Abstract
In the modern era of next-generation genomics and Fourth Industrial Revolution, there is a growing demand for translational research that brings about not only impactful research but also potential commercialisation of R- and D-based products. Advancement of metabolic engineering and synthetic biology has put forward a viable and innovative biotechnological platform for bioproduct development especially using microbial chassis. In this chapter, readers will be introduced on the concepts of metabolic engineering, synthetic biology and microbial chassis and the applications of these biological engineering (BioE) components in the advancement of industrial and agricultural biotechnology. Main strategies in employing BioE platform are discussed especially for waste bioconversion and value-added product development. More importantly, this chapter will also discuss current endeavours in integrating systems and synthetic biology for microbial production of natural products by introducing flavonoid biosynthesis genes of Polygonum minus, a medicinally important tropical plant in engineered yeast.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Metabolic engineering
- Synthetic biology
- Microbial chassis
- Biological engineering
- Industrial biotechnology
6.1 Introduction
Genetic modification is central to global industrial practices be it in the traditional cross-breeding and mutagenesis means or recombinant DNA technology-inspired targeted genetic improvement. The common goal of these approaches is to improve biochemical reactions and obtain desirable traits or product titre from genetically modified organisms. Employment of genetic technologies is the hallmark of modern biotechnology that enables researchers to make changes at the DNA and protein levels for acquiring knowledge and creating new products and technologies to solve concurrent problems in healthcare, agriculture and environmental sectors. The advent of genomics and high-throughput biology has accelerated the expansion of molecular biology tools and advanced genetic engineering techniques far-reaching than single-disciplinary recombinant protein expression and functional studies of individual gain-of-function and loss-of-function genetic mutants.
As discussed in the previous chapters, complex biological mechanisms elucidated using omics-driven systems biology platforms served as the basis and in-depth information on gene regulation, metabolic pathways and network modelling that enabled further applications and explorations using biotechnological- and engineering-based methodologies. Principally, metabolic engineering and synthetic biology are the expansion of the broad fields of molecular biology and represent the sophisticated versions of genetic engineering research areas that are built on recombinant DNA technology principles. The rapid progress of metabolic engineering, systems biology and synthetic biology is dovetailed with the emergence of revolutionary technologies such as RNA interference (RNAi) and clustered regularly interspaced short palindromic repeats (CRISPR)-based genome editing tools and booming bio-based industries and healthcare sectors in developed and developing countries. In this chapter, a particular emphasis is placed on the implementation of biological engineering (BioE) platform comprising of microbial-based metabolic engineering and synthetic biology approaches, as the enabling technology for integrating data-driven systems biology input essential in developing sustainable and value-added biotechnological applications.
6.1.1 Metabolic Engineering
The metabolic engineering era that started in the 1990s has led to many exciting research discoveries and accelerated an advanced genetic engineering approach that aimed towards investigating broader aspects of metabolic and biochemical interactions . Metabolic engineering is principally defined as the directed modulation of metabolic pathways using recombinant DNA technology for overproducing fuels, chemicals and pharmaceutical products, which generated greater attention and consequently rapid expansion due to its industrial relevance [1, 2]. Metabolic engineering somehow differs from the generic genetic engineering field with its virtue in the multilevel investigation of metabolic pathways and gene regulatory networks as compared to individual studies of genes and enzymes, especially the rate-limiting enzymes.
Fundamental underlying aspects of metabolic engineering involve genetic construction , performance analysis, pathway engineering and optimisation of metabolic pathways for attaining desirable products using techniques ranging from recombinant protein expression to biochemical analysis of flux, kinetics and thermodynamics of the engineered cells or proteins. In essentiality, metabolic engineering focuses on broader impacts of genetic modifications in the engineered cells by investigating stoichiometric balance, pathway regulation and network modelling, in systematic means that in a way precedes systems biology in understanding the biological mechanisms of complex metabolic perturbance at systems level [3]. This multidisciplinary research field has brought about seminal research findings such as the production of biofuels from amino acids [4], synthesis of antimalarial drug precursors [5], production of nonnatural chemicals such as 1,4-butanediol [6] and, more recently, complete biosynthesis of opiods using metabolically engineered microbes [7]. These landmark research have brought about emerging bio-based industries that employ metabolically engineered microbes for the production of biofuels, chemicals and pharmaceuticals with sustainable and green technology business models culminating in the creation of a new market segment for industrial biotechnology.
