Abstract
The phylum Dictyoglomi consists a single genus, Dictyoglomus, with two type strains and several related pure culture isolates. All isolates are thermophilic anaerobic Gram-type negative rods. A major distinguishing phenotypic feature is the formation of spherical bodies in late stationary phase of growth, the function of which is not understood. Most isolates are fermentative using a range of simple carbohydrates, but some isolates are able to grow on crystalline cellulose and chemolithotrophy using carbon monoxide as energy source has been reported for one pure culture. There have been relatively few applications for Dictyoglomus enzymes as a result of a number of factors. Although many of their kinetic properties are exceptional, they have had to be cloned and expressed in standard fermentation strains as hosts, and their low G:C content has required significant genetic manipulation to provide expression. Some of the main applications have required inexpensive enzymes in bulk (e.g., pulp bleaching in paper manufacture), and they have had to be regarded as a replacement for well-established enzymes currently used in the industry. The major applications have involved glycosyl hydrolases, but new uses in value-added products involving drug precursor transformations have been reported recently.
Access provided by Autonomous University of Puebla. Download reference work entry PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Taxonomy: Historical and Current
The phylum Dictyoglomi consists of a single genus Dictyoglomus. The genus consists of two type species (Dictyoglomus thermophilum and Dictyoglomus turgidum synonym turgidus) and several related pure culture isolates. These are:
Dictyoglomus thermophilum type species H-6-12 | Saiki et al. (1985) |
Dictyoglomus thermophilum strain Rt46.B1 | Patel et al. (1987) |
Dictyoglomus strain 1521-1 | Kublanov et al. (2009) |
Dictyoglomus strain 1507-9 | Kublanov et al. (2009) |
Dictyoglomus turgidum type species Z-1310 | Svetlichnii and Svetlichnaya (1988) |
Dictyoglomus strain 1512 | Kochetkova et al. (2011) |
Dictyoglomus strain B1 | Mathrani and Ahring (1991) |
Dictyoglomus strain B4 | Mathrani and Ahring (1992) |
The exact phylogenetic position has been uncertain since different single-gene phylogenies have produced conflicting results, assigning the type species to the phyla Thermotogae (Love et al. 1993), Synergistetes (Rees et al. 1997), Firmicutes (Takai et al. 1999), or Chloroflexi (Wagner and Wiegel 2008). A more recent whole-genome comparison indicated a close phylogenetic relationship with the phylum Thermotogae (Nishida et al. 2011) and that together with Coprothermobacter they form a monophyletic group, which diverged from a common ancestor at an early stage of evolution. This conclusion was supported by a gene content comparison-based analysis and sequence comparison of 44 orthologous proteins. However, a 16S rRNA sequence comparison was unable to clearly differentiate the phylogenetic relationships at the early stage of bacterial evolution due to low bootstrap support and could not clearly differentiate relationships between the phyla Dictyoglomi, Firmicutes, Synergistetes, and Thermotogae.
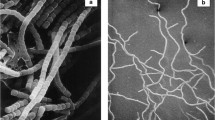
A clear distinguishing phenotypic feature of all species and isolates of Dictyoglomus is the formation of rotund or spherical bodies as the culture enters the late logarithmic phase of growth. Spherical body formation is initiated by aggregation of small numbers of cells, which then arrange themselves over the surface of a rotund body, or sphere, enclosed by a common outer cell membrane. Spherical bodies are quite large structures of between 30 and 100 μm in diameter and may contain up to 100 cells in a single layer around the circumference. As the culture ages, cells in the spherical bodies become disorganised and lyse. The body appears as a large sphere enclosing cell remnants. No function has been attributed to the formation of spherical bodies, and they are presumed to be an artifact of laboratory culture. Morphologically, they are similar to the rotund bodies described for cultures of Thermus aquaticus (Brock and Edwards 1970), but the quite different growth requirements for these organisms mean they are easily differentiated.
Molecular Analyses
The genome of both type strains has been sequenced and shows similarity in genome size and gene content. The genome of the type strain D. thermophilum H-6-12 (ATCC 35947) contains 1,959,987 bases arranged in a single circular chromosome of which 93.66 % are gene coding, encompassing 1,964 genes. No plasmids, phage, or virions are present, and the genome has a G+C mol% of 33.74. Two 16S rRNA genes are present with identical nucleotide sequence. The genome of the type strain D. turgidum (DSM 6724) contains 1,855,560 bases arranged in a single chromosome of which 93.14 % are gene coding, encompassing 1,865 genes. No plasmids, phage, or virions are present, and the genome has a G+C mol% of 33.96. Two 16S rRNA genes are present with identical nucleotide sequence. Sequence homology between the 16S rRNA genes of the two type strains is 99 %. A neighbor joining phylogenetic tree for the Dictyoglomi is shown in Fig. 48.1 . The close affiliation with Thermotoga, which is indicated in phylogenetic analysis, is also evident in the incidence of putatively horizontally transferred genes in the Dictyoglomi genomes, which is a feature of the Thermotoga MSB8 genome. D. thermophilum and D. turgidum contain 9.98 % and 7.4 % of putatively horizontally transferred genes in their genomes, respectively. Species of Aquifex, Caldicellulosiruptor, and Thermotoga (thermophiles habiting similar environments) appear to be the best matched sources for the majority of these genes. Gene content by KEGG and COG categories is remarkably similar in the genomes of the two type strains, and as expected there is a high degree of synteny between the chromosomes. D. thermophilum shows enrichment in genes under the KEGG categories for glycan biosynthesis and metabolism and membrane transport.
