Abstract
Rather than the presence of unique metabolic pathways, it is the absence of many pathways that characterizes the metabolism of Cryptosporidium. In fact, this genus of parasites has lost its ability of synthesizing de novo virtually all nutrients such as amino acids, nucleotides and fatty acids, thus relying on a large number of transporters to scavenge nutrients from the host. Members of this genus lack an apicoplast and associated pathways that are present in other apicomplexans. They lack cytochrome-based respiration, and rely mainly on glycolysis for energy production. Core metabolic pathways are highly streamlined, and redundancy is rare. These features make Cryptosporidium different from other apicomplexans. This chapter summarizes these features based on the analysis of genome sequences and published biochemical data in the context of drug targets and drug development.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Parasitophorous Vacuole Membrane
- Uridine Kinase
- Purine Salvage Pathway
- Very Long Chain
- Sodium Dehydrocholate
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
The genus Cryptosporidium comprises many species with different host specificities. C. parvum and C. hominis are the major human pathogens. The genomes of C. parvum and C. hominis have been reported (Abrahamsen et al. 2004; Xu et al. 2004), that of C. muris has been sequenced but remains to be published, and many more are being sequenced by an NIH-funded “Comparative Genomics of Cryptosporidium Species” project (http://www.genome.gov/26525388). The three sequenced genomes are accessible at GenBank and EuPathDB (http://www.EuPathDB.org). The availability of these genome sequences has greatly contributed to our understanding of parasite biology, and provided new opportunities to study Cryptosporidium metabolism. Currently, our knowledge on Cryptosporidium metabolism is mainly derived from genome annotations and analyses. A growing but limited number of enzymes have been investigated for their functions, biochemical features and potentials as therapeutic targets. However, systematic studies at the pathway level are still limited. Two reviews on Cryptosporidium biochemistry and metabolism were published as a book chapter in 2008 and a journal article in 2010 (Rider and Zhu 2010; Zhu 2008), These publications were mainly based on the C. parvum and C. hominis genomes and biochemical data available then. In this chapter, we will highlight the major metabolic features in Cryptosporidium with the addition of the newly sequenced C. muris genome and up-to-date biochemical data.
2 General Features of Cryptosporidium Metabolism
Phylogenetic and phylogenomic analyses have consistently placed Cryptosporidium at the base of the Apicomplexa with a closer relationship to gregarines than coccidia and haematozoa (Barta and Thompson 2006; Templeton et al. 2010; Zhu et al. 2000a). This view differs from the conventional taxonomy that considers Cryptosporidium as a sister group to the intestinal coccidia. The divergence of cryptosporidia from coccidia is also supported at the genomic and metabolic level. Cryptosporidium species have highly compact genomes (i.e., ~10 Mb), which are 3–5 times smaller than those of other apicomplexans and feature short intergenic sequences and very few introns (Abrahamsen et al. 2004; Xu et al. 2004). More importantly, this genus of parasites has lost the ability to synthesize virtually any nutrients de novo, such as amino acids, nucleotides and fatty acids. Cryptosporidium also lacks an apicoplast and mitochondrial genomes and associated metabolic pathways that are present in coccidia and haematozoa (Abrahamsen et al. 2004; Xu et al. 2004; Zhu et al. 2000b). Members of the genus possess a mitochondrial remnant that lacks the cytochrome-based respiratory chain, and retains only limited functions such as the assembly of ion-sulfur clusters (Lei et al. 2010; Kang et al. 2008; Keithly et al. 2005; Slapeta and Keithly 2004; Roberts et al. 2004; Riordan et al. 2003). Therefore, Cryptosporidium metabolism is extremely simplified. This feature differentiates these species from the evolutionarily more closely related gregarines that are able to synthesize many nutrients (e.g., Templeton et al. 2010).
Because Cryptosporidium cannot synthesize most nutrients de novo, it relies on a large family of transporters to scavenge nutrients such as amino acids (~11 transporters), sugars (~9), and nucleotides (at least one) (Abrahamsen et al. 2004; Xu et al. 2004). About 24 putative ATP-binding cassette (ABC) transporters can be identified in the C. parvum genome, but their substrates remain to be determined (Benitez et al. 2007; Li and Mun 2005; Bonafonte et al. 2004; Zapata et al. 2002; Perkins et al. 1999). ABC transporters are a large family of proteins with various substrate preferences including ions, sugars, amino acids, peptides, lipids, sterols and drugs. Therefore, many of them could be utilized by the parasite to scavenge nutrients. The parasite also possesses at least seven P-type ATPases (P-ATPases) that are mostly involved in cation transport. Among them, a putative Ca2+-ATPase and a heavy metal ATPase with binding specificity for reduced copper [Cu(I)] have been reported (LaGier et al. 2002, 2001; Zhu and Keithly 1997). One of the CpATPases belongs to the phospholipid transporter family and may be involved in lipid transport or membrane remodeling.
3 Carbohydrate and Energy Metabolisms
Carbohydrates are a source of energy and serve as building blocks for various biomolecules. Cryptosporidium is able to synthesize amylopectin as an energy storage polysaccharide. This biosynthetic capability is supported by earlier biochemical analyses and the presence of two glycogen branching enzymes (Harris et al. 2004; Zhang et al. 2012). The parasite can use polysaccharides, disaccharides or hexoses (e.g., glucose) to produce pyruvate and acetyl-CoA via the glycolytic pathway (Fig. 8.1). However, it employs two pyrophosphate-dependent phosphofructokinase (PPi-PFK) isoforms, rather than an ATP-PFK, to minimize the consumption of ATP. It also uses a unique pyruvate: NADP+ oxidoreductase comprised of pyruvate:ferredoxin oxidoreductase (PFO) and P450 reductase domains, rather than a pyruvate dehydrogenase complex to convert pyruvate into acetyl-CoA (Abrahamsen et al. 2004; Ctrnacta et al. 2006; Rotte et al. 2001). It has been speculated that NADP+ oxidoreductase is associated with the anti-cryptosporidial action of nitazoxanide (NTZ) that is currently the only drug approved by the FDA to treat cryptosporidial infections in immune-competent patients (Coombs and Muller 2002). Unlike classic inhibitor-enzyme interactions, NTZ may not act on NADP+ oxidoreductase. Rather, it is converted to a biotoxic free radical molecule by the PFO domain, similar to the reductive activation of 5-nitroimidazole metronidazole by PFO in the anaerobic protists Trichomonas and Giardia (Leitsch et al. 2011; Crossnoe et al. 2002; Yarlett et al. 1986). To better understand the enzymes in the carbohydrate pathway, protein crystals have been obtained for C. parvum LDH, glyceraldehyde 3-phosphate dehydrogenase (G3PDH), and pyruvate kinase (Nguyen et al. 2011; Cook et al. 2009; Senkovich et al. 2005). The crystal structures of pyruvate kinase and triosephosphate isomerase are resolved, and the structure of the active site of pyruvate kinase displays no obvious difference compared to the human homologue (Nguyen et al. 2011; Cook et al. 2009).