Metabolic engineers ultimately aim at improving production level while lowering energy burden and associated costs involved in product development and industrial commercialisation by debottlenecking, debugging and process optimisation of the engineered cells [8]. Greater understanding of the microbial genomes and metabolic pathways has accelerated genome-scale pathway engineering and systems metabolic engineering that utilise data-driven approaches including in silico and omics-based pathway prediction and gene selection tools for constructing, modulating and optimising of metabolic pathways and evolution of protein functions [9]. In silico-aided metabolic engineering approaches have sped up strain development for industrial production of amino acids and biochemicals that are attributed by the increased productivity and capacity of the engineered microbes for scale-up fermentation and bioprocessing [10]. The rapid progress of next-generation omics technologies and ever-expanding synthetic biology tools such as CRISPR interference (CRISPRi) shall further improve substrate utilisation and hyper-producing strain development via genome-wide analysis and high-throughput strain screening [11, 12].
6.1.2 Synthetic Biology
Synthetic biology is a rapidly emerging research discipline in the broad fields of molecular biology and has been interchangeably used in reference to metabolic engineering especially involving complicated genetic modification or alteration of living cells. The emergence of synthetic biology has been mostly associated with the lowering cost of high-throughput DNA synthesis and increasing interest in implementing engineering principles for modulating cellular and genetic systems. Synthetic biologists aim at standardising genetic tools and having greater defined control and modulation of the complex biological processes conferred by the genetic components based on abstraction hierarchy [13, 14]. Given the transdisciplinary aspects of this research field, the consensus definition of synthetic biology is the design and engineer of new biological parts, devices and systems as well as redesigning existing and natural biological system. In fact, synthetic biology shares overlapping characteristics of metabolic engineering particularly through the use of molecular biology and computational tools for DNA parts assembly, pathway engineering and use of well-studied model regulatory systems for genetic circuit designing and construction [2, 3]. Key defining events of this rapidly emerging field are dated back in the early 2000s where the first genetic counter and toggle switch were constructed and led to the development of various artificial genetic elements and control circuits including the use of logic gates for configurational control of gene expression [3, 15, 16]. Another important element of this field is the establishment of public collection and repositories such as Registry of Standard Biological Parts (RSBP) , Synthetic Biology Open Language (SBOL) and Addgene public plasmid repositories that greatly aided parts standardisation and resource sharing among the scientific communities [17, 18].
Essential genetic elements such as promoter, ribosome binding site (RBS), coding sequence (CDS) and transcription terminator are considered as biological parts for constructing standardised biological systems with desired behaviour and purposes. The designing and construction of artificial biological systems with well-defined genetic components have led to the significant breakthrough in constituting synthetic bacterial genome using chemical DNA synthesis and catalytic DNA assembly techniques such as Gibson isothermal assembly and in vivo homologous recombination [19,20,21]. Big progress made in Synthetic Yeast 2.0, the international Synthetic Yeast Genome project that aims at redesigning and constructing a synthetic eukaryotic genome, has been greatly facilitated by de novo biodesign tools and smart data-intensive technologies which will mark another important achievement for advancing microbial synthetic biology [22, 23]. The employment of synthetic genetic systems will allow greater control of the biological functions and outputs that are important in generating genetic design automation and customisation based on iterative design-build-test-learn model cycle [24, 25]. Programming and bioinformatics tools are instrumental in implementing the iterative circuit cycle that allows systematic means of designing, simulating, predicting and analysing the overall research scheme that can be constantly improved in high-throughput manners as compared to the conventional genetic engineering methods of build and test individual constructs.