Phylogenetic reconstruction of the family Dictyoglomaceae based on 16S rRNA and created using the neighbor-joining algorithm with the Jukes-Cantor correction. The sequence dataset and alignment were used according to the All-Species Living Tree Project (LTP) database (Yarza et al. 2010; http://www.arb-silva.de/projects/living-tree). The tree topology was stabilized with the use of a representative set of nearly 750 high quality type strain sequences proportionally distributed among the different bacterial and archaeal phyla. In addition, a 40 % maximum frequency filter was applied in order to remove hypervariable positions and potentially misplaced bases from the alignment. Scale bar indicates estimated sequence divergence
Phenotypic Analyses
All species and isolates of Dictyoglomus are obligately anaerobic non-sporing thermophiles fermenting a range of mono-, di-, and polysaccharides. Dictyoglomus isolate B1 is unusual in having a restricted substrate range with growth only recorded on beech wood or oat spelt xylan and no growth on soluble sugars, other polysaccharides, peptone, or yeast extract (Mathrani and Ahring 1991). Depending on the isolate, growth occurs over the temperature range 40–82 °C with optimum temperatures reported between 70 °C and 78 °C depending on the strain and culture conditions. Reported pH ranges for growth are from pH 4.5 to pH 9.0 with pH optimum for growth at pH 7.0–7.2. Doubling times vary with the type of culture method: in anaerobic tubes doubling times range from 4.3 h (D. thermophilum) to 10 h (isolate Rt46.B1), while in a minijar fermentor, doubling times of 2.5 h were obtained with D. thermophilum. On agar plates incubated at 60 °C, round nonpigmented colonies of between 2 and 4 mm diameter are produced after 3–5 days’ incubation. Cells are long, filamentous, nonmotile rods, typically 0.3 μm wide by 5-20 μm long. They occur singly, in pairs and in filaments, and increasingly as the culture ages, in bundles and spherical bodies. In late log phase of growth, cells aggregate in linear arrays with several cells sharing an outer wall membrane; these bundles continue cell division with cells enveloped in the common outer wall membrane until a spherical body of up to 100 μm diameter is formed containing up to 50 or more cells. Cells have a typical three-layered Gram-negative appearance under electron microscopy and stain Gram-type negative.
Carbohydrate fermentation results in production of acetate as the major end product together with lesser amounts of lactate. D. thermophilum produced 16 mM acetate, 3 mM lactate, 12 mM CO2, and a trace of H2 in anaerobic tube fermentation of soluble starch, whereas Dictyoglomus strain B.1 produced 55 mM acetate and only traces of CO2 and H2 when grown on xylan. Fermentation is by the Embden-Meyerhof pathway, but the key phosphofructokinase (PFK) enzyme in both type strains is pyrophosphate dependent, rather than more usual ATP-dependent enzyme (Ding et al. 1999). The PPi-PFK is a non-allosteric enzyme which makes the reaction more reversible, and glycolytic flow is dependent on metabolite concentration. Interestingly, the sequence for the pfp gene of D. thermophilum shows a closer relationship to the pfp gene of the archaeum Thermoproteus tenax (also a PPi-PFK enzyme) than that of Thermotoga maritima, which is an ATP-dependent PFK (Ding et al. 2000). Fermentable substrates cover a range of mono-,di-, and polysaccharides including starch and xylan (Table 48.1 ). Fermentable carbohydrates are an obligate requirement for growth; in their absence either no growth (D. thermophilum) or poor growth (isolate Rt46.B1) is recorded. Dictyoglomi have been associated with environments containing cellulosic materials, and this might reflect their capacity to ferment cellobiose mannose, pectin, and xylose. Genes for xylan degradation, in particular, have been cloned and overexpressed and the enzymes investigated for applicability to pulp treatment (see “Applications of Dictyoglomus enzymes”).
A significant difference in metabolism between the two type strains is their ability to utilize cellulose as a substrate. D. turgidum shows good growth on crystalline cellulose, while D. thermophilum and isolates Rt46.B1and B1 are unable to grow on any cellulosic substrates, even though the annotated genome sequence indicates putative genes normally required for its utilization (including B-glucosidase, endoglucanases, and several glycosyl hydrolases). In an extensive survey of enrichment isolates from hot springs in the Uzon caldera, Kublanov et al. (2009) reported the isolation of two pure cultures of Dictyoglomi (strains 1521-1 and 1507-9) whose 16S rRNA sequences most closely matched that of D. thermophilum strain Rt 46 B1 yet were able to hydrolyze microcrystalline cellulose and agarose, respectively. D. turgidum, which was also isolated from Uzon caldera (Svetlichnii and Svetlichnaya 1988), is able to grow on crystalline cellulose as the major fermentable carbon source, but surprisingly, in contrast to the genomes of most cellulolytic bacteria, the genome of D. turgidum contained a relatively small number of annotated cellulose genes. Brumm et al. (2011a) have cloned 12 novel glycosyl hydrolase genes and on the basis of their study suggested that D. turgidum does not appear to degrade cellulose using either conventional soluble enzymes or a cellulosomal degradation system. For example, a celA gene coded for an endoglucanase which had a broad substrate range, possessing both endo and exo activity on soluble and insoluble β-(1,4)-linked glucose-containing substrates as well as endo activity on soluble and insoluble β-(1,4)-linked mannose-containing substrates. The 312-amino acid protein showed low homology to proteins outside the genus Dictyoglomus and lacked a signal peptide (Brumm et al. 2011b). The types and quantities of glycosyl hydrolases and carbohydrate-binding modules present in the genome suggest that D. turgidum degraded cellulose via a mechanism similar to that used by Cytophaga hutchinsonii and Fibrobacter succinogenes. The novelty of the cellulose digestion system in D. turgidum has attracted the interest of commercial application. The original description of D. turgidum indicated an ability to degrade lignocellulose substrates and humic acid, but this has not been independently verified (Svetlichny and Svetlichnaya 1988), and no genes annotated from the genome have lignin-degrading or lignin-modifying properties.