Illustration of the carbohydrate metabolic pathway in Cryptosporidium, in which the core components are glycolytic and fermentative enzymes. Abbreviations: AceCS acetyl-CoA synthetase (also known as acetate-CoA ligase), ADH1 alcohol dehydrogenase 1 (monofunctional), ADH-E type E alcohol dehydrogenase (bifunctional), GDH glycerol phosphate dehydrogenase, GAPDH glyceraldehyde phosphate dehydrogenase, GBE glycogen branching enzyme, GDBE glycogen debranching enzyme, HK hexokinase, LDH lactate dehydrogenase, MDH malate dehydrogenase, ME malic-enzyme, PDC pyruvate decarboxylase, PEPCL phosphoenolpyruvate carboxylase, PGI phosphoglucose isomerase, PGluM phosphoglucose mutase, PGK phosphoglycerate kinase, PGM phosphoglycerate mutase, PK pyruvate kinase, PNO pyruvate: NADP+ oxidoreductase, PPi-PFK pyrophosphate-dependent phosphofructokinase, T6PS-TP trehalose-6-phosphate synthase-trehalose phosphatase, TIM triosephosphate isomerase, UGGP UDP-galactose/glucose pyrophosphorylase
There are at least two types of cryptosporidial mitochondrial remnants: the “intestinal-type” C. parvum and C. hominis lack both the tricarboxylic acid (TCA) cycle and the cytochrome-based respiratory chain (Abrahamsen et al. 2004; Xu et al. 2004); whereas the “gastric-type” C. muris similarly lacks the respiratory chain but, based on the C. muris genome annotation, retains a complete set of enzymes for the TCA cycle and a type II NADH dehydrogenase (unpublished). The absence of an electron transport chain in all three species indicates that their mitochondria are unlikely to be a major source of energy, although it is possible that a certain level of electron potential may be generated via the type II NADH dehydrogenase in C. muris. Without respiration, the TCA cycle is possibly used by C. muris to supply intermediate metabolites. Additionally, all three genomes encode a plant-type alternative oxidase, which may be involved in detoxification of oxygen and/or generating certain electron potential.
Cryptosporidium does not carry a mitochondrial genome and the machinery for the replication, transcription and translation of organellar genomes. In addition to the enzymes discussed above, their nuclear genomes also encode a number of other proteins with mitochondrial-targeting signals, which include several translocases of outer and inner membranes, heat-shock proteins (HSPs), solute carriers, nucleotide anti-porters, ferredoxin and ferredoxin reductase and a small set of enzymes involved in ion-sulfur [Fe-S] cluster assembly (Abrahamsen et al. 2004; Lei et al. 2010; Kang et al. 2008; Mogi and Kita 2010; LaGier et al. 2003). It is believed that the [Fe-S] cluster assembly is one of the core functions retained in the mitochondrial remnant.
Collectively, we may conclude that Cryptosporidium relies mainly on glycolysis to produce energy. This notion is also supported by the presence of fermentative enzymes for producing three organic end products to avoid the accumulation of pyruvate and acetyl-CoA at the end of glycolysis (Fig. 8.1). These include lactate produced by lactate dehydrogenase (LDH), acetic acid by acetyl-CoA synthetase (AceCS; also known as acetate-CoA ligase, AceCL) and alcohol by a type-E bifunctional alcohol dehydrogenase (ADH-E) from acetyl-CoA or monofunctional ADH coupled with pyruvate decarboxylase from pyruvate (Zhang et al. 2012). Among them, LDH and ADH are bacterial-type enzymes. Cryptosporidium LDH originated from malate dehydrogenase (MDH) by a relatively recent gene duplication event which occurred after this genus separated from other apicomplexans. In fact, all apicomplexan MDH and LDH are bacterial-type, derived from an α-proteobacterial MDH (Zhu and Keithly 2002; Madern et al. 2004). Interestingly, a recent microarray-based transcriptome analysis also revealed that LDH has the highest level of expression among all genes in C. parvum oocysts, suggesting that the parasite mainly depends on LDH to keep the glycolytic pathway unobstructed in the external environment (Zhang et al. 2012).
Cryptosporidium possesses a plant-type pathway for synthesizing trehalose, which is accomplished by UDP-glucose/galactose pyrophosphorylase (UGGP) and a bifunctional enzyme fusion containing trehalose-6P synthase and trehalose phosphatase (T6PS-TP) (Yu et al. 2010). The presence of trehalose in C. parvum oocysts has been confirmed biochemically (Yu et al. 2010). However, the genome lacks trehalase, suggesting that the parasite is unlikely to reuse trehalose as a carbon source, unless using reversed reactions by UGGP and T6PS-TP. This differs from the intestinal coccidian Eimeria that has a trehalase, but lacks UGGP and T6PS-TP. Instead, Eimeria possesses a mannitol cycle that is absent in most other apicomplexans, including Cryptosporidium (Coombs and Muller 2002; Schmatz 1989). Trehalose and mannitol are known to function as anti-desiccants, antioxidants or protein-stabilizing agents in microorganisms, plants and some invertebrates, thus likely playing an important role in protecting the parasite against environmental stress.
The glycolytic pathway also provides GDP-mannose derived from fructose-6P or mannose-6P for N-glycan biosynthesis. Cryptosporidium appears to have a complete set of enzymes for synthesizing N-glycans in the lumen of the endoplasmic reticulum (ER) (e.g., various asparagine-linked glycosylation [ALG] transferases and an oligosaccharidyl-lipid flippase RFT1). N-glycan synthesis is also connected to the GPI (glycosyl-phosphatidyl-inositol) anchor synthesis. Like other apicomplexans and protists, these parasites lack enzymes to make more complex N-glycans in the Golgi apparatus that are common in fungi and plants. The C. parvum genome encodes ~30 mucin-like proteins, many of which are (or are predicted to be) membrane or secretory proteins based on the presence of signal peptides (e.g., Cevallos et al. 2000a, b; Barnes et al. 1998; Chatterjee et al. 2010). Most of the mucins contain both N- and O-glycosylation sites, and at least four enzymes involved in mucin-type O-glycosylation have been identified in the C. parvum genome, including UDP-N-acetyl-D-galactosamine-polypetide N-acetyl-galactosaminyl transferases (Wanyiri and Ward 2006). The involvement of mucins in parasite attachment to, and invasion of, host cells is being actively investigated. Binding of some of the mucin-like proteins by antibodies can block or reduce infection in vitro and/or in vivo, suggesting that mucins may be targets for developing immunotherapeutics (Wanyiri and Ward 2006).
4 Amino Acid Metabolism
Cryptosporidium cannot synthesize any amino acids de novo, which differs from other apicomplexans that possess complete pathways to make at least some amino acids. Instead, it retains only enzymes to interconvert certain amino acids coupled with other metabolic pathways. These include: glutamine synthetase for recycling glutamate produced by GMP synthetase back to glutamine; serine hydroxymethyl transferase for converting glycine to serine within the folate cycle; asparagine synthetase for producing asparagine from aspartate (which may be required to recycle ammonia released by AMP-deaminase); S-adenosylmethionine (SAM or AdoMet) synthetase to catalyze the formation of SAM to serve as an important methyl donor for transmethylation; and S-adenosylhomocysteine (SAH) synthase (SAHS; also known as SAH hydrolase, SAHH) for converting SAH derived from SAM after transmethylation to homocysteine and adenosine.