The synthetic biology approaches are markedly useful for the production of biochemicals with complex biosynthetic pathways and gene clusters such as plant secondary metabolites and antibiotics through computer-aided design (CAD) tools and biosensor-based approaches for designing, constructing and screening of important and rate-limiting enzymes of the targeted pathway. CAD tools such as antiSMASH 3.0 [26] and RetroPath [27] have been employed for predicting, screening and, ultimately, producing the targeted compound using microbial chassis [28]. Rapid strain development with enhanced production activity and robustness has been aided using genetically encoded biosensors that enable precise gene control and high-throughput screening of targeted transcriptional, enzymatic and cellular activities based on colorimetric and fluorescence signals of the genetic constructs in whole cell or cell-free formats [29, 30].
6.1.3 Microbial Chassis for Metabolic Engineering and Synthetic Biology Endeavours
A chassis in synthetic biology refers to the host organism that fundamentally supports the genetic system and provides the essential cellular resources such as transcription and translation machinery for the genetic component to function as intended. Advancement of metabolic engineering and synthetic biology has been built on the fundamental genetics and mathematical modelling of microbial chassis specifically, Escherichia coli and Saccharomyces cerevisiae, considered as two of the most important model microbes for prokaryotes and eukaryotes, respectively. Important features of these model microbes include highly amenable to genetic manipulation, well-developed genetic tools and in-depth genome information and genetic control system. Both systems have been widely used as standard biological systems with an ever-expanding repertoire of synthetic biology toolbox being developed for parts/device assembly, modulated gene control, pathway construction and biomanufacturing of industrially important products.
Apart from these microbes, Gram-positive Bacillus subtilis and Corynebacterium glutamicum have also been well-characterised and well-utilised for fundamental and synthetic biology applications with an increasing catalogue of characterised biological parts that are interchangeable using suitable DNA assembly methods such as BioBrick™ and isothermal assembly modules . In general, bacterial chassis offers a simple nutrient requirement, rapid growth and expression system, while yeast chassis provides a better secretory mechanism and post-translational modification benefits especially for the production of eukaryotic proteins. Well-established DNA delivery methods and a more standardised and improved genetic toolkit are increasingly available for these microbes using modular DNA assembly techniques such as ePathBrick [31] and CoryneBrick [32] in addition to the homologous recombination-based in vivo DNA assembly in yeast. Detailed characteristics and advantages of using these microbial chassis for biotechnological applications can be found in these excellent reviews [33, 34].
6.2 Applications of Metabolic Engineering and Synthetic Biology for Industrial and Agricultural Biotechnology
In this section, biotechnological applications of metabolic engineering and synthetic biology are presented and proposed as sustainable and ‘greener’ approaches in addressing perennial problems in industrial and agricultural sectors. BioE platform using model microbial chassis was devised and demonstrated for three main thrusts: biosynthesis of value-added products, bioconversion of industrial waste and improved system for bioremediation. This section provides the proof of concept on the applications and feasibility of employing microbial strains specifically C. glutamicum, S. cerevisiae and E. coli as BioE chassis that can be further developed to meet industrial and environmental demands.
6.2.1 Biosynthesis of Value-Added Products
Metabolic engineering and synthetic biology approaches have been instrumental in developing biotechnological products with increased productivity and promoting sustainability in the product development . There is an extensive repertoire of biochemicals , biofuels and pharmaceuticals that are biosynthesised in engineered microbes using natural and, more importantly, nonnatural substrates by introducing an array of genes and regulatory elements suited for the production of desired products. Plasmid construction and genetic designs are critical in pathway engineering for directing the overproduction of the targeted products without heavily affecting the engineered cell fitness. In our efforts to develop high-performing strains, there are several important considerations in devising biological engineering strategies which are outlined in Fig. 6.1.
