Abstract
The conversion of low-priced glycerol to higher value products has been proposed as a way to improve the economic viability of the biofuels industry. In a previous study, the conversion of glycerol to ethanol in a metabolically engineered strain of Saccharomyces cerevisiae was accomplished by minimizing the synthesis of glycerol, the main by-product in ethanol fermentation processing. To further improve ethanol production, overexpression of the native genes involved in conversion of pyruvate to ethanol in S. cerevisiae was successfully accomplished. The overexpression of an alcohol dehydrogenase (adh1) and a pyruvate decarboxylase (pdc1) caused an increase in growth rate and glycerol consumption under fermentative conditions, which led to a slight increase of the final ethanol yield. The overall expression of the adh1 and pdc1 genes in the modified strains, combined with the lack of the fps1 and gpd2 genes, resulted in a 1.4-fold increase (about 5.4 g/L ethanol produced) in fps1Δgpd2Δ (pGcyaDak, pGupCas) (about 4.0 g/L ethanol produced). In summary, it is possible to improve the ethanol yield by overexpression of the genes involved in the conversion of pyruvate to ethanol in engineered S. cerevisiae using glycerol as substrate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The use of renewable waste substrates is an environmentally friendly practice that effectively reduces waste treatment costs and increases the economic value of by-products. Glycerol is a waste product derived from the transesterification step in biodiesel production, and large amounts of raw glycerol are formed as the main by-product, constituting approximately 10% of the total biodiesel generated. In addition to being cheap and abundant, glycerol can be bioconverted into high-value compounds through microbial fermentation, and the use of glycerol allows for higher yields of reduced chemicals due to its greater degree of reduction as compared to sugars. Thus, considerable efforts have been directed toward the development of methods to refine glycerol from a low-cost feedstock into industrially valuable materials including fuels, building blocks, and bioactive substances [1, 19].
Bioethanol is a combustible fuel that can be made using well-known fermentation technology from a wide range of carbohydrate feedstocks, although the technology required is not yet commercially available [4, 14]. High ethanol yield is becoming increasingly important in order to enhance the economic viability of the commercial process. This is likely to require a combination of both strain development and improved process technology. Industrial production of ethanol from carbohydrate feedstocks such as glycerol requires that the producing organism must not only tolerate and produce high levels of ethanol but also be able to convert the substrate directly to the end-product [2, 12].
The yeast Saccharomyces cerevisiae utilizes the general glycolytic pathway for the majority of its energy production. In this pathway, carbohydrates are converted to pyruvate, and production of energy (in the form of ATP) is coupled to the generation of intermediates and reducing power (in the form of NADH) for biosynthetic pathways. Ethanol is the major fermentation product in ethanologic microorganisms and is produced via pyruvate decarboxylation to acetaldehyde, followed by the reaction of acetaldehyde to form ethanol. These two steps are catalyzed by the enzymes pyruvate decarboxylase (PDC) and alcohol dehydrogenase (ADH), respectively. The pdc and adh genes play an important role in the ethanol fermentation process because both ethanol yield and fermentation efficiency are directly affected by the expression of these two genes [5, 11]. Therefore, these genes have become the focus of research, especially the construction of genetically engineered strains that can efficiently convert carbohydrate to ethanol. In S. cerevisiae, the structural genes pdc1, pdc5, and pdc6 each encode an active pyruvate decarboxylase. The pdc1 is the major isozyme, which is strongly expressed in actively fermenting yeast cells. The nearly identical pdc5 also have similar functions during glycolytic fermentation. However it is only expressed in the absence of pdc1 or under thiamine limitation [16]. The effects of deletion and overexpression of each pdc gene were studied. The decarboxylase activity in the pdcl null mutant was reduced to 72% of the wild-type strain and the isoamyl alcohol level fell to 69% of the wild type. In contrast, the decarboxylase activity was increased in the strain overexpressing pdc1, and the isoamyl alcohol yield was slightly enhanced. In S. cerevisiae, three related adh genes have been found. Adh1, which is constitutively expressed and located in the cytosol, is the major enzyme responsible for conversion of acetaldehyde to ethanol [16]. Adh1 is preferentially expressed in the presence of glucose, whereas adh2 is repressed under these conditions and is derepressed at the transcriptional level when cells are grown on respiratory carbon sources. Adh3 encodes the sole mitochondrial alcohol dehydrogenase present in S. cerevisiae, which seems to play a minor role in ethanol metabolism [17, 18].
