotoxins have a strong impact on human health because they inhibit protein synthesis. Potentially hazardous concentrations of the trichothecene mycotoxins can occur naturally in agricultural crops and commodities (Ueno 1989). Trichothecenes are a diverse family of sesquiterpenoid toxins produced by various fungi such as Fusarium spp., Myrothecium spp., Trichothecium spp., and Stachybotrys spp. In the trichothecenes biosynthetic pathway of Fusarium spp., the intermediate formation of trichodiene is the first committed step, and it is followed by several oxygenation steps. The subsequent C-2 hydroxylation of trichodiene is catalyzed by CYP58 (tri4). CYP58 has unique multifunctional properties and can catalyze four sequential reaction steps, of C-2 hydroxylation, C-12,13 epoxidation, C-11 hydroxylation, and C-3 hydroxylation, to produce isotrichotriol (Fig. 11.1) (Tokai et al. 2007). After the initial stage in the synthetic pathway, isotrichotriol is further modified by CYP65A1 (tri11; C-15 hydroxylase), CYP526 (tri13; C-4 hydroxylase), and CYP68 (tri1; C-8 hydroxylase) to produce the trichothecene mycotoxins (Kimura et al. 2007).
Multi-step reaction catalyzed by CYP58 (Kimura et al. 2007)
Apart from their mycotoxic effects, ascomycetous fungi have other biological impacts by, for example, producing plant hormones such as gibberellin . The gibberellins are diterpenoid acids and a group of phytohormones that influence many developmental processes in higher plants. Although gibberellin was first identified as a secondary metabolite of the rice pathogen Fusarium (previously Gibberella) fujikuroi, which causes the superelongation disease of rice shoots, they are ubiquitous in higher plants as endogenous growth regulators. The similar biosynthetic pathway in plants and fungus led to the suggestion of a possible evolutionary history in which the gene cluster for gibberellin biosynthesis in F. fujikuroi emerged by horizontal gene transfer from the host plant (Chapman and Ragan 1980). However, gene cloning of all the pathways in plants and in F. fujikuroi revealed dramatic differences in the character of the enzymes involved, suggesting that the gibberellin biosynthetic pathway had developed independently in plants and fungi (Tudzynski 2005). Both plants and fungi synthesize the cyclic diterpene, ent-kaurene, as an important intermediate in the initial stage of the gibberellin biosynthesis pathway, but, at the later stages, they employ different enzyme systems to produce gibberellin (Tudzynski 2005). After the intermediate formation of ent-kaurene, F. fujikuroi employs four different P450s (CYP68A1, CYP68B1, CYP69A1, and CYP503A1) to produce gibberellins. CYP68A1 (P450-1) was first cloned by differential cDNA screening. Subsequently, CYP68B1 (P450-2), CYP69A1 (P450-3), and CYP503A1 (P450-4) were identified from the gibberellin biosynthesis gene cluster. In particular, CYP68A1 and CYP503A1 are unique in their multifunctional properties ; CYP68A1 catalyzes four sequential steps in the gibberellin biosynthetic pathway and CYP503A1 catalyzes three oxidation steps between ent-kaurene and ent-kaurenoic acid.
2 Molecular Diversity of Fungal Cytochromes P450
2.1 Conservative Functions of Fungal P450s
In the fungal kingdom, only the housekeeping CYP51 and CYP61 gene families are evolutionarily conserved, and their functions are mandatory for ergosterol biosynthesis (Aoyama et al. 1984; Kelly et al. 1995). CYP51 was first purified from Saccharomyces cerevisiae in 1984 (Yoshida and Aoyama 1984; Aoyama et al. 1984), and orthologous genes from mammals (Trzaskos et al. 1986), plants (Kahn et al. 1996), and bacteria (Aoyama et al. 1998; Bellamine et al. 1999) were then discovered. The presence of its orthologues in all biological kingdoms suggests that CYP51 is the oldest of the known eukaryotic P450s. Fungal CYP51 catalyzes the oxidative removal of the 14α-methyl group of lanosterol and eburicol (24-methylene-24,25-dihydrolanosterol) to produce Δ14,15-desaturated intermediates in the ergosterol biosynthesis pathway. The presence of a second P450 in Saccharomyces cerevisiae was first demonstrated by disruption of CYP51 (Kelly et al. 1993), which was then sequenced and identified as CYP61 (Kelly et al. 1995; Skaggs et al. 1996). CYP61 is responsible for sterol C-22-desaturation at the later stage of the ergosterol biosynthesis pathway, suggesting that it may have evolved after a gene duplication of CYP51.