Recently, Kochetkova et al. (2011) described an isolate from a hot spring in Kamchatka (Dictyoglomus strain 1512) that exhibited hydrogenogenic carbon monoxide oxidation. The isolate grew on CO producing CO2 and H2 but had a requirement for yeast extract and pyruvate in the medium. Thus, the physiological range of the genus Dictyoglomus has been extended to include carboxydotrophic metabolism. This strain grew optimally under CO concentrations of 5 % at 75 °C and neutral pH; its 16S rRNA gene sequence showed 100 % homology to that of D. turgidum, but no further information is available on possible heterotrophic properties of the isolate.
Ecology, Isolation, Enrichment, and Maintenance Procedures
All isolations of Dictyoglomus have derived from hot pools or man-made hot environments. Dictyoglomus is widely distributed geographically, with isolates obtained from Japan (Tsuetate Hot Spring, Kumamoto), New Zealand (Kuirau Park, Rotorua), Russia (Oranzhevoe Pool, Uzon Caldera), and Iceland (pools in the Hveragerdi region), while 16S rDNA sequences from clone libraries with high homology to D. thermophilus have been reported from springs in Yellowstone National Park, USA; Kunming, China; and Taiwan. Together these indicate geographic associations; thus, the sequences from Yellowstone National Park cluster together and can be differentiated from the isolates from Uzon and Japan (Fig. 48.1 ). Interestingly, isolates from New Zealand have higher homology to the Japanese-type strain than other groups. However, within these geographic regions the organism is not universally distributed, and its occurrence does not reflect its optimum growth requirements in culture. As an example, Patel et al. (1987) recorded only three positive enrichments from 370 geothermal spring samples mainly from New Zealand, and two of these derived from the same geothermal area; other thermal areas in New Zealand have shown no enrichments for Dictyoglomus despite repeated attempts.
Surprisingly, those environments that have yielded enrichment are often outside the pH and temperature range for growth that have been established for the isolates in culture (Table 48.1 ). Isolation has involved enrichment culture under anaerobic conditions under a N2 atmosphere with an inoculum of either hot spring sediment and water or material from a hot tank from a papermaking plant and incubation at temperatures between 65 °C and 80 °C for periods of up to 1 week. A variety of different media have been used for the primary enrichment, but Dictyoglomus medium (DSMZ 381) supports the growth of all isolates as long as care is taken to supply the required major substrate; for Dictyoglomus strain B1 beech wood xylan is the only substrate to support growth. Following a primary enrichment, isolation proceeds by conventional endpoint dilution culture, or growth on medium solidified with agar and incubation for a longer period at lower temperatures until well-isolated colonies can be picked. For the isolation of D. turgidum, the incorporation of vancomycin (100 μg/mL) in medium dilution tubes was used as a selective agent. Enrichment and isolation can be aided by microscopic observation to ensure cells of the correct morphology are present and increase in numbers. Initially, long or filamentous cells of 0.3 μm diameter by 10–20 μm length are apparent, which in late stationary phase develop the characteristic multicellular aggregates that develop into rotund bodies.
Applications of Dictyoglomus Enzymes
There have been relatively few applications for Dictyoglomus enzymes as a result of a number of factors. Although many of their kinetic properties are exceptional, they have had to be cloned and expressed in standard fermentation strains as hosts, and their low G:C content has required significant genetic engineering skills to provide suitable recombinants for expression. Some of the main applications have required inexpensive enzymes in bulk (e.g., pulp bleaching in paper manufacture), and they have had to be regarded as a replacement for well-established but mesophilic enzymes currently used in the industry. The major applications have involved glucosyl hydrolases, but new uses in value-added products involving drug precursor transformations have been reported recently.
Pulp Bleaching
Xylanases have a number of industrial uses, particularly in baking, in modification of animal feed, and in pulp bleaching in the paper industry. In the last 10–12 years, there has been significant interest in xylanases with alkaline pH optima and stability under alkaline conditions. This interest primarily has been due to the proven utility of xylanases in kraft pulp bleaching where the pulp comes out of the kraft cook at high temperature and at an alkaline pH (Viikari et al. 1994). Several research groups have prospected for novel organisms producing xylanases in natural and man-made alkaline environments (e.g., Nakamura et al. 1993; Yang et al. 1995; Gessesse and Gashe 1997). A number of groups have attempted directed evolution of xylanases to increase the pH optimum of known xylanases (e.g., Chen et al. 2001; Shibuya et al. 2005; Gibbs et al. 2010a). One objective of these studies has been to better match the enzyme to industrial conditions encountered in pulp bleaching and to reduce reliance on chemicals in the bleaching process.
Mathrani et al. (1992) filed a patent describing Dictyoglomus B1 and its xylanase(s). They could not claim priority for the organism because of a prior publication by Patel et al. (1987). Their claims regarding pH range and temperature range were attributed to the presence of more than one enzyme but may have resulted from substrate limitation of the enzyme reactions shown in their examples under the conditions given.