Several metabolic engineering approaches have also been examined to reduce by-products and increase ethanol production in S. cerevisiae [20, 21]. Our previous study focused on the use of glycerol as a carbon source for ethanol production. To further increase ethanol production and evaluate fermentative performance, genes involved in the conversion of pyruvate to ethanol were overexpressed. These genes included PDC, which is involved in the decarboxylation of pyruvate and thus controls the first step in the production of ethanol from pyruvate, and ADH, which is the last enzyme in the ethanol production pathway. This study evaluated the potential of S. cerevisiae as a metabolic platform for the microbial conversion of glycerol to ethanol.
Materials and Methods
Strains and Media
The yeast strains used in this study are summarized in Table 1. Escherichia coli cultures were grown at 37 °C in Luria–Bertani broth (10 g/L tryptone, 5 g/L yeast extract, and 5 g/L sodium chloride) containing 50 μg/mL ampicillin. YPD (10 g/l yeast extract, 20 g/L peptone, and 20 g/L glucose) was used as a rich medium. Synthetic dextrose (SD) (20 g/L glucose, 6.7 g/L yeast nitrogen base without amino acids, and 0.77 g/L complete supplement mixture (Difco/Becton Dickinson Co., Sparks, MD, USA)) was used as a minimal medium. For the selection of yeast transformants, G418, kanamycin or zeocin were added to final concentrations of 500 μg/mL, 500 μg/mL, or 250 μg/mL, respectively.
Construction of Plasmids
The plasmids used in this study are illustrated in Fig. 2. The pdc1 gene was amplified from S. cerevisiae genomic DNA by PCR with the oligonucleotides 5′ - ACTAGTCCCGCCGCCACCAAGGAGATGTCTGAAATTACTTTGGG-3′, containing a SpeI restriction site and a Kozak translation initiation sequence, and 5′ - ACTAGTTTATTGCTTAGCGTTGGTAGCA - 3′, containing a SpeI restriction site. The adh1 gene was amplified from S. cerevisiae genomic DNA by PCR with the oligonucleotides 5′ - GAGCTCCCCGCCGCCACCAAGGAGATGTCTATCCCAGAAACTCA - 3′, containing a SacI restriction site and a Kozak translation initiation sequence, and 5′ - GAGCTCTTATTTAGAAGTGTCAACAACG - 3′, containing a SacI restriction site. The PCR product was cloned into pGcyaDak [20] collinear to the galactose-inducible GAL1 promoter. Transformation of the plasmid into S. cerevisiae was carried out using the lithium acetate method with a YEASTMAKER yeast transformation system (Clontech Laboratories, Inc., Palo Alto, CA). Yeast transformants were selected after growth on SD agar plates for 2–3 days. The condition of fermentation and induction were as described previously [20, 21].
Enzyme Activities
The PDC and ADH enzymatic activities were assayed as described previously [8, 9]. One unit of PDC/ADH activity is defined as the generation of 1 μmol NAD+/NADH per min under the conditions specified. Protein concentration was determined using the Bio-Rad Protein assay dye with BSA as a standard.
Metabolite Analysis
Metabolites were analyzed as described previously [20]. Cell growth was monitored by measuring the optical density at 550 nm. The condition of ethanol analysis was as described previously [20, 21].
Results and Discussion
Overexpression of PDC1 and ADH1 for Ethanol Fermentation Using Glycerol
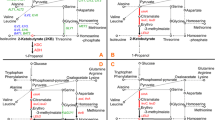
Recently, S. cerevisiae was successfully manipulated for ethanol production using glycerol by improving the carbon flux to the target metabolite [20]. To further develop the strain, the glycerol production genes were deleted, which eventually led to a decrease in glycerol content with an increase in ethanol yield [20, 21]. However, double deletion of the glycerol production genes caused a slight decrease in growth rate and an increase in sensitivity to osmotic stress. In the present study, a further increase in the growth rate and ethanol yield was accomplished by applying a different metabolic engineering strategy (Fig. 1).