Main aspects of biological engineering (BioE) strategies for value-added product development by engineered microbial hosts. (a) Selection of product and corresponding biosynthetic pathway/gene circuit . (b) Selection of prokaryotic and/or eukaryotic host. (c) Acquiring a suitable type of plasmid vector based on selection marker, replication mode and copy number. (d) Designing and constructing biological parts for desirable gene expression performance. Abbreviations: CDS coding sequence, RBS ribosome binding site
Using the BioE platform, C. glutamicum has been successfully engineered to overproduce 5-aminolevulinic acid (ALA) by combining pathway engineering and metabolic perturbation workflow. ALA is a non-proteinogenic amino acid that is in high demand for its use as biofertiliser (agriculture), photosensitiser (medical) and acne treatment (cosmetics). For industrial applications, ALA is currently produced using non-sustainable chemical synthesis which implicates the overall costs of the product development. To this end, C. glutamicum bacterium, which naturally synthesises high amount of glutamate, was engineered to overproduce ALA using glutamate as the building block of the ALA backbone [35]. C. glutamicum is well-known as the industrial producer of amino acids with well-established pathways of TCA cycle at the pyruvate node and glutamate node.
To establish a high-performing expression system in C. glutamicum, a strong constitutive Trc promoter was used in place of the weak p-Out promoter and signal sequence of the original pMT1s expression plasmid [36]. The promoter sequence includes new RBS that was designed to be high in purines and within 7–9 bp distance from the CDS start codon. To convert glutamate to ALA, two genes from the C5 pathway, glutamyl-tRNA reductase HemA and glutamate 1-semialdehyde aminotransferase HemL were co-expressed in the engineered C. glutamicum. A feedback-deregulated HemA was created by introducing two lysine (KK) residues at the third position of HemA variants from several selected bacterial strains. Following ALA production screening, mutated HemA from Salmonella typhimurium was selected and co-expressed with E. coli BL21 (DE3) HemL on pMT-Trc plasmid yielding pHemAL construct. Addition of penicillin and 2,2′-dipyridyl was carried out to improve ALA production based on increasing glutamate flux and lowering HemA-limiting heme formation, respectively. In shake-flask fermentation, engineered C. glutamicum produced about 1.1 g/L ALA and 2.2 g/L in respective HemA and HemAL operon-expressing strains that represent 10.7-fold and 22-fold improvement from the control strain. Overall, the specially designed bioengineering strategies succeeded in improving ALA production in engineered C. glutamicum that can be used as the platform strain for higher-scale synthesis of ALA for potential commercial applications.
6.2.2 Bioconversion of Agricultural Waste
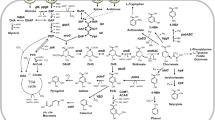
Metabolic engineering and synthetic biology are considered as the enabling technologies for waste-to-wealth concept especially through the development of bioengineered microbial strains. These powerful approaches have enabled and extended the range of carbon sources utilisation by engineered microbes using natural and nonnatural substrates mainly from cellulosic biomass waste made up of cellulose, hemicellulose and inhibitory amounts of acetic acid. Metabolic pathway designing often focuses on maintaining stoichiometric balance and cell growth of the microbial strains when grown on the waste substrates. Figure 6.2 illustrates an overview of the metabolic pathways and key enzymes involved in the utilisation of cellulosic biomass waste and glycerol waste using S. cerevisiae as the model microbe for waste-to-biofuel bioconversion.