The Dictyoglomus strain B1 xylanase was produced under anaerobic conditions in a continuous fermentor using the individual parameters determined from batch culture trials, and 73 °C, pH 8, and a dilution rate of 0.112 h−1 gave optimal xylanase activity in supernatant (Adamson et al. 1995). This strain had low cellulase activity and was isolated initially from a pulp and paper mill. The enzyme preparation isolated from the culture supernatant was probably a mixture of family 10 and 11 enzymes as deduced from the amount of free xylose determined by HPLC after pulp treatment. The preparation gave a brightness increase of two ISO units using peroxide-bleached pine pulp (Rāttö et al. 1994). Gibbs et al. (1995) cloned the xynA gene from a λ Zap library of D. thermophilum Rt46.B1 and expressed it in E. coli using an overexpression plasmid vector. The apparent optimal temperature at pH 6.5 was 85 °C, at the time the most thermophilic xylanase reported and had half-life at that temperature of 24 h. The partially purified enzyme was able to release color from unbleached Pinus radiata pulp and broke down soluble xylan into xylobiose and xylotriose. An account of the cloning and expression of xynA and xynB genes from D. thermophilum Rt46.B1 was published by Morris et al. (1996) who demonstrated that both XynA and XynB released reducing sugars from Pinus radiata kraft pulp. It was clear that another xylanase gene was present on genome from the analysis of a λZap library of D. thermophilum Rt46.B1 genomic DNA. This gene coded for XynB, a family 11 xylanase that was found to contain a catalytic domain and a xylan-binding domain. A more thermostable derivative, XynB6 was constructed and overexpressed in E. coli, and the recombinant enzyme was examined for its ability to bleach Eucalyptus kraft-oxygen-treated pulp in elemental and total chlorine-free procedures, proving superior to XynA and two commercial xylanase preparations in trials (Morris et al. 1998; Bergquist et al. 1998). At the lowest chemical charge, pulp treated with the xylanase had a brightness 6–8 % ISO greater than the brightness of the untreated reference pulp (reviewed in Bergquist et al. 2003, 2004).
It became apparent that it was not economically feasible to prepare the enzyme in E. coli and considerable effort was expended on optimizing XynB expression in the high-secreting fungus Trichoderma reesei RUTC-30. The native gene was not expressed in the fungus until the codon sage was modified to resemble that of Trichoderma (T’eo et al. 2000). Early experiments improving the yeilds of D. thermophilum XynB and construction of the shuttle vector producing sufficient plasmid DNA in E. coli for transformation into Trichoderma where the DNA is integrated into a chromosome were reviewed by Bergquist et al. (2002). Accordingly, the vector was larger than typical bacterial vectors and had provision for Trichoderma regulatory and termination sites. The xynB gene was fused to the catalytic core of the Trichoderma cbh1 gene. Several promoters were trialled for increased expression in Trichoderma to allow expression and secretion of the enzyme into the culture supernatant (Bergquist et al. 2002, 2003). Around the same time, Walsh and Bergquist (1997) expressed the xynA gene in Kluyveromyces lactis. This yeast secretes few homologous proteins and does not hyperglycosylate heterologous enzymes. A high-copy-number plasmid vector was modified to include the signal sequence of the K. lactis killer toxin and a strong, inducible promoter. The xynA gene was ligated as an in-frame fusion into the vector, and it was transformed into K. lactis strain CBS1065. The yield obtained of approximately 90 % pure XynA was greater than that produced in E. coli and showed activity in pulp bleaching but was expressed at a lower level than commercially viable.
A number of papers have reported on the activity of various xylanases at alkaline pH. Process conditions in kraft pulp bleaching generally favor an enzyme that is active at high pH values. The activities of several glycosyl hydrolase family 11 xylanases reported to be active under alkaline conditions were determined under optimal conditions and found to have optima in the pH 5–6 range (Gibbs et al. 2010b). Only one enzyme tested, BadX from Bacillus agaradhaerens, was shown to have an alkaline pH optimum. Significant activity at pH values higher than 8 appears often to be the result of excess enzyme added to the reaction mixtures so that substrate is limiting. The different nature of laboratory and industrial substrates needs to be taken into consideration in designing assay conditions. In some cases, significant differences were observed in pH profiles generated using a small molecule substrate when compared to those generated using xylan (Poon et al. 2003; Gibbs et al. 2010b). Nearly all microbial xylanases characterized exhibit acidic or neutral pH optima (Gibbs et al. 2010b). The gene for the enzyme XynB from Dictyoglomus thermophilum Rt46.B.1 was cloned, and the recombinant protein product showed a superior performance to a family 10 xylanase from the same organism in the bleaching of kraft pulp (Bergquist et al. 2003). The enzyme was anticipated to have greater commercial potential if its performance under alkaline conditions could be improved. A gene shuffling procedure was developed in an attempt to use directed evolution to achieve a better performance at alkaline pH [Degenerate Oligonucleotide Gene Shuffling, DOGS: Gibbs et al. 2001; Bergquist et al. 2005]. An extensive examination of a library generated by family shuffling did not allow detection of any progeny with improved pH characteristics. This result was explained later when it was shown that the measurements of alkaline pH activity of the other enzymes in the shuffling were artifactual (Gibbs et al. 2010b). Gene shuffling experiments between D. thermophilum xynB and B. agaradhaerens badX genes demonstrated that alkaline activity was determined by a number of changed amino acids distributed throughout the shuffled sequence and was not the result of a single mutated site.
Most of the applications of Dictyoglomus enzymes that have been suggested require the proteins to be produced cheaply as bulk chemical products. Transfer of the genes into the usual “cell factory” strains normally used for bulk products has turned out to be difficult for the expression of genes from other sources (heterologous proteins). This area has been reviewed by Nevalainen et al. (2005) who provide a commentary on the difficulties in getting expression of heterologous proteins in filamentous fungi using D. thermophilum XynB as a model gene product. Alterations to the vector sequence to provide suitable processing site, tags for the secretion of the protein, and problem upregulation of proteases attendant on the expression of the heterologous gene have been recognized and discussed. Knowledge of fermentation techniques coupled with an understanding of the physiology of the organism is emphasized as a necessary requirement for the enhanced production of heterologous proteins.
Other Applications of Glycosyl Hydrolases
The gene for the β-mannanase from D. thermophilum Rt46.B1, ManA, was isolated from a λ Zap library as for the xylanase genes from this organism. It was cloned into a heat-inducible overexpression vector, and the enzyme was produced in E. coli. The purified enzyme had an apparent temperature optimum of 80 °C and a pH optimum of 5.0 under the conditions used. It was particularly active on locust bean gum, and examination of the products of digestion suggested that ManA only acted on substituted mannans (Reeves et al. 1999). This enzyme may have applications in oil recovery techniques (Comfort et al. 2004).