The 14α-demethylation by CYP51 is a more complex reaction that is dissimilar from the common P450-dependent monooxygenation reaction. The 14α-demethylation involves three sequential reaction steps, each requiring one molecule of oxygen and one molecule of NADPH (Waterman and Lepesheva 2005; Lepesheva and Waterman 2007; Yoshida 1992; Shyadehi et al. 1996). The first and second reactions follow a typical P450 reaction mechanism in which the 14α-methyl group is oxidized to the related alcohol and then aldehyde. The 14α-aldehyde group is released as formic acid, and a C14–C15 double bond is introduced in the third step. Although the catalytic properties of CYP51 are well conserved, the protein sequences are considerably diverse. The average amino acid sequence identity of CYP51 within the fungal kingdom is about 41 %, which is significantly lower than in animals (77 %), plants (76 %), and bacteria (77 %). Phylogenetic analysis revealed the presence of three clades (type A, type B, type C) of the CYP51 family in the fungal kingdom (Becher et al. 2011). In Aspergillus fumigatus, two different CYP51s, classified into type A and type B, are present. Neither the type-A nor type-B CYP51 variant is individually essential for fungal growth and virulence, but inactivation of both genes is lethal (Hu et al. 2007), indicating either of the proteins could probably replace the other. In contrast, only type A but not type B of CYP51 is required for conidiation of virulence of Magnaporthe oryzae (Yan et al. 2011).
Besides the housekeeping genes, the CYP53 family is also distributed widely in both ascomycetous and basidiomycetous fungi . CYP53 was first discovered from Aspergillus niger (van Gorcom et al. 1990) and the functions of this family are well characterized; they catalyze the para-hydroxylation of benzoic acid and its derivatives, which are naturally occurring antifungal plant metabolites (Amborabe et al. 2002). Because the phenolic compounds produced by CYP53 are further metabolized and degraded via the β-ketoadipate pathway (Harwood and Parales 1996), the CYP53 family was presumed to be crucial for competitive plant defenses. The numbers of CYP53 genes vary in the genomes of wood-rotting basidiomycetes, and the phylogenetic diversity of this family in Postia placenta is clear. Interestingly, CYP53D2 is capable of catalyzing the O-demethylation of stilbene derivatives, indicating its possible roles in degrading plant-based aromatic compounds, as well as benzoate derivatives, during the decomposition of woody biomass. A unique reaction mechanism might be involved in the O-demethylation reaction catalyzed by CYP53D2 because the CYP53 family has generally been considered to exhibit substrate specificity for benzoate; indeed, the carboxyl group in benzoate is known to be essential for enzyme–substrate binding (Matsuzaki and Wariishi 2005; Podobnik et al. 2008).
The CYP505 family is also common in ascomycetes and basidiomycetes. The first enzyme from this family, CYP505 (P450foxy), was isolated from Fusarium oxysporum and its catalytic activity, the subterminal hydroxylation of fatty acids, was demonstrated (Nakayama et al 1996). The characteristics of CYP505 resemble those of bacterial CYP102 in that both enzymes are self-sufficient flavocytochrome P450s composed of an amino-terminal P450 domain and a carboxy-terminal reductase domain in a single polypeptide chain. It was hypothesized that fungal CYP505 evolved from bacterial CYP102 by horizontal gene transfer (Kitazume et al. 2000), but the opposite transfer pathway (fungi to bacteria) is also possible (Moktali et al. 2012). The majority of fungal species encode multiple CYP505 genes and, currently, the white-rot basidiomycetes Phanerochaete chrysosporium has the largest number of isoforms (seven genes) in its genome. CYP505s are known to catalyze fatty acid hydroxylation, although the full physiological functions of this family have not yet been established. Notably, it has been demonstrated that CYP505B1 from Fusarium verticilloides (Gibberella moniliformis) is involved in the fumonisin (a mycotoxin ) synthesis pathway, where it catalyzes polyketide-amino acid hydroxylation (Proctor et al. 2003). Thus, the molecular evolution of the CYP505 family was required to meet the need for either fatty acid degradation or secondary metabolite biosynthesis.
2.2 Molecular Diversity of Fungal P450s
Phanerochaete chrysosporium is the first basidiomycetous fungus for which the whole-genome sequence was determined. The P. chrysosporium genome project provided a glimpse into the molecular diversity of basidiomycetous fungi and identified more than 100 novel P450s (Martinez et al. 2004). The discovery of more than 100 novel P450s abolished the earlier notion that fungal P450s might show limited diversity because only three isoforms had been found in the yeast (S. cerevisiae) genome and only a modest number of P450 families (51–69 families) were found in the pre-genomic era. With the increasing amount of information that is available as a result of the many genome projects, the current fungal cytochrome P450 database has enlarged exponentially and continues to increase (Moktali et al. 2012). To date, at least 2,784 species of fungal P450s have been identified from 53 sequenced genomes and assigned to 399 families by the P450 nomenclature committee (Nelson 2011); however, the vast majority of their functions and biological roles are still obscure. The compilation of fungal P450 sequences has highlighted a unique feature of the P450 family in the fungal kingdom: it is strikingly diverse. Consequently, CYP51 –CYP69, CYP501–CYP 699, and CYP5001–CYP6999 are allocated to fungal P450s. These findings suggested that the diversification of P450s occurred more vigorously in ascomycetous and basidiomycetous fungi than in yeast (Cornell et al. 2007).