Simplified scheme of metabolic pathways and key enzymes involved in waste-to-ethanol bioconversion by engineered S. cerevisiae. Nonnatural substrates and corresponding enzymes involved in the biocatalysis are highlighted in the same colour code. Acetate utilisation is coupled with xylose conversion from pretreated hemicellulosic waste, and cellulose bioconversion involves multiple enzymatic pretreatments and subsequent uptake of cellobiose extracellularly or intracellularly using transporter protein [37]. Glycerol utilisation requires corresponding glycerol uptake and conversion enzymes. An innovative approach that integrates carbon dioxide (CO2) fixation with the endogenous ethanol fermentation pathway using non-native enzymes in xylose-fermenting S. cerevisiae is also highlighted [38]. Solid and dotted black arrows indicate metabolic reaction with single and multiple intermediates, respectively. Light blue and green boxes denote metabolic node for pentose phosphate (PP) and glycolytic pathway, respectively. Abbreviations for enzymes are as follows: ACS acetyl coenzyme A synthetase, AADH acetylating acetaldehyde dehydrogenase, XR xylose reductase, XDH xylitol dehydrogenase, XK xylulose kinase, PRK phosphoribulokinase, RuBisCO ribulose-1,5-bisphosphate carboxylase/oxygenase, GK (Gut1) glycerol kinase, GDH (Gcy) glycerol dehydrogenase, PDC pyruvate decarboxylase, and ADH, alcohol dehydrogenase. DHAP denotes dihydroxyacetone phosphate; P and BP abbreviate for intermediate compound with phosphate and biphosphate group, respectively
Glycerol is a major by-product of the biodiesel industry and represents an inexpensive feedstock for bio-based product development. Bioconversion of waste by S. cerevisiae is of huge biotechnological interest due to the yeast bioprocessing capacity as an industrial workhorse for bioenergy (e.g. first-generation bioethanol) and biochemical (e.g. antimalarial drug precursor) production. However, wild-type yeast S. cerevisiae could not grow on glycerol as the sole substrate, hence preventing direct waste conversion using wild-type strains. Thus, to confer high-rate glycerol-utilising capability, S. cerevisiae YPH499 strain was engineered to overexpress glycerol dehydrogenase (Gcy) and dihydroxyacetone kinase (Dak) , the key enzymes in converting glycerol to dihydroxyacetone phosphate, a glycolytic pathway intermediate in addition to the expression of glycerol uptake/transporter protein (Gup1) for improved glycerol uptake and utilisation [39, 40]. The engineered S. cerevisiae expressing Gcy, Dak and Gup1 formed the starting platform for glycerol bioconversion to bioethanol that was further enhanced via additional expression of alcohol dehydrogenase (Adh1), pyruvate decarboxylase (Pdc1) , SAGA (Spt-Ada-Gcn5-acetyltransferase) complex genes and reduction of endogenous glycerol production Fps1 and Gpd2 genes [41, 42]. Overall, the specially designed bioengineering strategies successfully led to improved ethanol bioconversion up to 8.1 g/L of ethanol produced by engineered S. cerevisiae strains using glycerol as the main substrate.
Similar bioengineering strategies were employed for bioconversion of triacylglycerol (TAG) , a microbial oil feedstock for biodiesel production using engineered S. cerevisiae. The overproduction of TAG from glycerol in S. cerevisiae was attained by introducing TAG biosynthetic genes diacylglycerol acyltransferase (DGA1) and phospholipid diacylglycerol acyltransferase (LRO1) in tandem with glycerol kinase (GUT1) for enhancing glycerol utilisation [43]. The engineered yeast produced about 8.2% overall TAG content, a 2.3-fold enhancement from the wild-type strain (3.6%). The microbial oil synthesised via bioengineering approaches offers an alternative and highly abundant nonfood feedstock for microdiesel and fatty acid-based third-generation biofuel development in the growing bioenergy sectors.
6.2.3 Improved System for Bioremediation
Modulation of metabolic pathways and genetic circuits is the cornerstone of metabolic engineering and synthetic biology approaches that provide researchers with the means for creating a finely tuned genetic system. Pathway engineering has been important in debottlenecking the targeted pathway for improving the flux and providing the precursors and cofactors required for optimal performance of the engineered cells. As depicted in Fig. 6.3, the overall performance of the genetic system in bioengineered chassis can be improved using pathway engineering strategies that focused on increasing the catalytic activities of key enzymes in the targeted metabolic reactions.