Among these enzymes, the general molecular and biochemical features of C. parvum SAHH (CpSAHH) has been characterized. Its inhibitors D-eritadenine and 9-(S)-(2, 3-dihydroxypropyl)adenine [(S)-DHPA] display efficacy at low micromolar levels against growth of C. parvum in vitro (Ctrnacta et al. 2007, 2010).
5 Nucleotide Metabolism
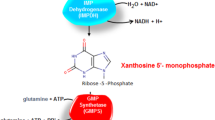
Most apicomplexans scavenge purines, but are capable of synthesizing pyrimidines de novo. However, Cryptosporidium lacks synthetic pathways for both purines and pyrimidines. The highly simplified purine salvage pathway starts with the uptake of adenosine by a nucleoside transporter. Adenosine is converted to AMP by adenosine kinase, IMP by AMP deaminase, XMP by IMP dehydrogenase (IMPDH), and GMP by GMP synthase (Fig. 8.2) (Striepen and Kissinger 2004). IMPDH genes in C. parvum and C. hominis were acquired from ε-proteobacteria, and differ from the eukaryotic type IMPDHs found in humans, animals and other apicomplexans (Striepen et al. 2002). However, an IMPDH gene has yet to be identified in the current version of the C. muris genome. Based on the essential role of this enzyme, C. muris needs IMPDH unless it directly scavenges GMP from the host. The C. parvum and C. hominis IMPDH genes are located at the end of chromosome 6. As this region is not represented in the sequenced C. muris genome, it is likely that a C. muris IMPDH gene is located in an unsequenced region.
Streamlined purine and pyrimidine salvaging pathways and folate cycle in Cryptosporidium. Abbreviations: AK adenosine kinase, AMPDA AMP deaminase, CTPS CTP synthase, dCMPDA dCMP deaminase, DHFR dihydrofolate reductase, GMPS GMP synthase (glutamine-hydrolyzing), IMPDH IMP dehydrogenase, RNR ribonucleoside-diphosphate reductase, SHMT serine hydroxymethyl transferase, TK thymidine kinase, TS thymidylate synthase, UDPK UDP/CDP kinase, UK uridine kinase, UMPK UMP/CMP kinase, UPRT uracil phosphoribosyltransferase
Because Cryptosporidium lacks hypoxanthine-xanthine-guanine phosphoribosyl transferase to serve as an alternative purine salvaging pathway, blocking this AMP-GMP pathway can effectively kill the parasite. The bacterial-type IMPDH that is highly divergent from humans and animals has been considered an attractive drug target. Its protein structure has been determined, and a number of potent inhibitors have been identified and are being evaluated for drug development (Johnson et al. 2013; Gorla et al. 2012; Sharling et al. 2010; Umejiego et al. 2004, 2008).
For pyrimidine salvaging, Cryptosporidium may utilize uracil, uridine and cystidine by converting them to UMP and CMP by uridine kinase (UK) and a bifunctional enzyme with UK fused to uracil phosphoribosyltransferase (UK-UPRT) (Fig. 8.2). Thymidine can also be used and converted to dTMP by a bacterial-type thymidine kinase (TK), which is subsequently converted to dUMP by thymidylate synthase (TS) coupled with the folate cycle, and to dCMP by dCMP deaminase. dCMP, CMP and UMP can be further converted to dCDP/dCTP, CDP/CTP and UDP/UTP by a multi-functional UMP kinase and UDP kinase. The three pyrimidine nucleotide pathways are interconnected by a ribonucleoside-diphosphate reductase enzyme. Therefore, inhibition of a single pathway may be insufficient to block the pyrimidine synthesis, unless interconvertion is restricted by the presence of rate-limiting enzymes and/or if the supply of any single source of precursors is limited. Although the pyrimidine salvaging pathways appear to be redundant, a recent study has shown that TK-mediated pro-drug activation may be utilized as an effective strategy for treating cryptosporidiosis (Sun et al. 2010).
In the folate cycle, dihydrofolate reductase and thymidylate synthase (DHFR-TS) are fused into a bifunctional enzyme in apicomplexans and some other protists (Vasquez et al. 1996). The linker between the DHFR and TS domains in Cryptosporidium is unique as it contains an 11-residue α-helix with extensive interactions with the opposite DHFR-TS monomer of the homodimeric enzyme (O’Neil et al. 2003). The active site of C. parvum DHFR contains unique residues that are analogous to the point mutations associated with antifolate resistance in other DHFRs, suggesting CpDHFR may be intrinsically resistant to some antifolate inhibitors (Vasquez et al. 1996). However, several novel CpDHFR inhibitors have been identified using a yeast complementation system and structure-based virtual screens, but their anti-cryptosporidial activity in vitro or in vivo remains to be determined (Senkovich et al. 2009; Martucci et al. 2009; Bolstad et al. 2008; Popov et al. 2006; Anderson 2005; Lau et al. 2001; Brophy et al. 2000).
6 Lipid Metabolism
Fatty acids are a source of energy in many organisms and major components of all biomembranes. However, Cryptosporidium is unable to use fatty acids as an energy source due to the absence of the β-oxidation pathway. It also lacks an apicoplast and its associated pathways such as isoprenoid synthesis and the Type II fatty acid synthase (FAS) system. Therefore, the parasite cannot synthesize fatty acids (Zhu 2004). However, Cryptosporidium possesses a 25-kb intronless Type I FAS gene that resembles bacterial polyketide synthase (PKS) and predicts a ~900 kDa megasynthase comprised of at least 21 enzymatic domains (Zhu et al. 2000c). The basic biochemical features have been studied using recombinant proteins. Its N-terminal loading unit containing an acyl-ligase (AL) and an acyl-carrier protein (ACP) has a substrate preference towards long chain fatty acids (LCFAs), indicating that CpFAS1 functions as a fatty acid “elongase” rather than synthesizing fatty acids (Zhu et al. 2004). This notion is further supported by functional analysis of its C-terminal reductase domain that is only active with very long chain (VLC) fatty acyl-CoAs (i.e., >C20:0) (Zhu et al. 2010). The reductase domain-catalyzed reductive reaction may release final products as fatty acyl aldehydes or fatty acyl alcohol, which differs from classic Type I FAS (in humans and animals) and Type II FAS (in prokaryotes, plants and plastid-containing apicomplexans) that use thioesterase to release acyl chains as fatty acids by hydrolysis (Zhu 2004). Between the loading unit and the reductase domain are three internal acyl elongation modules, each consisting of a complete set of 6 enzymes: (1) ACP for carrying acyl chains; (2) acyl-transferase (AT) for loading malonyl-CoA and transferring an acyl-chain from the previous module to ACP; (3) ketoacyl-ACP synthase (KS) for condensing a two-carbon (C2) unit from malonyl-CoA into the acyl-chain by a carboxylation reaction; (4) ketoacyl-ACP reductase for the reduction of a keto group; (5) hydroxyacyl-ACP dehydrase for the dehydration of a hydroxyl group; and (6) enoyl-ACP reductase for the reduction of double bonds (Zhu 2004; Zhu et al. 2000c, 2004, 2010). Therefore, at least three C2 units can be added into the acyl precursors (e.g., C16:0 palmitic acid) to form very long fatty acyl chains (e.g., C22:0) that are released as fatty acyl aldehydes or alcohol. Because aldehydes are biotoxic, fatty acyl alcohols are likely the final products that can be produced by two series of reductive reactions.