Fungal P450s show marked divergence even though the fungi are taxonomically similar. Aspergillus oryzae, for instance, has 155 P450 genes (including 13 pseudogenes), which is significantly more than the 111 P450 genes in Aspergillus nidulans (Nazir et al. 2010; Kelly et al. 2009). These differences reflect the evolution of the Aspergillus species, in which A. oryzae increased the number of genes by horizontal gene transfer and duplication (Machida et al. 2005). There are 87 and 89 P450 families in A. oryzae and A. nidulans, respectively. The ascomycetous fungi Fusarium graminearum and Magnaporthe grisea are also known to have extremely varied P450 families (Deng et al. 2007). The diversity of the P450 family in ascomycetous fungi is considerably higher than its diversity in basidiomycetous fungi ; for example, only 42 P450 families in Postia placenta and 31 P450 families in P. chrysosporium. Fungal P450s appear to have diversified continuously after generic separation, and perhaps also after speciation. In fact, A. oryzae and A. flavus exhibit strikingly similar genetic features including similar P450 divergence (155 P450 genes in A. oryzae and 167 P450 genes in A. flavus), although 16 P450 genes were found in A. oryzae and not in A. flavus.
The large-scale diversity of basidiomycetous P450s was first uncovered by the white-rot P. chrysosporium genomic project, which identified about 150 isoforms. The importance of P450s in the decomposition of plant materials and in the detoxification of environmental pollutants is also true in the brown-rot basidiomycetes ; in fact, P. placenta possesses a larger number of P450s than P. chrysosporium and at least 250 isoforms, including 60 allelic variants, were identified in the whole genome of this fungus (Ide et al. 2012). Currently, the molecular diversity of P450s in both white-rot and brown-rot basidiomycetes has become clearer because of the availability of the whole-genome sequences (Floudas et al. 2012). Comparative phylogenetic analyses of the P450s in P. chrysosporium and P. placenta have revealed interesting aspects; for example, that the large CYP5027, CYP53 50, and CYP5348 families in P. placenta are absent in P. chrysosporium. In contrast, the CYP51 44 family has been enlarged in P. chrysosporium but not in P. placenta. Another marked difference in the distribution of the P450 family of genes was demonstrated by a comparative analysis of the white-rot basidiomycetes , Bjerkandera sp., Ganoderma sp., and Phlebia sp. (Syed et al. 2013a). In basidiomycetous fungi , P450 gene expansion seemed to be achieved by tandem gene duplication , and 18–37 % of P450 genes were estimated to have appeared as a result of these events (Syed et al. 2013a; Suzuki et al. 2012). The tandem duplication events most likely occurred continuously even after speciation because 266 P450 genes were identified in the Phanerochaete carnosa genome, which is considerably higher than the number in the P. chrysosporium genome (Suzuki et al. 2012).
3 Genomics to Functions
3.1 How We Can Determine P450 Functions
A thorough understanding of the sequence–structure–function relationships of the P450s is a challenging task even in a postgenomic era. Experimental screening remains essential to determine the catalytic potentials of individual P450s. In particular, there is a paucity of literature relating to the biochemical and functional characterization of fungal P450s, making it difficult to predict their functions based on sequence homology. Under such circumstances, transcriptome studies have helped uncover novel P450 functions. For example, it has been demonstrated that basidiomycetous P450s are differently regulated in response to culture conditions, exogenous chemicals, and growth stages (Ichinose et al. 2002; Doddapaneni and Yadav 2005; Chigu et al. 2010). Syed et al. (2010) employed a two-step strategy to identify the specific P450 involved in PAH metabolism in Phanerochaete chrysosporium. First, P450s that were upregulated in response to exogenously added PAHs were identified, and then their catalytic functions were determined using recombinant enzymes expressed in Pichia pastoris. Consequently, CYP51 36A2, CYP5136A3, CYP5142A3, CYP5144A5, CYP5144A7, and CYP5145A3 were shown to be capable of oxidizing PAHs. Later, the same research group functionally expressed and characterized CYP63A2 (Syed et al. 2011, 2013b). Recently, in Phanerochaete carnosa it was shown that various P450s were induced when the fungus was grown on woody substrates, suggesting that some P450s play roles in the decomposition of woody biomass (MacDonald et al. 2011; Suzuki et al. 2012).
The development of a rapid functional screening system will open the door for advanced studies of fungal P450s. Recently, functionomic studies were conducted using a functional screening system in which 425 isoforms of fungal P450s from the basidiomycetous fungi P. chrysosporium and P. placenta, and from koji mold A. oryzae, were coexpressed with yeast NADPH-P450 reductase in S. cerevisiae (Hirosue et al. 2011; Nazir et al. 2011; Ide et al. 2012). A functionomic survey resulted in the discovery of novel catalytic potentials of the fungal P450s (Table 11.1), which provided new insight into their biology and demonstrated their potential for use in the biotechnology sector. CYP51 45A3, CYP5136A1, CYP5136A3, CYP5141C1, and CYP5150A2, which exhibit versatile functions with broad substrate profiles against a series of aromatic compounds, have been discovered from Phanerochaete chrysosporium. CYP5150D1, CYP5027B1, and CYP53 50B2v1 from Postia placenta also show multifunctional properties against PAH s such as anthracene, carbazole, phenanthrene, and pyrene (Ide et al. 2012). The multifunctional properties of a single P450 species could be physiologically linked to the xenobiotic metabolism system in basidiomycetes, similar to the drug-metabolizing P450s in mammals. A number of basidiomycetous fungi have an expanded CYP512 family in their genome, and the P450s in this family from P. chrysosporium and P. placenta exhibit catalytic activities against testosterone and progesterone. CYP512N and CYP512P convert both testosterone and dehydroabietic acid, which emphasizes the structural similarity between steroids and abietane diterpenoids. Considering the structural similarity of mammalian steroids and fungal terpenoids, the hypothesis that the CYP512 family is likely to be involved in triterpenoid biosynthesis was proposed (Ide et al. 2012; Chen et al. 2012). Although further biochemical studies are needed to elucidate the physiological functions of the CYP512 family in fungi, the provisional substrates were first demonstrated from functionomic approaches. This example illustrates how comprehensive and non-target-driven screening was able to provide interesting results.