Pathway engineering strategies for improving the performance of bioengineered cells and targeted output. (a) Increasing flux and precursor supply via gene expression enhancement . (b) Eliminating negative feedback regulation of rate-limiting enzyme via protein engineering or mutagenesis. (c) Increasing endogenous cofactor supply and regeneration via pathway modulation/optimisation. (d) Reducing the effects of competing enzymes via gene downregulation and extracellular product secretion. WT denotes for wild type and BioE represents bioengineered system
Bioremediation is one of the promising applications of metabolic engineering and synthetic biology especially for degrading recalcitrant compounds in the environment. Natural microbes and enzymes have been extensively investigated for their uses in treating contaminants, but as often the case, process optimisation and associated costs hampered further in field applications. Biocatalysis by dye-decolourising peroxidase (DyP) is of great biotechnological interest due to its catalytic ability in degrading xenobiotics and lignin derivative compounds, hence offering a non-chemical approach for bioremediation and bioenergy applications. To date, development of DyP-based bioremediation is limited by the costly supply of precursors specifically ALA and heme chemicals that are important for functionality and catalytic activities of recombinant heme-containing DyP.
To this end, a bioengineering-inspired recombinant expression system was developed for producing recombinant DyP in engineered E. coli [44]. The focus was to enhance endogenous supply of heme cofactor for increasing the amount of holo-DyP protein via C5 biosynthesis pathway modulation. The need for exogenous precursor addition was offset by co-expressing a synthetic HemAL operon that increased ALA and heme content in the recombinant E. coli. The ALA-overproducing HemAL operon and recombinant Dyp from Bacillus subtilis were expressed using T7 expression systems to yield pHemAL-DyP plasmid construct. When compared with other systems, specifically DyP only and DyP added with hemin, the peroxidase activity of pHemAL-DyP was markedly increased of up to 66.7 U/mg in comparison with 39.0 and 43.4 U/mg peroxidase activity for pDyP and pDyP + hemin, respectively. Importantly, the pHemAL-DyP construct demonstrated an increased dye-decolourising activity by giving out the highest decolourisation percentage of 84.7% when tested on Reactive Blue 19 dye as compared to respective recombinant DyP only (69.9%) and DyP supplied with exogenous hemin (72.8%). The improved catalytic activities of the recombinant DyP will aid in developing crude DyP-based bioremediation of recalcitrant dyes and lignin residues in wastewater and other fields. Therefore, BioE-mediated pathway engineering represents a feasible and effective approach for improving genetic system performance and lowering the production costs by eliminating the need to supply exogenous chemical in the bacterial fermentation.
6.3 Case Study: Integrating Systems and Synthetic Biology for Omics-Driven Microbial Production of Natural Products
As discussed in the previous chapters, multidisciplinary systems biology research has been actively pursued in the context of multiple omics including transcriptomics, proteomics and metabolomics platforms. Using P. minus as the focal point, fundamental aspects of aromatic and bioactive properties of the medicinally important tropical herb were unravelled at molecular and systems levels [45, 46]. Despite its popular use in delicacy and folk medicine, potential commercialisation of single mixture and purified bioactive compounds of P. minus remains untapped due to the lack of enabling technologies other than conventional chemical extraction techniques that beset with poor yields and low productivity.
Consequently, metabolic engineering and synthetic biology approaches were undertaken to complement and utilise biological information gathered from the multi-omics analysis of P. minus. Considering the natural abundance and high antioxidant properties of the herbal extracts, flavonoid was chosen for its targeted production in bioengineered yeast by introducing a total of six biosynthesis genes for naringenin formation via native L-phenylalanine metabolic route. Figure 6.4 demonstrates a schematic representation of the microbial production of flavonoid using systems and synthetic biology research platforms.