In parallel to Type I FAS, the Cryptosporidium genome also encodes a giant 45-kb intronless PKS, which represents the first PKS discovered in a protist (Zhu et al. 2002). Molecular and biochemical analysis reveals that CpPKS1 is similarly structured as CpFAS1, but contains seven internal acyl elongation modules that lack one or more of the five enzymatic domains. Therefore, elongated acyl chains will contain keto groups, hydroxyl groups and/or double bonds, which are characteristic of polyketides. The CpPKS1 loading unit also displays substrate preference towards LCFAs, suggesting the final product(s) may contain 30 or more carbons (Fritzler and Zhu 2007).
The ACP domains in all types of FAS and PKS systems require a post-translational modification by phosphopantetheinyl transferase (PPT) to add a prosthetic phosphopantetheine to a serine residue to become a functional holo-ACP. There are two types of PPT with different substrate preferences: SFP-type for activating Type I ACPs and ACPS-type for Type II ACPs. In fact, the types of PPT present in various apicomplexans match well with the types of FAS systems. For example, Cryptosporidium possesses only Type I FAS and SFP-PPT, Plasmodium has only Type II FAS and ACPS-PPT, while Toxoplasma contains both types I and II FAS and both SFP-PPT and ACPS-PPT. The activation of CpFAS1-ACP domains by SFP-PPT has been biochemically characterized and demonstrated (Cai et al. 2005).
In addition to type I FAS and PKS, Cryptosporidium possesses another set of enzymes capable of elongating fatty acyl chains, in which the substrates are fatty acyl-CoA thioesters, rather than acyl-ACP. The hallmark enzyme is a long chain acyl elongase (LCE). The biochemical features of CpLCE1 have been characterized using recombinant protein expressed in HEK-293 T cells as its expression in bacterial systems was found to be difficult (Fritzler et al. 2007). CpLCE1 is a membrane protein localized on the surface of sporozoites and the parasitophorous vacuole membrane (PVM). Localization on the surface contrasts with CpFAS1 and CpPKS1 that are mainly cytosolic. CpLCE1 is able to add a single C2 unit to LCFAs with substrate preference towards C14:0 myristoyl-CoA and C16:0 palmitoyl-CoA (Fritzler et al. 2007).
Long chain fatty acyl-CoA synthetase (ACS; aka fatty acid-CoA ligase, ACL) is another family of important enzymes in lipid metabolism. It catalyzes the first reaction in all fatty acid metabolisms by activating free fatty acids to form fatty acyl-CoA thioesters except for the Type I and II FAS/PKS systems. In fact, AL domains in Type I FAS/PKS systems share similar molecular and biochemical properties with ACS, and can also catalyze the formation of fatty acyl-CoA (Fritzler and Zhu 2007). Cryptosporidium possesses three ACS genes, which are under investigation in our laboratory. Our data have shown that CpACS enzymes prefer LCFAs as substrates and their inhibitors could inhibit the growth of C. parvum, suggesting that ACS can be explored as a novel therapeutic target in the parasite (unpublished observation).
Cryptosporidium also possesses a long-type fatty acyl-CoA binding protein (ACBP) that is responsible for restraining the movement of fatty acyl-CoA and/or forming an acyl-CoA pool in cells. This function is important, as free fatty acyl-CoA may be harmful to cellular membranes due to its “detergent effect” if it is not restrained or immediately routed into other metabolic pathways. CpACBP1 is also a membrane protein localized to PVM. It prefers binding to LC and VLC fatty acyl-CoA thioesters (Zeng et al. 2006). More recently, a fluorescence assay was developed for CpACBP1 and used to screen 1,040 known drugs, from which 28 drugs displayed inhibitory effects on CpACBP1 at sub-micromolar concentrations. Among them, four drugs (i.e., broxyquinoline, cloxyquin, cloxacillin sodium and sodium dehydrocholate) displayed efficacies against the growth of C. parvum with ID50 values at low micromolar levels. This observation raises hopes for potential repurposing of known drugs to treat cryptosporidiosis (Fritzler and Zhu 2012).
Two distinct oxysterol binding protein (OSBP)-related proteins (ORPs) have also been identified in C. parvum, designated as CpORP1 and CpORP2 (Zeng and Zhu 2006). The short-type CpOPR1 contains only a ligand binding domain, while the long-type CpORP2 contains Pleckstrin homology and ligand-binding domains. Lipid–protein overlay assays have revealed that CpORP1 and CpORP2 could specifically bind to phosphatidic acid, various phosphatidylinositol phosphates (PIPs), and sulfatide, but not to other types of lipids with simple heads. However, cholesterol was not a ligand for these two proteins. Like CpLCE1 and ACBP, CpORP1 also localized to the PVM, while CpORP2 localized only in intracellular merozoites (Zeng and Zhu 2006).
Due to the incapability of synthesizing fatty acids de novo, Cryptosporidium needs to scavenge fatty acids/lipids from the host. However, it is unclear how the parasite scavenges lipids as no specific fatty acid or lipid transporters have been identified or experimentally validated. A recent study has provided strong evidence that C. parvum is able to scavenge cholesterol from host cells and from the intestinal lumen (Ehrenman et al. 2013). Among lipoproteins, LDL is an important source of cholesterol, and C. parvum can obtain cholesterol that is incorporated into micelles and internalized into enterocytes by the NPC1L1 transporter. Pharmacological blockage of NPC1L1 function by ezetimibe or moderate down-regulation of NPC1L1 expression decreases parasite infectivity (Ehrenman et al. 2013).
The presence of acyl-CoA binding protein, long-chain acyl elongase, acyl-CoA synthetase and OSBP-related protein in PVM indicates that this unique membrane structure is involved in lipid metabolism including transport, activation and/or elongation of fatty acids in Cryptosporidium. Some ACS proteins in bacteria and yeast are also known to function as fatty acid transporters (Black and DiRusso 2007; DiRusso and Black 1999). Based on the most recent data, we have formulated a working hypothesis that fatty acids may be directly transported into the parasite via an undefined pathway(s) as free fatty acids, or by a PVM-specific ACS coupled with the formation of fatty acyl-CoA (Fig. 8.3). Free fatty acids may be elongated by the Type I FAS or PKS, or activated by ACS within the parasite to form fatty acyl-CoA. All fatty acyl-CoA thioesters may be immediately routed into subsequent metabolic pathways (such synthesis of complex lipids and biomembranes, or protein palmitoylation), undergo chain elongation by LCE, or bound to ACBP to form an acyl-CoA pool before entering subsequent pathways. Cryptosporidium may also scavenge phospholipids as implied by the presence of a type IV P-ATPase (cgd7_1760) with predicted substrate affinity towards phospholipids. However, it remains to be determined if this transporter is truly involved in lipid scavenging or is simply responsible for intracellular trafficking of phospholipids in parasite cells.