Nazir et al. (2011) conducted functional screening to discover potential P450s for industrial applications and demonstrated that CYP57B3 from the koji mold Aspergillus oryzae converted genistein to 8-hydroxy-, 6-hydroxy-, and 3′-hydroxygenistein, all of which have known biological and pharmacological potential (Fig. 11.2). Although hydroxylated genistein can be isolated from natural products including fermented products, the supply of these natural compounds is limited, making it problematic for practical use. Because the synthesis of the isoflavones remains an important objective, the use of CYP57B3 for the production of these value-added rare isoflavonoids from genistein would be of great interest.
3.2 Mechanistic Investigations In Vitro
As well as compiling information about the catalytic potentials of P450s, it is also of great importance to thoroughly understand the biochemical features of individual P450s. Although the biochemical characterization and mechanistic understanding of basidiomycetous P450s are still in their infancy compared with the rich history of these studies in other organisms, some researchers have reported some unique aspects of the reaction mechanisms of the basidiomycetous P450s. The catalytic potentials of CYP51 47A1 and CYP5136A1 were first discovered using a functionomic approach, and their kinetic properties in the 3′-hydroxylation of flavone and O-deethylation of 7-ethoxycoumarin were investigated intensively using a microsomal fraction of recombinant S. cerevisiae (Kasai et al. 2010). Based on the kinetic studies, it was suggested that CYP5147A1 and CYP5136A1 accommodate two substrates in their active sites. Molecular modeling of CYP5147A1 and a docking study of flavone to its active site supported the proposed kinetic properties (Kasai et al. 2010).
The catalytic properties of CYP51 50A2 were investigated intensively using the purified enzyme and CYP5150A2 was shown to have high affinity for 4-pentylbenzoic acid (Ichinose and Wariishi 2012). Notably, CYP5150A2 is unique in exhibiting substantial activity with redox partners cytochrome b5 (Cyt-b5) and NADH-dependent Cyt-b5 reductase (CB5R), even in the absence of cytochrome P450 oxidoreductase (CPR). These results showed that a combination of CB5R and Cyt-b5 may be capable of donating both the first and the second electrons required for the monooxygenation reaction (Fig. 11.3). An alternative electron-transfer pathway in the fungal P450 system associated with CB5R and Cyt-b5 was also demonstrated in vitro using CYP51 from Candida albicans and CYP63A2 from P. chrysosporium (Lamb et al. 1999; Syed et al. 2011). Disruption of the CPR gene in S. cerevisiae was not lethal, and the yeast could still synthesize ergosterol (Sutter and Loper 1989; Venkateswarlu et al. 1998), indicating that the alternative electron-transfer pathway was also active in vivo. The alternative electron-transfer pathway is most likely active in gibberellin production by Fusarium fujikuroi (Troncoso et al. 2008). Thus, it may be quite common that some eukaryotic P450s, including CYP5150A2, can be activated by alternative redox partners.
4 Conclusion
The data generated by the genomic projects have revealed the massive scale of P450 diversity in the fungal kingdom. The tremendous variations among the P450s imply that fungi have vigorously diversified P450 functions to meet novel metabolic needs, and unique aspects of this genomic feature may be related to evolutionary histories in which each fungal species has diversified differently and expanded the range of its P450 enzymes. In the postgenomic era, the numbers of “omics” studies have increased, which has contributed to the increased genome-wide understanding and advanced applications of fungal P450s. In addition to building sequence compilations, it is of great importance to explore the catalytic potentials of individual P450s. Many researchers have developed experimental approaches to investigate thoroughly the sequence–structure–function relationships of fungal P450s. Although the biochemical characterization and mechanistic understanding of basidiomycetous P450s are still in the early stages compared with the large amount of information known about P450s in other organisms, snapshots of the unique functions of the fungal P450s are already providing new insights into their fascinating biology and possible uses. Further systematic and functional compilations of fungal P450s will increase our understanding of these enzymes and give rise to new applications for them in the biotechnology sector.