Schematic representation of the microbial production of flavonoid using systems and synthetic biology approaches. Flavonoid biosynthetic pathway genes identified from transcriptomics analysis of P. minus were selected for heterologous pathway construction for natural products biosynthesis in engineered S. cerevisiae. Episomal plasmid vectors with different selection markers were employed in the pathway construction. BioE represents bioengineered yeast system. Abbreviations for enzymes are as follows: PAL phenylalanine ammonia lyase, C4H cinnamate-4-hydroxylase, CPR cytochrome P450 reductase, 4CL 4-coumarate-CoA ligase, CHS chalcone synthase, CHI chalcone isomerase
Using S. cerevisiae as the microbial chassis, the integrative systems and synthetic research efforts were mainly aimed at synthesising bioactive natural products in the engineered yeast by introducing P. minus flavonoid biosynthetic pathway. Transcriptome dataset of P. minus was utilised in the mining of flavonoid biosynthetic genes including PAL, C4H, CPR, 4CL, CHS and CHI to constitute a complete naringenin production pathway from endogenous L-phenylalanine in engineered S. cerevisiae. To facilitate a rapid construction of plasmid with multiple genes, a homology-based joining of rate-limiting enzyme C4H and yeast regulatory elements Tef1 promoter and ADH1 terminator was successfully carried out using isothermal assembly methods. Further plasmid construction was employed using modular Gateway and pCEV series plasmid vectors to direct the production of phenylpropanoid compounds in the engineered yeast strains. Successful implementation of this project shall represent an enabling technological platform for tropical plant-based natural product development via the integration of systems and synthetic biology approaches. In summary, further development of bioengineered systems for natural product biosynthesis using tropical genetic resources is envisaged owing to the rapid progression of data-driven high-throughput biologics and next-generation sequencing technologies that are available to systems and synthetic biology research communities.
References
Bailey JE (1991) Toward a science of metabolic engineering. Science 252:1668
Nielsen J, Keasling JD (2011) Synergies between synthetic biology and metabolic engineering. Nat Biotechnol 29:693
Stephanopoulos G (2012) Synthetic biology and metabolic engineering. ACS Synth Biol 1:514–525
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86
Ro DK et al (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940–943
Yim H et al (2011) Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol 7:445
Galanie S, Thodey K, Trenchard IJ, Interrante MF, Smolke CD (2015) Complete biosynthesis of opioids in yeast. Science 349:1095–1100
Keasling JD (2010) Manufacturing molecules through metabolic engineering. Science 330:1355
Chae TU, Choi SY, Kim JW, Ko Y-S, Lee SY (2017) Recent advances in systems metabolic engineering tools and strategies. Curr Opin Biotechnol 47:67–82
Lee SY, Kim HU (2015) Systems strategies for developing industrial microbial strains. Nat Biotechnol 33:1061
Mougiakos I, Bosma EF, Ganguly J, van der Oost J, van Kranenburg R (2018) Hijacking CRISPR-Cas for high-throughput bacterial metabolic engineering: advances and prospects. Curr Opin Biotechnol 50:146–157
Donohoue PD, Barrangou R, May AP (2018) Advances in industrial biotechnology using CRISPR-Cas systems. Trends Biotechnol 36:134–146
Endy D (2005) Foundations for engineering biology. Nature 438:449–453
Canton B, Labno A, Endy D (2008) Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol 26:787–793
Elowitz MB, Leibler S (2000) A synthetic oscillatory network of transcriptional regulators. Nature 403:335–338
Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 403:339–342
Heinemann M, Panke S (2006) Synthetic biology—putting engineering into biology. Bioinformatics 22:2790
Cameron DE, Bashor CJ, Collins JJ (2014) A brief history of synthetic biology. Nat Rev Microbiol 12:381–390
Gibson DG et al (2010) Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329:52–56
Gibson DG et al (2008) One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci U S A 105:20404–20409
Gibson DG, Venter JG (2014) Synthetic biology: construction of a yeast chromosome. Nature 509:168–169
Richardson SM et al (2017) Design of a synthetic yeast genome. Science 355:1040