Working hypothesis on the fatty acid metabolism associated with the parasitophorous vacuole membrane (PVM) in Cryptosporidium. Abbreviations: ACBP fatty acyl-CoA binding protein, ACC acetyl-CoA carboxylase, ACS Fatty acyl-CoA synthetase, FAS1 Type I FAS, LCE long chain fatty acyl-elongase, LCFA-CoA long chain fatty acyl-CoA, PKS1 Type I PKS
Cryptosporidium has a small set of enzymes involved in synthesis of complex lipids. In glycerolipid synthesis, it only retains enzymes for conversions between 1, 2-diacyl-sn-glycerol-3-phosphate, 1,2-diacyl-sn-glycerol and triacylglycerol by phosphatidate phosphatase, diacylglycerol kinase, and diacylglycerol acyltransferase 1, indicating that the parasite may store fatty acids in the form of triacylglycerol. Phosphatidyl-ethanolamine may be synthesized from diacyl-sn-glycerol and CDP-ethanolamine (produced from phospho-ethanolamine) by ethanolamine-phosphotransferase (ETHPT), or via the diacyl-sn-glycerol-3-phosphate to CDP-diacyl-glycerol to phosphatidyl-L-serine to phosphatidyl-entholamine pathway by CDP-diacylglycerol synthase (CDS), phosphatidylserine synthase and phosphatidylserine decarboxylase (PSDC). However, a phosphatidylserine synthase gene has not be identified in the Cryptosporidium genomes. Additionally, enzymes involved in synthesizing lipoproteins and glycolipids are present in the Cryptosporidium genomes, which include up to nine DHHC family palmitoyl transferases for post-translational S-palmitoylation of proteins, and PIG-A, PIG-C, PIG-H, PIG-P, PIG-M and PIG-U involved in GPI anchor synthesis.
7 Stress-Related Pathways
Cryptosporidium must face various external and internal environmental stresses, including UV irradiation, temperature changes and dehydration in the natural environment, as well as hazardous chemicals, free radical molecules, drugs and host immune responses at various life cycle stages. The parasite genome encodes several classes of proteins that are important in handling these stresses, including heat shock proteins (HSPs) and various anti-oxidant molecules. HSPs are well known stress proteins that are generally up-regulated as part of the stress responses by participating in protein folding, maintaining proper protein conformation, and monitoring cellular proteins. The expression of the CpHSP70 gene has been found to be highly up-regulated in response to chlorine-based oxidants and heat treatment (Bajszar and Dekonenko 2010).
Cryptosporidium possesses a number of anti-oxidant molecules, including superoxide dismutase (SOD), glutathione S-transferase, glutathione peroxidase, thioredoxin reductase, and a number of thioredoxin related proteins (e.g., Kang et al. 2008; Zhang et al. 2012; Yoon et al. 2012). In C. parvum oocysts, all three glutaredoxin-associated genes and five out of 13 thioredoxin-associated genes are expressed at various levels. Upon UV treatment, three putative t-complex protein 1 (TCP-1) subunits and several thioredoxin-associated genes are up-regulated in oocysts, whereas various HSP/DNAj family members, SOD and glutaredoxin-related genes are not up-regulated or are even down-regulated, suggesting that different stress proteins play different roles in responses to different stresses (Zhang et al. 2012).
DNA damage may occur more frequently in the natural environment than during DNA replication in the host cell due to the exposure of oocysts to UV irradiation. Cryptosporidium possesses a machinery for DNA excision repair. Several genes encoding excision repair enzymes were found to be up-regulated upon UV treatment, including the replication protein large subunit 1B (CpRPA1B) (Zhang et al. 2012; Rider and Zhu 2008; Rochelle et al. 2005). There are two types of RPA1 proteins in Cryptosporidium (i.e., RPA1A and RPA1B) which are involved in DNA replication, repair and recombination (Rider and Zhu 2008; Zhu et al. 1999; Millership and Zhu 2002). Several studies have indicated that RPA1A is mainly responsible for general DNA replication in the parasite, whereas RPA1B may play a role in DNA recombination and repair (Rider and Zhu 2008; Rider et al. 2005).
Trehalose synthesis is another important anti-stress pathway as described in Sect. 8.3 (Yu et al. 2010). Despite Cryptosporidium lacking amino acid synthetic pathways, a single standalone bacterial-type tryptophan synthase β-subunit gene is present in the C. parvum and C. hominis genomes. It is known that tryptophan starvation is one of the innate immunity’s strategies in humans and animals to kill cells infected with certain pathogens including Toxoplasma and probably Cryptosporidium by activating the tryptophan degradation pathway (MacKenzie et al. 2007; Habara-Ohkubo et al. 1993). It is possible that Cryptosporidium may use tryptophan synthase β-subunit to synthesize tryptophan from indole present in the gut to evade tryptophan depletion.
8 Conclusions
Cryptosporidium is extremely well adapted to a parasitic life style. These parasites rely on the host to supply virtually all nutrients for their highly streamlined metabolic pathways. The insensitivity of Cryptosporidium to many anti-apicomplexan drugs is explained by the absence of the drug targets which are common in other apicomplexans. Examples of such pathways are de novo biosynthetic pathways and the cytochrome-based respiratory chain. Evolutionary divergence from other apicomplexans (e.g., bacterial type IMPDH and DHFR-TS with unusual amino acids at the active site) was also observed and may be relevant to drug design. The availability of whole genome sequences provides opportunities not only to better understand the unique metabolic features of these parasites, but also to identify key enzymes and study their biochemical features of interest to drug development. Indeed, a number of potential drug targets have been proposed and/or are currently pursued by various laboratories, including protein kinases and enzymes in the carbohydrate, energy, nucleotide, and fatty acid metabolic pathways.
Currently, research on Cryptosporidium is still hampered by the lack of genetic tools and by technical difficulties in manipulating of the parasite in the laboratory. Therefore, recombinant protein-based biochemical analysis is still the best approach to study the metabolism and characterize potential drug targets. On the other hand, although gene knockout or knockdown tools are not available to validate drug targets, we can still effectively predict potential drug targets in the streamlined core metabolic pathways that are essential to the parasite. Drug development against cryptosporidial infection has been progressing slowly, but promising new data are being reported (see also Chap. 11). Research will be accelerated by new knowledge generated by biochemical analysis, target-based high-throughput screening of drugs, and structure-based analysis of protein-inhibitor interactions.