References
Amborabe BE, Fleurat-Lessard P, Chollet JF, Roblin G (2002) Antifungal effects of salicylic acid and other benzoic acid derivatives towards Eutypa lata: structure–activity relationship. Plant Phys Biochem 40:1051–1060
Aoyama Y, Yoshida Y, Sato R (1984) Yeast cytochrome P-450 catalyzing lanosterol 14 alpha-demethylation. II. Lanosterol metabolism by purified P-450(14)DM and by intact microsomes. J Biol Chem 259:1661–1666
Aoyama Y, Horiuchi T, Gotoh O, Noshiro M, Yoshida Y (1998) CYP51 -like gene of Mycobacterium tuberculosis actually encodes a P450 similar to eukaryotic CYP51. J Biochem (Tokyo) 124:694–696
Becher R, Weihmann F, Deising HB, Wirsel SG (2011) Development of a novel multiplex DNA microarray for Fusarium graminearum and analysis of azole fungicide responses. BMC Genomics 12:52
Bellamine A, Mangla AT, Nes WD, Waterman MR (1999) Characterization and catalytic properties of the sterol 14alpha-demethylase from Mycobacterium tuberculosis. Proc Natl Acad Sci USA 96:8937–8942
Blackwell M (2011) The Fungi: 1, 2, 3 … 5.1 million species? Am J Bot 98:426–438
Blanchette RA (1991) Delignification by wood-decay fungi. Annu Rev Phytopathol 29:381–398
Chapman DJ, Ragan MA (1980) Evolution of biochemical pathways: evidence from comparative biochemistry. Annu Rev Plant Physiol 31:639–678
Chen S, Xu J, Liu C et al (2012) Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat Commun 3:913
Chigu NL, Hirosue S, Nakamura C, Teramoto H, Ichinose H, Wariishi H (2010) Cytochrome P450 monooxygenases involved in anthracene metabolism by the white-rot basidiomycete Phanerochaete chrysosporium. Appl Microbiol Biotechnol 87:1907–1916
Cornell MJ, Alam I, Soanes DM, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Talbot NJ, Oliver SG (2007) Comparative genome analysis across a kingdom of eukaryotic organisms: specialization and diversification in the fungi. Genome Res 17:1809–1822
Deng J, Carbone I, Dean RA (2007) The evolutionary history of cytochrome P450 genes in four filamentous Ascomycetes. BMC Evol Biol 7:30
Doddapaneni H, Yadav JS (2005) Microarray-based global differential expression profiling of P450 monooxygenases and regulatory proteins for signal transduction pathways in the white rot fungus Phanerochaete chrysosporium. Mol Genet Genomics 274:454–466
Ehrlich KC, Chang PK, Yu J, Cotty PJ (2004) Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation. Appl Environ Microbiol 70:6518–6524
Eriksson K-EL, Blanchette RA, Ander P (1990) Biodegradation of lignin. In: Timell TE (ed) Microbial and enzymatic degradation of wood and wood components. Springer, Berlin, pp 225–333
Fahr K, Wetzstein HG, Grey R, Schlosser D (1999) Degradation of 2,4-dichlorophenol and pentachlorophenol by two brown rot fungi. FEMS Microbiol Lett 175:127–132
Ferrer-Sevillano F, Fernández-Cañón JM (2007) Novel phacB-encoded cytochrome P450 monooxygenase from Aspergillus nidulans with 3-hydroxyphenylacetate 6-hydroxylase and 3,4-dihydroxyphenylacetate 6-hydroxylase activities. Eukaryot Cell 6:514–520
Floudas D, Binder M, Riley R, Barry K, Blanchette RA et al (2012) The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719
Golan-Rozen N, Chefetz B, Ben-Ari J, Geva J, Hadar Y (2011) Transformation of the recalcitrant pharmaceutical compound carbamazepine by Pleurotus ostreatus: role of cytochrome P450 monooxygenase and manganese peroxidase. Environ Sci Technol 45:6800–6805
Gold MH, Wariishi H, Valli K (1989) Extracellular peroxidases involved in lignin degradation by the white rot basidiomycete Phanerochaete chrysosporium. In: Whitaker JR, Sonnet PE (eds) Biocatalysis in agricultural biotechnology. American Chemical Society Symposium Series, vol 389. American Chemical Society, Washington, DC, pp 127–140
Harwood CS, Parales RE (1996) The beta-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol 50:553–590
Hawksworth DL (2001) The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol Res 105:1422–1432
Hirosue S, Tazaki M, Hiratsuka N, Yanai S, Kabumoto H, Shinkyo R, Arisawa A, Sakaki T, Tsunekawa H, Johdo O, Ichinose H, Wariishi H (2011) Insight into functional diversity of cytochrome P450 in the white-rot basidiomycete Phanerochaete chrysosporium: involvement of versatile monooxygenase. Biochem Biophys Res Commun 407:118–123
Hu W, Sillaots S, Lemieux S, Davison J, Kauffman S, Breton A, Linteau A, Xin C, Bowman J, Becker J, Jiang B, Roemer T (2007) Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog 3:e24
Ichinose H, Wariishi H (2012) Heterologous expression and mechanistic investigation of a fungal cytochrome P450 (CYP51 50A2): involvement of alternative redox partners . Arch Biochem Biophys 518:8–15