Nielsen et al. (2007) used cattle manure as a substrate for a two-step anaerobic fermentation at a biogas plant in Denmark. The lignocellulosic fibers are incompletely digested in the manure. Caldicellulosiruptor lactoaceticus and Dictyoglomus strain B4a were tested for their ability to increase the yield of methane. Strain B4a only uses xylan and xylose as a substrate for growth, but it gave lower yields of methane than Caldicellulosiruptor which has enzymes for the hydrolysis of a greater variety of polysaccharides.
Genomics
A thermostable amylase gene was cloned from a HindIII library from D. thermophilum H-6-12 and expressed in E. coli by Fukusumi et al. (1988). No applications were described and the gene (amyA) was not annotated in the full genome sequence of the thermophile. The protein was significantly larger than the other two amylases subsequently cloned from D. thermophilum H-6-12 but did not appear to be a precursor for either AmyB or AmyC. A further paper reports on the cloning of two more amylases from this organism. These two papers describe the cloning and expression of three thermostable amylase genes from Dictyoglomus thermophilum H-6-12 in Escherichia coli. It was suggested that the enzymes may be useful in starch degradation, but no tests were reported apart from a TLC analysis of the saccharides produced, on hydrolysis of starch which differed between the three enzymes. The authors noted the unusually low G:C content of the DNA of the genes encoding these enzymes (Horinouchi et al. 1988). D. thermophilum H-6-12 has been sequenced and only two amylases have been recorded with AmyB classified as belonging to family 57 and AmyC to family 13 (hamap.expasy.org/proteomes/DICT6.html).
Mathrani and Ahring (1991) described the isolation and characterization of a strictly xylan-degrading Dictyoglomus from a man-made, thermophilic anaerobic environment in a pulp mill. Their largely taxonomic description did draw attention to the fact that the G:C content of the genomic DNA was low but was higher than the type species, D. thermophilum with the implication that it was a distinct species. They described thermophilic and alkalophilic xylanases from several Dictyoglomus isolates including an Icelandic culturec B4, type strain D. thermophilum and Dictyoglomus B1 (described earlier). Crude enzymes from culture supernatants were used. The pH assays most probably were substrate limited as they showed very broad peaks and they were all reported as % of maximal activity. The temperature assays similarly appeared to be substrate limited. They claimed to be the first to report on alkalophilic, thermophilic xylanases but the results may have been due to substrate limitation (Gibbs et al. 2010b).
McCarthy et al. (2000) reported on the crystal structure of the β-1,4-xylanase from Dictyoglomus thermophilum Rt46B.1 The structure was solved at 1.8 Å resolution and showed that the enzyme had a single-domain fold characteristic of family 11 xylanases composed of a jelly roll of two twisted β-sheets that create a deep substrate-binding cleft containing the two catalytic residues Glu90 and Glu180. It is one of the most thermostable xylanases for which the structure has been solved and has a slightly extended C-terminus in comparison to mesophilic equivalents. This structure allowed the modelling of mutants created by directed evolution. An X-ray structure of a xylanase from a Streptomyces species with similarities to XynB was reported by Wouters et al. (2001) that was placed in the same subclass as Rt46.B1 XynB since it was claimed to be active in alkaline conditions. They noted that the three cysteines in Rt46.B.1 do not form disulfide bridges and that XynB has a longer C-terminus that may contribute to its thermostability. Detailed methods for the isolation, purification, and assay of XynA (family 10) and XynB (family 11) from D. thermophilum Rt46.B1 were published by Bergquist et al. (2001).
A GH3 β-glucosidase was isolated apparently by direct cloning from the D. turgidum sequenced genome. The enzyme produced in E. coli was characterized and found to be a dimer of 200 mDa molecular mass. It was able to digest spent coffee grounds isoflavones, particularly converting diazdin to diazdein. It was suggested that the enzyme may be useful for adding value to a product largely used in compost since isoflavone aglycones have established therapeutical applications and are pharmaceutically superior to isoflavone glycosides (Kim et al. 2011).
d-Lyxose is a pharmaceutical precursor for antitumor and immunostimulatory agents and can be produced from d-xylulose by d-lyxose isomerase. The gene was isolated from D. turgidum by similar methods employed by the authors for the β-glucosidase from the same organism (Choi et al. 2012). The enzyme had a greater apparent optimal temperature, thermostability, and productivity compared to the mesophilic homologs that have been evaluated and may be useful in biochemical transformations involved with therapeutic agent production.
One of the few studies of Dictyoglomus enzymes that did not focus on glucosyl hydrolases or the pyrophosphate-dependent phosphofructokinase was the report on a DNA polymerase from D. thermophilum Rt46.B1 by Shandilya et al. (2004). The gene for DNA polymerase was isolated by a genomic walking technique using consensus primers as part of a study of Taq-like polymerases with reverse transcriptase activity. The polymerase was very thermostable but showed little RT activity. However, it had as high an apparent optimal temperature as Thermus DNA polymerases and good thermostability. It was only tested comprehensively for RT-PCR ability and showed little activity.
The use of a yeast intron as a translational terminator in a plasmid shuttle vector was described by Gibbs et al. (2004). The Kluyveromyces PLAC promoter has several transcriptional sites that are recognized in E. coli, and leakage could reduce or prevent large quantities of plasmid DNA being produced for subsequent transfer into yeast as the promoter and related sequences are recognized in the bacterium and allow expression of the heterologous gene. They inserted a yeast intron into the vector that cannot be processed in E. coli and reduced transcription of the heterologous gene, in this case, Dictyoglomus xynB to a low level. The vector has potential for allowing lethal but non-transcribed genes in E. coli to be shut down so sufficient plasmid DNA can be produced for transformation into a suitable eukaryotic host.