References
Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA et al (2004) Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304(5669):441–445. doi:10.1126/science.1094786
Anderson AC (2005) Targeting DHFR in parasitic protozoa. Drug Discov Today 10(2):121–128. doi:10.1016/S1359-6446(04)03308-2
Bajszar G, Dekonenko A (2010) Stress-induced Hsp70 gene expression and inactivation of Cryptosporidium parvum oocysts by chlorine-based oxidants. Appl Environ Microbiol 76(6):1732–1739. doi:10.1128/AEM.02353-09
Barnes DA, Bonnin A, Huang JX, Gousset L, Wu J, Gut J et al (1998) A novel multi-domain mucin-like glycoprotein of Cryptosporidium parvum mediates invasion. Mol Biochem Parasitol 96(1–2):93–110
Barta JR, Thompson RC (2006) What is Cryptosporidium? Reappraising its biology and phylogenetic affinities. Trends Parasitol 22(10):463–468. doi:10.1016/j.pt.2006.08.001
Benitez AJ, McNair N, Mead J (2007) Modulation of gene expression of three Cryptosporidium parvum ATP-binding cassette transporters in response to drug treatment. Parasitol Res 101(6):1611–1616. doi:10.1007/s00436-007-0701-x
Black PN, DiRusso CC (2007) Vectorial acylation: linking fatty acid transport and activation to metabolic trafficking. Novartis Found Symp 286:127–138, discussion 38–41, 62–3, 96–203
Bolstad DB, Bolstad ES, Frey KM, Wright DL, Anderson AC (2008) Structure-based approach to the development of potent and selective inhibitors of dihydrofolate reductase from Cryptosporidium. J Med Chem 51(21):6839–6852. doi:10.1021/jm8009124
Bonafonte MT, Romagnoli PA, McNair N, Shaw AP, Scanlon M, Leitch GJ et al (2004) Cryptosporidium parvum: effect of multi-drug reversing agents on the expression and function of ATP-binding cassette transporters. Exp Parasitol 106(3–4):126–134. doi:10.1016/j.exppara.2004.03.012
Brophy VH, Vasquez J, Nelson RG, Forney JR, Rosowsky A, Sibley CH (2000) Identification of Cryptosporidium parvum dihydrofolate reductase inhibitors by complementation in Saccharomyces cerevisiae. Antimicrob Agents Chemother 44(4):1019–1028
Cai X, Herschap D, Zhu G (2005) Functional characterization of an evolutionarily distinct phosphopantetheinyl transferase in the apicomplexan Cryptosporidium parvum. Eukaryot Cell 4(7):1211–1220. doi:10.1128/EC.4.7.1211-1220.2005
Cevallos AM, Bhat N, Verdon R, Hamer DH, Stein B, Tzipori S et al (2000a) Mediation of Cryptosporidium parvum infection in vitro by mucin-like glycoproteins defined by a neutralizing monoclonal antibody. Infect Immun 68(9):5167–5175
Cevallos AM, Zhang X, Waldor MK, Jaison S, Zhou X, Tzipori S et al (2000b) Molecular cloning and expression of a gene encoding Cryptosporidium parvum glycoproteins gp40 and gp15. Infect Immun 68(7):4108–4116
Chatterjee A, Banerjee S, Steffen M, O’Connor RM, Ward HD, Robbins PW et al (2010) Evidence for mucin-like glycoproteins that tether sporozoites of Cryptosporidium parvum to the inner surface of the oocyst wall. Eukaryot Cell 9(1):84–96. doi:10.1128/EC.00288-09
Cook WJ, Senkovich O, Chattopadhyay D (2009) An unexpected phosphate binding site in glyceraldehyde 3-phosphate dehydrogenase: crystal structures of apo, holo and ternary complex of Cryptosporidium parvum enzyme. BMC Struct Biol 9:9. doi:10.1186/1472-6807-9-9
Coombs GH, Muller S (2002) Recent advances in the search for new anti-coccidial drugs. Int J Parasitol 32(5):497–508
Crossnoe CR, Germanas JP, LeMagueres P, Mustata G, Krause KL (2002) The crystal structure of Trichomonas vaginalis ferredoxin provides insight into metronidazole activation. J Mol Biol 318(2):503–518. doi:10.1016/S0022-2836(02)00051-7
Ctrnacta V, Ault JG, Stejskal F, Keithly JS (2006) Localization of pyruvate: NADP+ oxidoreductase in sporozoites of Cryptosporidium parvum. J Eukaryot Microbiol 53(4):225–231. doi:10.1111/j.1550-7408.2006.00099.x
Ctrnacta V, Stejskal F, Keithly JS, Hrdy I (2007) Characterization of S-adenosylhomocysteine hydrolase from Cryptosporidium parvum. FEMS Microbiol Lett 273(1):87–95. doi:10.1111/j.1574-6968.2007.00795.x
Ctrnacta V, Fritzler JM, Surinova M, Hrdy I, Zhu G, Stejskal F (2010) Efficacy of S-adenosylhomocysteine hydrolase inhibitors, D-eritadenine and (S)-DHPA, against the growth of Cryptosporidium parvum in vitro. Exp Parasitol 126(2):113–116. doi:10.1016/j.exppara.2010.04.007
DiRusso CC, Black PN (1999) Long-chain fatty acid transport in bacteria and yeast. Paradigms for defining the mechanism underlying this protein-mediated process. Mol Cell Biochem 192(1–2):41–52
Ehrenman K, Wanyiri JW, Bhat N, Ward HD, Coppens I (2013). Cryptosporidium parvum scavenges LDL-derived cholesterol and micellar cholesterol internalized into enterocytes. Cell Microbiol 15(7):1182–1197. doi:10.1111/cmi.12107
Fritzler JM, Zhu G (2007) Functional characterization of the acyl-[acyl carrier protein] ligase in the Cryptosporidium parvum giant polyketide synthase. Int J Parasitol 37(3–4):307–316. doi:10.1016/j.ijpara.2006.10.014
Fritzler JM, Zhu G (2012) Novel anti-Cryptosporidium activity of known drugs identified by high-throughput screening against parasite fatty acyl-CoA binding protein (ACBP). J Antimicrob Chemother 67(3):609–617. doi:10.1093/jac/dkr516
Fritzler JM, Millership JJ, Zhu G (2007) Cryptosporidium parvum long-chain fatty acid elongase. Eukaryot Cell 6(11):2018–2028. doi:10.1128/EC.00210-07
Gorla SK, Kavitha M, Zhang M, Liu X, Sharling L, Gollapalli DR et al (2012) Selective and potent urea inhibitors of Cryptosporidium parvum inosine 5′-monophosphate dehydrogenase. J Med Chem 55(17):7759–7771. doi:10.1021/jm3007917
Habara-Ohkubo A, Shirahata T, Takikawa O, Yoshida R (1993) Establishment of an antitoxoplasma state by stable expression of mouse indoleamine 2,3-dioxygenase. Infect Immun 61(5):1810–1813
Harris JR, Adrian M, Petry F (2004) Amylopectin: a major component of the residual body in Cryptosporidium parvum oocysts. Parasitology 128(Pt 3):269–282
Johnson CR, Gorla SK, Kavitha M, Zhang M, Liu X, Striepen B et al (2013) Phthalazinone inhibitors of inosine-5′-monophosphate dehydrogenase from Cryptosporidium parvum. Bioorg Med Chem Lett 23(4):1004–1007. doi:10.1016/j.bmcl.2012.12.037