Ichinose H, Wariishi H, Tanaka H (1999) Biotransformation of recalcitrant 4-methyldibenzothiophene to water-extractable products using lignin-degrading basidiomycete Coriolus versicolor. Biotechnol Prog 15:706–714
Ichinose H, Wariishi H, Tanaka H (2002) Identification and heterologous expression of the cytochrome P450 oxidoreductase from the white-rot basidiomycete Coriolus versicolor. Appl Microbiol Biotechnol 59:658–664
Ide M, Ichinose H, Wariishi H (2012) Molecular identification and functional characterization of cytochrome P450 monooxygenases from the brown-rot basidiomycete Postia placenta. Arch Microbiol 194:243–253
Itoh N, Yoshida M, Miyamoto T, Ichinose H, Wariishi H, Tanaka H (1997) Fungal cleavage of thioether bond found in yperite. FEBS Lett 412:281–284
Kahn RA, Bak S, Olsen CE, Svendsen I, Moller BL (1996) Isolation and reconstitution of the heme-thiolate protein obtusifoliol 14alpha-demethylase from Sorghum bicolor (L.) Moench. J Biol Chem 271:32944–32950
Kamada F, Abe S, Hiratsuk N, Wariish H, Tanaka H (2002) Mineralization of aromatic compounds by brown-rot basidiomycetes -mechanisms involved in initial attack on the aromatic ring. Microbiology 148:1939–1946
Kasai N, Ikushiro S, Hirosue S, Arisawa A, Ichinose H, Uchida Y, Wariishi H, Ohta M, Sakaki T (2010) Atypical kinetics of cytochromes P450 catalysing 3′-hydroxylation of flavone from the white-rot fungus Phanerochaete chrysosporium. J Biochem (Tokyo) 147:117–125
Kelly SL, Lamb DC, Baldwin BC, Kelly DE (1993) Benzo(a)pyrene hydroxylase activity in yeast is mediated by P450 other than sterol 14 alpha-demethylase. Biochem Biophys Res Commun 197:428–432
Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Parks LW, Kelly DE (1995) Purification and reconstitution of activity of Saccharomyces cerevisiae P450 61, a sterol Δ22-desaturase. FEBS Lett 377:217–220
Kelly DE, Kraševec N, Mullins J, Nelson DR (2009) The CYPome (cytochrome P450 complement) of Aspergillus nidulans. Fungal Genet Biol 46:S53–S61
Kensler TW, Roebuck BD, Wogan GN, Groopman JD (2011) Aflatoxin: a 50-year odyssey of mechanistic and translational toxicology. Toxicol Sci 120(suppl 1):S28–S48
Kerem Z, Bao W, Hammel KE (1998) Rapid polyether cleavage via extracellular one-electron oxidation by a brown-rot basidiomycete. Proc Natl Acad Sci USA 95:10373–10377
Kimura M, Tokai T, Takahashi-Ando N, Ohsato S, Fujimura M (2007) Molecular and genetic studies of fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci Biotechnol Biochem 71:2105–2123
Kirk TK, Adler E (1970) Methoxy-deficient structural elements in lignin of sweetgum decayed by brown-rot fungus. Acta Chem Scand 24:3379–3390
Kirk TK, Farrell RL (1987) Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol 41:465–505
Kitazume T, Takaya N, Nakayama N, Shoun H (2000) Fusarium oxysporum fatty-acid subterminal hydroxylase (CYP505 ) is a membrane-bound eukaryotic counterpart of Bacillus megaterium cytochrome P450BM3. J Biol Chem 275:39734–39740
Lamb DC, Kelly DE, Manning NJ, Kaderbhai MA, Kelly SL (1999) Biodiversity of the P450 catalytic cycle: yeast cytochrome b5 /NADH cytochrome b5 reductase complex efficiently drives the entire sterol 14-demethylation (CYP51 ) reaction. FEBS Lett 462:283–288
Lepesheva GI, Waterman MR (2007) Sterol 14alpha-demethylase cytochrome P450 (CYP51 ), a P450 in all biological kingdoms. Biochim Biophys Acta 1770:467–477
MacDonald J, Doering M, Canam T, Gong Y, Guttman DS, Campbell MM, Master ER (2011) Transcriptomic responses of the softwood-degrading white-rot fungus Phanerochaete carnosa during growth on coniferous and deciduous wood. Appl Environ Microbiol 77:3211–3218
Machida M, Asai K, Sano M et al (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature (Lond) 438:1157–1161
Marco-Urrea E, Pérez-Trujillo M, Caminal G, Vicent T (2009) Dechlorination of 1,2,3- and 1,2,4-trichlorobenzene by the white-rot fungus Trametes versicolor. J Hazard Mater 166:1141–1147
Martinez D, Larrondo LF, Putnam N et al (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22:695–700
Masaphy S, Levanon D, Henis Y, Venkateswarlu K, Kelly SL (1995) Microsomal and cytosolic cytochrome P450 mediated benzo[a]pyrene hydroxylation in Pleurotus pulmonarius. Biotechnol Lett 17:969–974
Masaphy S, Levanon D, Henis Y, Venkateswarlu K, Kelly SL (1996) Evidence for cytochrome P-450 and P-450-mediated benzo(a)pyrene hydroxylation in the white rot fungus Phanerochaete chrysosporium. FEMS Microbiol Lett 135:51–55