Glycosyl hydrolase genes from D. turgidum were isolated by Brumm et al. (2011a, b) after library production in a plasmid vector and by direct cloning from genomic DNA based on whole sequence annotation. They identified 12 novel glycosyl hydrolases by shotgun and direct PCR-amplification techniques. This microorganism has had its genomic DNA completely sequenced and annotated, allowing correlation between in silico and isolation results. The most outstanding feature was the paucity of cellulase genes, and alternative pathway for cellulose digestion for this strain was suggested. Two of the enzymes have been commercialized for research purposes (Lucigen Corpn., Middleton, WI, USA).
Applications in General Biochemistry
Zhang et al. (2009) examined the influence of temperature on the fluorescence intensity and enzyme activity of the fusion protein of GFP and D. thermophilum Rt46.B1 xylanase B. They fused GFP to the N-terminus with a linker to maintain flexibility between the two proteins to assist folding and found that the refolding properties of the gfp portion of the fusion were more temperature dependent than that of the XynB portion. They suggested that gfp fusion proteins could aid in the understanding of the folding properties of proteins.
Zhang et al. (2010) reported on the expression and characterization of the Dictyoglomus thermophilum Rt46B.1 xylanase xynB gene in Bacillus subtilis. Their results were essentially a confirmation of the work of Morris et al. (1998), but the enzyme was expressed in a different bacterial host. They mistakenly attributed the origin of the strain to the deep Atlantic Trench, but it was isolated from a geothermal pool in the Rotorua region of New Zealand. The enzyme was secreted into the supernatant by the host, and there were statements about its possible use in pulp bleaching but no demonstration of its applicability. The equivalent enzyme expressed in E. coli had been shown to give a good bleaching performance by Morris et al. (1998).
In an attempt to reduce pretreatment of lignocellulose, D. thermophilum Rt46.B1 xynA and xynB genes were synthesized with codon optimization for plants and were expressed constitutively from the CaMV 35S promoter for transient expression in tobacco and stable expression in Arabidopsis. The biochemical characteristics of both enzymes were retained, and active enzyme could be assayed from dried stem tissue (Borkhadt et al. 2010). It was proposed by the authors that as the temperature of plant growth was well below the apparent temperature optima of the enzymes, the enzymes could be activated after harvest and would contribute to biomass breakdown under relatively mild conditions.
Sunna et al. (2013) characterized a linker peptide with high affinity towards materials containing silica. A recombinant form of D. thermophilum XynB was fused to the specific C-terminal linker sequence and bound to zeolite, retaining enzyme activity. These experiments indicated that a fusion protein plus matrix could provide a method for enzyme concentration from dilute solutions and a reusable immobilized enzyme system for reuse in industrial processes.
References
Adamson AK, Lindhagen J, Ahring BK (1995) Optimization of extracellular xylanase production by Dictyoglomus sp. B1 in continuous culture. Appl Microbiol Biotechnol 44:327–332
Bergquist PL, Gibbs MD, Saul DJ, Reeves RA, Morris DD, Te’o VSJ (1998) Isolation and expression of genes for hemicellulases from extremely thermophilic culturable and unculturable bacteria. In: Eriksson K-E, Cavaco-Paulo A (eds) Enzyme applications in fiber processing, vol 687, American chemical society symposium series. American Chemical Society, Washington, DC, pp 168–177
Bergquist PL, Gibbs MD, Morris DD, Thompson DR, Uhls AH, Daniel RM (2001) Hyperthermophilic xylanases. Methods Enzymol 330:301–308
Bergquist PL, Te’o VSJ, Gibbs MD, Cziferszky ACE, de Faria FP, Azevedo MO, Nevalainen KMH (2002) Production of enzymes from thermophilic microorganisms in fungal hosts. Extremophiles 6:177–184
Bergquist P, Te’o V, Gibbs M, Curach N, Nevalainen K (2003) Recombinant bleaching enzymes from thermophiles expressed in fungal hosts. In: Mansfield SD, Saddler JN (eds) Applications of enzymes to lignocellulosics, vol 855, American Chemical Society symposium series. American Chemical Society, Washington, DC, pp 435–445
Bergquist PL, Nevalainen KMH, Te’o VSJ, Gibbs MD, Curach NC (2004) Recombinant enzymes from thermophilic microorganisms produced in fungal hosts. Biochem Soc Trans 32:293–297
Bergquist PL, Reeves RA, Gibbs MD (2005) Degenerate oligonucleotide gene shuffling (DOGS) and random drift mutagenesis (RNDM): two complementary techniques for enzyme evolution. Biomol Eng 22:63–72
Borkhardt B, Harholt J, Ulvskov P, Ahring BK, Jorgensen B, Brinch-Petersen H (2010) Autohydrolysis of plant xylans by apoplastic expression of thermophilic bacterial endo-xylanases. Plant Biotechnol J 8:363–374
Brock TD, Edwards MR (1970) Fine structure of Thermus aquaticus, an extreme thermophile. J Bacteriol 104:509–517