Kang JM, Cheun HI, Kim J, Moon SU, Park SJ, Kim TS et al (2008) Identification and characterization of a mitochondrial iron-superoxide dismutase of Cryptosporidium parvum. Parasitol Res 103(4):787–795. doi:10.1007/s00436-008-1041-1
Keithly JS, Langreth SG, Buttle KF, Mannella CA (2005) Electron tomographic and ultrastructural analysis of the Cryptosporidium parvum relict mitochondrion, its associated membranes, and organelles. J Eukaryot Microbiol 52(2):132–140. doi:10.1111/j.1550-7408.2005.04-3317.x
LaGier MJ, Zhu G, Keithly JS (2001) Characterization of a heavy metal ATPase from the apicomplexan Cryptosporidium parvum. Gene 266(1–2):25–34
LaGier MJ, Keithly JS, Zhu G (2002) Characterisation of a novel transporter from Cryptosporidium parvum. Int J Parasitol 32(7):877–887
LaGier MJ, Tachezy J, Stejskal F, Kutisova K, Keithly JS (2003) Mitochondrial-type iron-sulfur cluster biosynthesis genes (IscS and IscU) in the apicomplexan Cryptosporidium parvum. Microbiology 149(Pt 12):3519–3530
Lau H, Ferlan JT, Brophy VH, Rosowsky A, Sibley CH (2001) Efficacies of lipophilic inhibitors of dihydrofolate reductase against parasitic protozoa. Antimicrob Agents Chemother 45(1):187–195. doi:10.1128/AAC.45.1.187-195.2001
Lei C, Rider SD Jr, Wang C, Zhang H, Tan X, Zhu G (2010) The apicomplexan Cryptosporidium parvum possesses a single mitochondrial-type ferredoxin and ferredoxin: NADP+ reductase system. Protein Sci 19(11):2073–2084. doi:10.1002/pro.487
Leitsch D, Burgess AG, Dunn LA, Krauer KG, Tan K, Duchene M et al (2011) Pyruvate:ferredoxin oxidoreductase and thioredoxin reductase are involved in 5-nitroimidazole activation while flavin metabolism is linked to 5-nitroimidazole resistance in Giardia lamblia. J Antimicrob Chemother 66(8):1756–1765. doi:10.1093/jac/dkr192
Li LC, Mun YF (2005) Partial characterization of genes encoding the ATP-binding cassette proteins of Cryptosporidium parvum. Trop Biomed 22(2):115–122
MacKenzie CR, Heseler K, Muller A, Daubener W (2007) Role of indoleamine 2,3-dioxygenase in antimicrobial defence and immuno-regulation: tryptophan depletion versus production of toxic kynurenines. Curr Drug Metab 8(3):237–244
Madern D, Cai X, Abrahamsen MS, Zhu G (2004) Evolution of Cryptosporidium parvum lactate dehydrogenase from malate dehydrogenase by a very recent event of gene duplication. Mol Biol Evol 21(3):489–497. doi:10.1093/molbev/msh042
Martucci WE, Udier-Blagovic M, Atreya C, Babatunde O, Vargo MA, Jorgensen WL et al (2009) Novel non-active site inhibitor of Cryptosporidium hominis TS-DHFR identified by a virtual screen. Bioorg Med Chem Lett 19(2):418–423. doi:10.1016/j.bmcl.2008.11.054
Millership JJ, Zhu G (2002) Heterogeneous expression and functional analysis of two distinct replication protein A large subunits from Cryptosporidium parvum. Int J Parasitol 32(12):1477–1485
Mogi T, Kita K (2010) Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium. Parasitol Int 59(3):305–312. doi:10.1016/j.parint.2010.04.005
Nguyen TN, Abendroth J, Leibly DJ, Le KP, Guo W, Kelley A et al (2011) Structure of triosephosphate isomerase from Cryptosporidium parvum. Acta Crystallogr Sect F Struct Biol Cryst Commun 67(Pt 9):1095–1099. doi:10.1107/S1744309111019178
O’Neil RH, Lilien RH, Donald BR, Stroud RM, Anderson AC (2003) Phylogenetic classification of protozoa based on the structure of the linker domain in the bifunctional enzyme, dihydrofolate reductase-thymidylate synthase. J Biol Chem 278(52):52980–52987. doi:10.1074/jbc.M310328200
Perkins ME, Riojas YA, Wu TW, Le Blancq SM (1999) CpABC, a Cryptosporidium parvum ATP-binding cassette protein at the host-parasite boundary in intracellular stages. Proc Natl Acad Sci U S A 96(10):5734–5739
Popov VM, Chan DC, Fillingham YA, Atom Yee W, Wright DL, Anderson AC (2006) Analysis of complexes of inhibitors with Cryptosporidium hominis DHFR leads to a new trimethoprim derivative. Bioorg Med Chem Lett 16(16):4366–4370. doi:10.1016/j.bmcl.2006.05.047
Rider SD Jr, Zhu G (2008) Differential expression of the two distinct replication protein A subunits from Cryptosporidium parvum. J Cell Biochem 104(6):2207–2216. doi:10.1002/jcb.21784
Rider SD Jr, Zhu G (2010) Cryptosporidium: genomic and biochemical features. Exp Parasitol 124(1):2–9. doi:10.1016/j.exppara.2008.12.014
Rider SD Jr, Cai X, Sullivan WJ Jr, Smith AT, Radke J, White M et al (2005) The protozoan parasite Cryptosporidium parvum possesses two functionally and evolutionarily divergent replication protein A large subunits. J Biol Chem 280(36):31460–31469. doi:10.1074/jbc.M504466200
Riordan CE, Ault JG, Langreth SG, Keithly JS (2003) Cryptosporidium parvum Cpn60 targets a relict organelle. Curr Genet 44(3):138–147. doi:10.1007/s00294-003-0432-1
Roberts CW, Roberts F, Henriquez FL, Akiyoshi D, Samuel BU, Richards TA et al (2004) Evidence for mitochondrial-derived alternative oxidase in the apicomplexan parasite Cryptosporidium parvum: a potential anti-microbial agent target. Int J Parasitol 34(3):297–308. doi:10.1016/j.ijpara.2003.11.002
Rochelle PA, Upton SJ, Montelone BA, Woods K (2005) The response of Cryptosporidium parvum to UV light. Trends Parasitol 21(2):81–87. doi:10.1016/j.pt.2004.11.009
Rotte C, Stejskal F, Zhu G, Keithly JS, Martin W (2001) Pyruvate : NADP+ oxidoreductase from the mitochondrion of Euglena gracilis and from the apicomplexan Cryptosporidium parvum: a biochemical relic linking pyruvate metabolism in mitochondriate and amitochondriate protists. Mol Biol Evol 18(5):710–720
Schmatz DM (1989) The mannitol cycle–a new metabolic pathway in the Coccidia. Parasitol Today 5(7):205–208
Senkovich O, Speed H, Grigorian A, Bradley K, Ramarao CS, Lane B et al (2005) Crystallization of three key glycolytic enzymes of the opportunistic pathogen Cryptosporidium parvum. Biochim Biophys Acta 1750(2):166–172. doi:10.1016/j.bbapap.2005.04.009
Senkovich O, Schormann N, Chattopadhyay D (2009) Structures of dihydrofolate reductase-thymidylate synthase of Trypanosoma cruzi in the folate-free state and in complex with two antifolate drugs, trimetrexate and methotrexate. Acta Crystallogr D Biol Crystallogr 65(Pt 7):704–716. doi:10.1107/S090744490901230X
Sharling L, Liu X, Gollapalli DR, Maurya SK, Hedstrom L, Striepen B (2010) A screening pipeline for antiparasitic agents targeting Cryptosporidium inosine monophosphate dehydrogenase. PLoS Negl Trop Dis 4(8):e794. doi:10.1371/journal.pntd.0000794