Matsuzaki F, Wariishi H (2004) Functional diversity of cytochrome P450s of the white-rot fungus Phanerochaete chrysosporium. Biochem Biophys Res Commun 324:387–393
Matsuzaki F, Wariishi H (2005) Molecular characterization of cytochrome P450 catalyzing hydroxylation of benzoates from the white-rot fungus Phanerochaete chrysosporium. Biochem Biophys Res Commun 334:1184–1190
Moktali V, Park J, Fedorova-Abrams ND, Park B, Choi J, Lee YH, Kang S (2012) Systematic and searchable classification of cytochrome P450 proteins encoded by fungal and oomycete genomes. BMC Genomics 13:525
Nakayama N, Takemae A, Shoun H (1996) Cytochrome P450foxy, a catalytically self-sufficient fatty acid hydroxylase of the fungus Fusarium oxysporum. J Biochem (Tokyo) 119:435–440
Nazir KHMNH, Ichinose H, Wariishi H (2010) Molecular characterization and isolation of cytochrome P450 genes from the filamentous fungus Aspergillus oryzae. Arch Microbiol 192:395–408
Nazir KHMNH, Ichinose H, Wariishi H (2011) Construction and application of a functional library of cytochrome P450 monooxygenases from the filamentous fungus Aspergillus oryzae. Appl Environ Microbiol 77:3147–3150
Nelson DR (2011) Progress in tracing the evolutionary paths of cytochrome P450. Biochim Biophys Acta 1814:14–18
Niemenmaa O, Uusi-Rauva A, Hatakka A (2008) Demethoxylation of [O14CH3]-labelled lignin model compounds by the brown-rot fungi Gloeophyllum trabeum and Poria (Postia) placenta. Biodegradation 19:555–565
Podobnik B, Stojan J, Lah L, Krasevec N, Seliskar M, Rizner TL et al (2008) CYP53 A15 of Cochliobolus lunatus, a target for natural antifungal compounds. J Med Chem 51:3480–3486
Prieto R, Woloshuk CP (1997) ord1, an oxidoreductase gene responsible for conversion of O-methylsterigmatocystin to aflatoxin in Aspergillus flavus. Appl Environ Microbiol 63:1661–1666
Prieto A, Möder M, Rodil R, Adrian L, Marco-Urre E (2011) Degradation of the antibiotics norfloxacin and ciprofloxacin by a white-rot fungus and identification of degradation products. Bioresour Technol 102:10987–10995
Proctor RH, Brown DW, Plattner RD, Desjardins AE (2003) Co-expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet Biol 38:237–249
Shyadehi AZ, Lamb DC, Kelly SL, Kelly DE, Schunck WH, Wright JN, Corina D, Akhtar M (1996) The mechanism of the acyl-carbon bond cleavage reaction catalyzed by recombinant sterol 14 alpha- demethylase of Candida albicans (other names are lanosterol 14 alpha-demethylase, P-45014DM, and CYP51 ). J Biol Chem 271:12445–12450
Skaggs BA, Alexander JF, Pierson CA, Schweitzer KS, Chun KT, Koegel C, Barbuch R, Bard M (1996) Cloning and characterization of the Saccharomyces cerevisiae C-22 sterol desaturase gene, encoding a second cytochrome P-450 involved in ergosterol biosynthesis. Gene (Amst) 169:105–109
Subramanian V, Yadav JS (2009) Role of P450 monooxygenases in the degradation of the endocrine-disrupting chemical nonylphenol by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol 75:5570–5580
Sutherland JB, Selby AL, Freeman JP, Evans FE, Cerniglia CE (1991) Metabolism of phenanthrene by Phanerochaete chrysosporium. Appl Environ Microbiol 57:3310–3316
Sutter TR, Loper JC (1989) Disruption of the Saccharomyces cerevisiae gene for NADPH-cytochrome P450 reductase causes increased sensitivity to ketoconazole. Biochem Biophys Res Commun 160:1257–1266
Suzuki H, MacDonald J, Syed K, Salamov A, Hori C et al (2012) Comparative genomics of the white-rot fungi, Phanerochaete carnosa and P. chrysosporium, to elucidate the genetic basis of the distinct wood types they colonize. BMC Genomics 13:444
Syed K, Doddapaneni H, Subramanian V, Lam YW, Yadav JS (2010) Genome-to-function characterization of novel fungal P450 monooxygenases oxidizing polycyclic aromatic hydrocarbon s (PAH s). Biochem Biophys Res Commun 399:492–497
Syed K, Kattamuri C, Thompson TB, Yadav JS (2011) Cytochrome b5 reductase-cytochrome b 5 as an active P450 redox enzyme system in Phanerochaete chrysosporium: atypical properties and in vivo evidence of electron transfer capability to CYP63A2. Arch Biochem Biophys 509:26–32
Syed K, Nelson DR, Riley R, Yadav JS (2013a) Genome-wide annotation and comparative genomics of cytochrome P450 monooxygenases (P450s) in the polyporale species Bjerkandera adusta, Ganoderma sp. and Phlebia brevispora. Mycologia 105:1445–1455
Syed K, Porollo A, Lam YW, Grimmett PE, Yadav JS (2013b) CYP63A2, a catalytically versatile fungal P450 monooxygenase capable of oxidizing higher-molecular-weight polycyclic aromatic hydrocarbon s, alkylphenols, and alkanes. Appl Environ Microbiol 79:2692–2702