Brumm P, Hermanson S, Hochstein B, Boyum J, Hermersmann N, Gowda K, Mead D (2011a) Mining Dictyoglomus turgidum for enzymatically active carbohydrases. Appl Biochem Biotechnol 163:205–214
Brumm PJ, Hermanson S, Luedtke J, Mead DA (2011b) Identification, cloning and characterization of Dictyoglomus turgidum CelA, an endoglucanase with cellulose and mannanase activity. J Life Sci 5:488–496
Chen Y-L, Tang T-Y, Cheng K-J (2001) Directed evolution to produce an alkalophilic variant from a Neocallimastix patriciarum xylanase. Can J Microbiol 47:1088–1094
Choi J-G, Hong S-H, Kim Y-S, Kim K-R, Oh D-K (2012) Characterization of a recombinant thermostable d-lyxose isomerase from Dictyoglomus turgidum that produced d-lyxose from d-xylulose. Biotechnol Lett 34:1079–1085
Comfort DA, Chhabra SR, Connors SB, Chou C-J, Epting KL, Johnson MR, Jones KI, Sehgal AC, Kelly RM (2004) Strategic biocatalysis with hyperthermophilic enzymes. Green Chem 6:459–465
Ding Y-HR, Ronimus RS, Morgan HW (1999) Purification and properties of the pyrophosphate-dependent phosphofructokinase from Dictyoglomus thermophilum Rt46 B.1. Extremophiles 3:131–137
Ding Y-HR, Ronimus RS, Morgan HW (2000) Sequencing, cloning and high level expression of the pfp gene encoding a PPi-dependent phosphofructokinase from the extremely thermophilic eubacterium Dictyoglomus thermophilum. J Bacteriol 182:4661–4666
Fukusumi S, Kamizono A, Horinouchi S, Beppu T (1988) Cloning and nucleotide sequence of a heat-stable amylase gene from an anaerobic thermophile, Dictyoglomus thermophilum. Eur J Biochem 174:15–21
Gessesse A, Gashe BA (1997) Production of an alkaline xylanase by an alkaliphilic Bacillus sp. isolated from an alkaline soda lake. J Appl Microbiol 83:402–406
Gibbs MD, Reeves RA, Bergquist PL (1995) Cloning, sequencing and expression of a xylanase gene from the extreme thermophile Dictyoglomus Rt46B.1 and the activity of the enzyme on fiber-bound substrate. Appl Environ Microbiol 61:4403–4408
Gibbs MD, Nevalainen KHM, Bergquist PL (2001) Degenerate oligonucleotide gene shuffling (DOGS): a method for enhancing the frequency of recombination with family shuffling. Gene 271:13–20
Gibbs MD, Reeves RA, Sunna A, Bergquist PL (2004) A yeast intron as a translational terminator in a plasmid shuttle vector. FEMS Yeast Res 4:573–577
Gibbs MD, Reeves RA, Choudhary PR, Bergquist PL (2010a) Alteration of the pH optimum of a family 11 xylanase, XynB6 of Dictyoglomus thermophilum. New Biotechnol 27:803–806
Gibbs MD, Reeves RA, Choudhary PR, Daniel RM, Bergquist PL (2010b) The activity of family 11 xylanases at alkaline pH. New Biotechnol 27:795–802
Horinouchi S, Fukusumi S, Oshima Y, Beppu T (1988) Cloning and expression in Escherichia coli of two additional amylase genes of a strictly anaerobic thermophile, Dictyoglomus thermophilum, and their nucleotide sequences with extremely low guanine-plus-cytosine contents. Eur J Biochem 176:243–253
Kim Y-S, Yeom S-J, Oh D-K (2011) Characterization of a GH3 family β-glucosidase from Dictyoglomus turgidum and its application to the hydrolysis of isoflavone glycosides in spent coffee grounds. J Agric Food Chem 59:1182–11818
Kochetkova TV, Rusanov II, Pimenov NV, Kolganova TV, Lebedinsky AV, Bonch-Osmolovskya EA, Sokolova TG (2011) Anaerobic transformation of carbon monoxide by microbial communities of Kamchatka hot springs. Extremophiles 15:319–325
Kublanov IV, Perevalova AA, Slobodkina GB, Lebedinsky AV, Bidzhieva SK, Kolganova TV, Kaliberda EN, Rumsh LD, Haertie T, Bonch-Osmolovskya EA (2009) Biodiversity of thermophilic prokaryotes with hydrolytic activities in hot springs of Uzon caldera, Kamchatka (Russia). Appl Environ Microbiol 75:286–291
Love CA, Patel BKC, Ludwigm W, Stackebrandt E (1993) The phylogenetic position of Dictyoglomus thermophilum based on 16S rRNA sequence analysis. FEMS Microbiol Lett 107:317–320
Mathrani IM, Ahring BK (1991) Isolation and characterization of a strictly xylan-degrading Dictyoglomus from a man-made, thermophilic anaerobic environment. Arch Microbiol 157:13–17
Mathrani IM, Ahring BK (1992) Thermophilic and alkalophilic xylanases from several Dictyoglomus isolates. Appl Microbiol Biotechnol 38:23–37
Mathrani IM, Ahring BK, Anker E (1992) Novel microorganism and novel enzymes. Patent WO92/18612
McCarthy AA, Morris DD, Bergquist PL, Baker EN (2000) Crystal structure of a highly thermostable β-1,4-xylanase from Dictyoglomus thermophilum Rt46B.1 at 1.8 Å resolution. Acta Crystallogr D 56:1367–1375
Morris DD, Gibbs MD, Bergquist PL (1996) Cloning of a family G xylanase gene (xynB) from the extremely thermophilic bacterium Dictyoglomus thermophilum and activity of the gene product on kraft pulp. In: Jeffries TJ, Viikari L (eds) Enzymes for pulp and paper processing, vol 655, American Chemical Society symposium series. American Chemical Society, Washington, DC, pp 101–115
Morris DD, Gibbs MD, Chin CW, Koh MH, Wong KKY, Allison RW, Nelson PJ, Bergquist PL (1998) Cloning of the xynB gene from Dictyoglomus thermophilum strain Rt46B.1 and action of the gene-product on kraft pulp. Appl Environ Microbiol 64:1759–1765