Slapeta J, Keithly JS (2004) Cryptosporidium parvum mitochondrial-type HSP70 targets homologous and heterologous mitochondria. Eukaryot Cell 3(2):483–494
Striepen B, Kissinger JC (2004) Genomics meets transgenics in search of the elusive Cryptosporidium drug target. Trends Parasitol 20(8):355–358. doi:10.1016/j.pt.2004.06.003
Striepen B, White MW, Li C, Guerini MN, Malik SB, Logsdon JM Jr et al (2002) Genetic complementation in apicomplexan parasites. Proc Natl Acad Sci U S A 99(9):6304–6309. doi:10.1073/pnas.092525699
Sun XE, Sharling L, Muthalagi M, Mudeppa DG, Pankiewicz KW, Felczak K et al (2010) Prodrug activation by Cryptosporidium thymidine kinase. J Biol Chem 285(21):15916–15922. doi:10.1074/jbc.M110.101543
Templeton TJ, Enomoto S, Chen WJ, Huang CG, Lancto CA, Abrahamsen MS et al (2010) A genome-sequence survey for Ascogregarina taiwanensis supports evolutionary affiliation but metabolic diversity between a Gregarine and Cryptosporidium. Mol Biol Evol 27(2):235–248. doi:10.1093/molbev/msp226
Umejiego NN, Li C, Riera T, Hedstrom L, Striepen B (2004) Cryptosporidium parvum IMP dehydrogenase: identification of functional, structural, and dynamic properties that can be exploited for drug design. J Biol Chem 279(39):40320–40327. doi:10.1074/jbc.M407121200
Umejiego NN, Gollapalli D, Sharling L, Volftsun A, Lu J, Benjamin NN et al (2008) Targeting a prokaryotic protein in a eukaryotic pathogen: identification of lead compounds against cryptosporidiosis. Chem Biol 15(1):70–77. doi:10.1016/j.chembiol.2007.12.010
Vasquez JR, Gooze L, Kim K, Gut J, Petersen C, Nelson RG (1996) Potential antifolate resistance determinants and genotypic variation in the bifunctional dihydrofolate reductase-thymidylate synthase gene from human and bovine isolates of Cryptosporidium parvum. Mol Biochem Parasitol 79(2):153–165
Wanyiri J, Ward H (2006) Molecular basis of Cryptosporidium-host cell interactions: recent advances and future prospects. Future Microbiol 1(2):201–208. doi:10.2217/17460913.1.2.201
Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG et al (2004) The genome of Cryptosporidium hominis. Nature 431(7012):1107–1112. doi:10.1038/nature02977
Yarlett N, Yarlett NC, Lloyd D (1986) Ferredoxin-dependent reduction of nitroimidazole derivatives in drug-resistant and susceptible strains of Trichomonas vaginalis. Biochem Pharmacol 35(10):1703–1708
Yoon S, Park WY, Yu JR (2012) Recombinant thioredoxin peroxidase from Cryptosporidium parvum has more powerful antioxidant activity than that from Cryptosporidium muris. Exp Parasitol 131(3):333–338. doi:10.1016/j.exppara.2012.04.018
Yu Y, Zhang H, Zhu G (2010) Plant-type trehalose synthetic pathway in Cryptosporidium and some other apicomplexans. PLoS One 5(9):e12593. doi:10.1371/journal.pone.0012593
Zapata F, Perkins ME, Riojas YA, Wu TW, Le Blancq SM (2002) The Cryptosporidium parvum ABC protein family. Mol Biochem Parasitol 120(1):157–161
Zeng B, Zhu G (2006) Two distinct oxysterol binding protein-related proteins in the parasitic protist Cryptosporidium parvum (Apicomplexa). Biochem Biophys Res Commun 346(2):591–599. doi:10.1016/j.bbrc.2006.05.165
Zeng B, Cai X, Zhu G (2006) Functional characterization of a fatty acyl-CoA-binding protein (ACBP) from the apicomplexan Cryptosporidium parvum. Microbiology 152(Pt 8):2355–2363. doi:10.1099/mic.0.28944-0
Zhang H, Guo F, Zhou H, Zhu G (2012) Transcriptome analysis reveals unique metabolic features in the Cryptosporidium parvum oocysts associated with environmental survival and stresses. BMC Genomics 13:647. doi:10.1186/1471-2164-13-647
Zhu G (2004) Current progress in the fatty acid metabolism in Cryptosporidium parvum. J Eukaryot Microbiol 51(4):381–388
Zhu G (2008) Biochemistry. In: Fayer R, Xiao L (eds) Cryptosporidium and cryptosporidiosis, 2nd edn. CRC Press, Boca Raton, pp 57–77
Zhu G, Keithly JS (1997) Molecular analysis of a P-type ATPase from Cryptosporidium parvum. Mol Biochem Parasitol 90(1):307–316
Zhu G, Keithly JS (2002) Alpha-proteobacterial relationship of apicomplexan lactate and malate dehydrogenases. J Eukaryot Microbiol 49(3):255–261
Zhu G, Marchewka MJ, Keithly JS (1999) Cryptosporidium parvum possesses a short-type replication protein A large subunit that differs from its host. FEMS Microbiol Lett 176(2):367–372
Zhu G, Keithly JS, Philippe H (2000a) What is the phylogenetic position of Cryptosporidium? Int J Syst Evol Microbiol 50(Pt 4):1673–1681
Zhu G, Marchewka MJ, Keithly JS (2000b) Cryptosporidium parvum appears to lack a plastid genome. Microbiology 146(Pt 2):315–321
Zhu G, Marchewka MJ, Woods KM, Upton SJ, Keithly JS (2000c) Molecular analysis of a Type I fatty acid synthase in Cryptosporidium parvum. Mol Biochem Parasitol 105(2):253–260
Zhu G, LaGier MJ, Stejskal F, Millership JJ, Cai X, Keithly JS (2002) Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase. Gene 298(1):79–89
Zhu G, Li Y, Cai X, Millership JJ, Marchewka MJ, Keithly JS (2004) Expression and functional characterization of a giant Type I fatty acid synthase (CpFAS1) gene from Cryptosporidium parvum. Mol Biochem Parasitol 134(1):127–135
Zhu G, Shi X, Cai X (2010) The reductase domain in a Type I fatty acid synthase from the apicomplexan Cryptosporidium parvum: restricted substrate preference towards very long chain fatty acyl thioesters. BMC Biochem 11:46. doi:10.1186/1471-2091-11-46
Acknowledgements
We thank Dr. J. M. Fritzler at Weber State University and Dr. S. D. Rider at Wright State University for their critical reading of the manuscript. Studies derived from the author’s laboratory have been mainly supported by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag Wien
About this chapter
Cite this chapter
Zhu, G., Guo, F. (2014). Cryptosporidium Metabolism. In: Cacciò, S., Widmer, G. (eds) Cryptosporidium: parasite and disease. Springer, Vienna. https://doi.org/10.1007/978-3-7091-1562-6_8
Download citation
DOI: https://doi.org/10.1007/978-3-7091-1562-6_8
Published:
Publisher Name: Springer, Vienna
Print ISBN: 978-3-7091-1561-9
Online ISBN: 978-3-7091-1562-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)