Teramoto H, Tanaka H, Wariishi H (2004a) Degradation of 4-nitrophenol by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Microbiol Biotechnol 66:312–317
Teramoto H, Tanaka H, Wariishi H (2004b) Fungal cytochrome P450s catalyzing hydroxylation of substituted toluenes to form their hydroxymethyl derivatives. FEMS Microbiol Lett 234:255–260
Tokai T, Koshino H, Takahashi-Ando N, Sato M, Fujimura M, Kimura M (2007) Fusarium Tri4 encodes a key multifunctional cytochrome P450 monooxygenase for four consecutive oxygenation steps in trichothecene biosynthesis. Biochem Biophys Res Commun 353:412–417
Troncoso C, Cárcamo J, Hedden P, Tudzynski B, Rojas MC (2008) Influence of electron transport proteins on the reactions catalyzed by Fusarium fujikuroi gibberellin monooxygenases. Phytochemistry 69:672–683
Trzaskos J, Kawata S, Gaylor JL (1986) Microsomal enzymes of cholesterol biosynthesis. Purification of lanosterol 14 alpha-methyl demethylase cytochrome P-450 from hepatic microsomes. J Biol Chem 261:14651–14657
Tudzynski B (2005) Gibberellin biosynthesis in fungi: genes, enzymes, evolution, and impact on biotechnology. Appl Microbiol Biotechnol 66:597–611
Tudzynski B, Rojas MC, Gaskin P, Hedden P (2002) The gibberellin 20-oxidase of Gibberella fujikuroi is a multifunctional monooxygenase. J Biol Chem 277:21246–21253
Ueno Y (1989) Trichothecene mycotoxin s : mycology, chemistry, and toxicology. Adv Nutr Res 3:301–353
van Gorcom RF, Boschloo JG, Kuijvenhoven A, Lange J, van Vark AJ, Bos CJ, van Balken JA, Pouwels PH, van den Hondel CA (1990) Isolation and molecular characterisation of the benzoate-para-hydroxylase gene (bphA) of Aspergillus niger: a member of a new gene family of the cytochrome P450 superfamily. Mol Gen Genet 223:192–197
Venkateswarlu K, Lamb DC, Kelly DE, Manning NJ, Kelly SL (1998) The N-terminal membrane domain of yeast NADPH-cytochrome P450 (CYP) oxidoreductase is not required for catalytic activity in sterol biosynthesis or in reconstitution of CYP activity. J Biol Chem 273:4492–4496
Wang J, Hirai H, Kawagishi H (2012) Biotransformation of acetamiprid by the white-rot fungus Phanerochaete sordida YK-624. Appl Microbiol Biotechnol 93:831–835
Waterman MR, Lepesheva GI (2005) Sterol 14 alpha-demethylase, an abundant and essential mixed-function oxidase. Biochem Biophys Res Commun 338:418–422
Wetzstein HG, Stadler M, Tichy HV, Dalhoff A, Karl W (1999) Degradation of ciprofloxacin by basidiomycetes and identification of metabolites generated by the brown rot fungus Gloeophyllum striatum. Appl Environ Microbiol 65:1556–1563
Xiao P, Mori T, Kondo R (2011a) Biotransformation of the organochlorine pesticide trans-chlordane by wood-rot fungi. N Biotechnol 29:107–115
Xiao P, Mori T, Kamei I, Kondo R (2011b) A novel metabolic pathway for biodegradation of DDT by the white rot fungi, Phlebia lindtneri and Phlebia brevispora. Biodegradation 22:859–867
Yan X, Ma WB, Li Y, Wang H, Que YW, Ma ZH, Talbot NJ, Wang ZY (2011) A sterol 14α-demethylase is required for conidiation, virulence and for mediating sensitivity to sterol demethylation inhibitors by the rice blast fungus Magnaporthe oryzae. Fungal Genet Biol 48:144–153
Yelle DJ, Wei D, Ralph J, Hammel KE (2011) Multidimensional NMR analysis reveals truncated lignin structures in wood decayed by the brown rot basidiomycete Postia placenta. Environ Microbiol 13:1091–1100
Yoshida Y (1992) Sterol biosynthesis. In: Omura T, Oshimura Y, Fujii-Kuriyama Y (eds) Cytochrome P450, 2nd edn. Kodansha, Tokyo, pp 93–101
Yoshida Y, Aoyama Y (1984) Yeast cytochrome P-450 catalyzing lanosterol 14 alpha-demethylation. I. Purification and spectral properties. J Biol Chem 259:1655–1660
Yu J, Chang PK, Cary JW, Wright M, Bhatnagar D, Cleveland TE, Payne GA, Linz JE (1995) Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl Environ Microbiol 61:2365–2371
Yu J, Chang PK, Ehrlich KC, Cary JW, Bhatnagar D, Cleveland TE, Payne GA, Linz JE, Woloshuk CP, Bennett JW (2004) Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol 70:1253–1262
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Japan
About this chapter
Cite this chapter
Ichinose, H. (2014). Metabolic Diversity and Cytochromes P450 of Fungi. In: Yamazaki, H. (eds) Fifty Years of Cytochrome P450 Research. Springer, Tokyo. https://doi.org/10.1007/978-4-431-54992-5_11
Download citation
DOI: https://doi.org/10.1007/978-4-431-54992-5_11
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-54991-8
Online ISBN: 978-4-431-54992-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)