Nakamura S, Wakabayashi K, Nakai R, Horikoshi K (1993) Purification and some properties of an alkaline xylanase from alkaliphilic Bacillus sp. strain 41M-1. Appl Environ Microbiol 59:2311–2316
Nevalainen KMH, T’eo VSJ, Bergquist PL (2005) Heterologous protein expression in filamentous fungi. Trends Biotechnol 23:468–474
Nielsen HB, Mladenovska Z, Ahring BK (2007) Bioaugmentation of a two-stage thermophilic (68 °C/55 °C) anaerobic digestion concept for improvement of the methane yield from cattle manure. Biotechnol Bioeng 97:1638–1643
Nishida H, Beppu T, Ueda K (2011) Whole-genome comparison clarifies close phylogenetic relationship between the phyla Dictyoglomi and Thermotogae. Genomics 98:370–375
Patel BKC, Morgan HW, Wiegel J, Daniel RM (1987) Isolation of an extremely thermophilic chemoorganotrophic anaerobe similar to Dictyoglomus thermophilum from New Zealand hot springs. Arch Microbiol 147:21–24
Poon DKY, Webster P, Withers SG, McIntosh LP (2003) Characterizing the pH-dependent stability and catalytic mechanism of the family 11 xylanase from the alkalophilic Bacillus agaradhaerens. Carbohydr Res 338:415–421
Rāttö M, Mathrani IM, Ahring B, Viikari L (1994) Application of thermostable xylanase of Dictyoglomus sp. in enzymatic treatment of kraft pulps. Appl Microbiol Biotechnol 41:130–133
Rees GN, Patel BKC, Grassia GS, Sheehy AJ (1997) Anaerobaculum thermoterrenum gen. nov., sp. nov., a novel thermophilic bacterium which ferments citrate. Int J Syst Bacteriol 47:150–154
Reeves RA, Sunna A, Gibbs MD, Bergquist PL (1999) Sequencing and expression of a β-mannanase from the extreme thermophile Dictyoglomus thermophilum Rt46B.1 and characteristics of the recombinant enzyme. Curr Microbiol 39:351–357
Saiki T, Kobayashi Y, Kawagoe K, Beppu T (1985) Dictyoglomus thermophilum gen. nov., sp. nov., a chemoorganotrophic, anaerobic, thermophilic bacterium. Int J Syst Bacteriol 35:253–259
Shandilya H, Flynn EK, Astatke M, Shih PJ, Gerard GF, Lee JE, Griffiths K, Gibbs MD, Bergquist PL (2004) Thermophilic bacterial DNA polymerases with reverse transcriptase activity. Extremophiles 8:243–251
Shibuya H, Kaneko S, Hayashi K (2005) A single amino acid substitution enhances the catalytic activity of Family 11 xylanase at alkaline pH. Biosci Biotechnol Biochem 69:1492–1497
Sunna A, Chi F, Bergquist P (2013) Characterization of a linker peptide with high affinity towards silica-containing materials. New Biotechnol 30: 485–492
Svetlichny VA, Svetlichnaya TP (1988) Dictyoglomus turgidus sp.nov., a new extremely thermophilic eubacterium isolated from hot springs of the Uzon volcanic caldera. Mikrobiologiya 57:364–369
Takai K, Inoue A, Horikoshi K (1999) Thermaerobacter marianensis gen.nov., sp. nov., an aerobic extremely thermophilic marine bacterium from the 11000 m deep Mariana Trench. Int J Syst Bacteriol 49:619–628
Te’o VSJ, Cziferszky AE, Bergquist PL, Nevalainen KMH (2000) Codon optimisation of the thermophile xylanase gene xynB from Dictyoglomus thermophilum for expression in Trichoderma reesei. FEMS Microbiol Lett 190:13–19
Viikari L, Kantelinen A, Sundquistand J, Linko M (1994) Xylanases in bleaching: from an idea to an industry. FEMS Microbiol Revs 13:335–350
Wagner ID, Wiegel J (2008) Diversity of thermophilic anaerobes. Ann N Y Acad Sci 1125:1–43
Walsh DJ, Bergquist PL (1997) Expression and secretion of a thermostable xylanase in Kluyveromyces lactis. Appl Environ Microbiol 63:3297–3300
Wouters J, Georis J, Engher D, Vandenhaute J, Dusart J, Frere JM, Depiereux E, Charlier P (2001) Crystallographic analysis of family 11 endo-β-1,4-xylanase Xyl1 from Streptomyces strain S38. Acta Crystallogr D57:1813–1819
Yang VW, Zhuang Z, Elegir G, Jeffries TW (1995) Alkaline-active xylanase produced by an alkaliphilic Bacillus sp. isolated from kraft pulp. J Ind Microbiol 15:434–441
Yarza P, Ludwig W, Euzeby, Amann R, Schleifer K-H, Glockner FO, Rossello-Mora R (2010) Update of the All-Species Living Tree project based on 16S and 23S rRNA sequence analyses. Syst Appl Microbiol 33:291–299
Zhang C, Liu MS, Xing XH (2009) Temperature influence on fluorescence intensity and enzyme activity of the fusion protein of GFP and hyperthermophilic xylanase. Appl Microbiol Biotechnol 84:511–517
Zhang W, Lou K, Li G (2010) Expression and characterization of the Dictyoglomus thermophilum Rt46B.1 xylanase gene (xynB) in Bacillus subtilis. Appl Biochem Biotechnol 160:1484–1495
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag Berlin Heidelberg
About this entry
Cite this entry
Bergquist, P.L., Morgan, H.W. (2014). The Phylum Dictyoglomi. In: Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F. (eds) The Prokaryotes. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-38954-2_123
Download citation
DOI: https://doi.org/10.1007/978-3-642-38954-2_123
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-38953-5
Online ISBN: 978-3-642-38954-2
eBook Packages: Biomedical and Life SciencesReference Module Biomedical and Life Sciences