Pretorius IS, Boeke JD (2018) Yeast 2.0—Connecting the dots in the construction of the world’s first functional synthetic eukaryotic genome. FEMS Yeast Res 18(4):foy032
Nielsen AA et al (2016) Genetic circuit design automation. Science 352:aac7341
Appleton E, Densmore D, Madsen C, Roehner N (2017) Needs and opportunities in bio-design automation: four areas for focus. Curr Opin Chem Biol 40:111
Weber T et al (2015) AntiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237
Carbonell P, Parutto P, Baudier C, Junot C, Faulon JL (2014) Retropath: automated pipeline for embedded metabolic circuits. ACS Synth Biol 3:565
Fehér T et al (2014) Validation of RetroPath, a computer-aided design tool for metabolic pathway engineering. Biotechnol J 9:1446
Rogers JK, Taylor ND, Church GM (2016) Biosensor-based engineering of biosynthetic pathways. Curr Opin Biotechnol 42:84–91
Jiang L, Zhao J, Lian J, Xu Z (2018) Cell-free protein synthesis enabled rapid prototyping for metabolic engineering and synthetic biology. Synth Sys Biotechnol 3:90–96
Xu P, Vansiri A, Bhan N, Koffas MAG (2012) EPathBrick: a synthetic biology platform for engineering metabolic pathways in E. coli. ACS Synth Biol 1:256
Kang MK et al (2014) Synthetic biology platform of CoryneBrick vectors for gene expression in Corynebacterium glutamicum and its application to xylose utilization. Appl Microbiol Biotechnol 98:5991
Kavšček M, Stražar M, Curk T, Natter K, Petrovič U (2015) Yeast as a cell factory: current state and perspectives. Microb Cell Factories 14:94
Calero P, Nikel PI (2018) Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non-traditional microorganisms. Microb Biotechnol [Epub ahead of print]
Ramzi AB, Hyeon JE, Kim SW, Park C, Han SO (2015) 5-Aminolevulinic acid production in engineered Corynebacterium glutamicum via C5 biosynthesis pathway. Enzyme Microb Technol 81:1–7
Hyeon JE, Jeon WJ, Whang SY, Han SO (2011) Production of minicellulosomes for the enhanced hydrolysis of cellulosic substrates by recombinant Corynebacterium glutamicum. Enzyme Microb Technol 48:371
Wei N, Oh EJ, Million G, Cate JHD, Jin YS (2015) Simultaneous utilization of cellobiose, xylose, and acetic acid from lignocellulosic biomass for biofuel production by an engineered yeast platform. ACS Synth Biol 4:707
Xia PF et al (2016) Recycling carbon dioxide during xylose fermentation by engineered Saccharomyces cerevisiae. ACS Synth Biol 6:276–283
Yu KO, Kim SW, Han SO (2010) Engineering of glycerol utilization pathway for ethanol production by Saccharomyces cerevisiae. Bioresour Technol 101:4157–4161
Yu KO, Kim SW, Han SO (2010) Reduction of glycerol production to improve ethanol yield in an engineered Saccharomyces cerevisiae using glycerol as a substrate. J Biotechnol 150:209–214
Yu KO et al (2012) Increased ethanol production from glycerol by Saccharomyces cerevisiae strains with enhanced stress tolerance from the overexpression of SAGA complex components. Enzyme Microb Technol 51:237–243
Yu KO et al (2012) Improvement of ethanol yield from glycerol via conversion of pyruvate to ethanol in metabolically engineered Saccharomyces cerevisiae. Appl Biochem Biotechnol 166:856–865
Yu KO et al (2013) Development of a Saccharomyces cerevisiae strain for increasing the accumulation of triacylglycerol as a microbial oil feedstock for biodiesel production using glycerol as a substrate. Biotechnol Bioeng 110:343–347
Ramzi AB, Hyeon JE, Han SO (2015) Improved catalytic activities of a dye-decolorizing peroxidase (DyP) by overexpression of ALA and heme biosynthesis genes in Escherichia coli. Process Biochem 50:1272–1276
Ahmad R et al (2018) Polygonumins A, a newly isolated compound from the stem of Polygonum minus Huds with potential medicinal activities. Sci Rep 8(4202):4202
Loke K-K et al (2017) Transcriptome analysis of Polygonum minus reveals candidate genes involved in important secondary metabolic pathways of phenylpropanoids and flavonoids. Peer J 5:e2938
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Ramzi, A.B. (2018). Metabolic Engineering and Synthetic Biology. In: Aizat, W., Goh, HH., Baharum, S. (eds) Omics Applications for Systems Biology. Advances in Experimental Medicine and Biology, vol 1102. Springer, Cham. https://doi.org/10.1007/978-3-319-98758-3_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-98758-3_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-98757-6
Online ISBN: 978-3-319-98758-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)