Pathways and genes involved in the metabolic engineering strategy. Relevant genes and corresponding enzymes are included. The names of the genes are shown beside the arrows. The abbreviations correspond to glycerol uptake protein (Gup1), glycerol dehydrogenase (Gcy1), dihydroxyacetone (DHA), dihydroxyacetone kinase (Dak), dihydroxyacetone phosphate (DHAP), glycerol-3-phosphate (G3p), glyceraldehydes 3-phosphate (GAP), glycerol 3-phosphate dehydrogenase (GPD2), glycerol facilitator (FPS1), pyruvate decarboxylase (pdc1), and alcohol dehydrogenase (adh1)
This study investigated whether the overexpression of genes encoding proteins involved in the conversion of pyruvate to ethanol would enhance ethanol production when glycerol was provided as a carbon source. To achieve this, a recombinant plasmid pGcyaDakAdhPdc (Fig. 2) was constructed in which expression of the genes pdc1 and adh1 was driven by the constitutive Gal promoter and terminated by the CYC terminator. To verify gene expression, the cell lysate of each recombinant S. cerevisiae strain was assayed for PDC and ADH activity. The activities of PDC and ADH were 112% and 176% higher, respectively, in the modified strain fps1Δgpd2Δ (pGcyaDakAdhPdc, pGupCas) compared to the control strain fps1Δgpd2Δ (pGcyaDak, pGupCas) (Table 2). The studies described above also provided high enzyme activities affected the ethanol production by expression of the genes pdc1 and adh1. To further improve the overall enzyme activity especially PDC due to its low enzyme activity, we tried to overexpress pdc5 individually or in tandem with pdc1. However, overexpression of pdc5 with and without pdc1 did not increase the pyruvate decarboxylase activity and consequent ethanol yield (data not shown). We tried to optimize the induction condition such as inducer concentration. As a result, we successfully increased the enzyme activities although the ethanol yield was no longer increased (Fig. 3). We suggested that the enzyme activity is enough to improve the fermentation condition and ethanol production. In Hong’s article [5, 11], the recombinant plasmids pYH-pdc-adhB was prepared, in which expression of the genes pdc and adhB from Zymomonas mobilis was driven by the constitutive GAPDH promoter and terminated by the AOX terminator for increased ethanol production. As a result, the ethanol yield was increased and with enzyme activities of PDC and AdhE were only 121% and 170% higher. Compared to this study, the activities of PDC and AdhE in engineered strains in this study showed similar numerical value [5, 11].
The effects of overexpression of adh1 and pdc1 were evaluated under fermentative conditions optimized for increased ethanol yield and enzyme activities. Additional overexpression of pGcyaDakAdhPdc led to a slight increase in glycerol consumption (Fig. 4). When the fps1Δgpd2Δ (pGcyaDak, pGupCas) and fps1Δgpd2Δ (pGcyaDakAdhPdc, pGupCas) strains were grown for 48 h in SD media, glycerol consumption in fps1Δgpd2Δ (pGcyaDakAdhPdc, pGupCas) was slightly higher than in fps1Δgpd2Δ (pGcyaDak, pGupCas), and the growth rate was also increased when 2% glycerol was used as a carbon source (Fig. 4). Based on these results, we suggest that the overexpression of genes involved in the conversion of pyruvate to ethanol was successfully accomplished. In our previous article, the knockout strain was osmotically sensitive, showed lower growth rate and redox imbalance compared to control strain. By overexpression of pdc1 and adh1, the growth rate was slightly increased as these enzymes play a critical role involving in not only ethanol formation and consumption but also the general cofactor balance mechanism.
The pdc and adh genes from ethanol-tolerant strains such as S. cerevisiae and Z. mobilis have been well studied [3], and the expression of these two genes in E. coli, Bacillus subtilis, and Lactobacillus casei has allowed for the successful production of ethanol in these organisms [6, 15]. Recently, the ethanol yield was also increased in the methylotrophic yeast Hansenula polymorpha, which was engineered to express the pdc and adhB genes from Z. mobilis [5, 7]. However, there is no report on the effect of overexpression of native pdc1 and adh1 in S. cerevisiae using glycerol for ethanol production.
Improved Ethanol Yield from Glycerol in Recombinant S. cerevisiae
In general, the main aims of engineering a microbial host are to create a production system that is faster in rate, better in titer, and cheaper in cost. Two approaches have been widely used by pathway engineers, the first of which is to expand the carbohydrate utilization capacities of hosts that are already efficient in converting carbohydrate to ethanol [13]. The next approach is to divert carbon flow from the endogenous fermentation products to ethanol in the hosts [6, 10]. This study successfully established the utilization of glycerol, a cheap carbon source, to produce elevated ethanol yield by overexpression of pyruvate to ethanol pathway.
The overexpression of adh1 and pdc1 led to increases in both growth rate and glycerol consumption under fermentative conditions, which led to a slight increase in the final ethanol yield. To understand the contribution of each single modification, we showed all intermediate strains. By overexpression of adh1 and pdc1individually, the ethanol yield was subsequently increased. The strain fps1Δgpd2Δ (pGcyaDakAdh, pGupCas) produced 4.9 g/L ethanol, whereas fps1Δgpd2Δ (pGcyaDakPdc, pGupCas) produced 4.4 g/L (Fig. 5). By overexpression of adh1, the ethanol yield is relatively higher compared to overexpression of pdc1 as ADH serves to regenerate the glycolytic NAD +, thereby restoring the redox balance, through the reduction of acetaldehyde to ethanol.
Figure 6 shows the complete profiles for strains fps1Δgpd2Δ (pGcyaDak, pGupCas) and fps1Δgpd2Δ (pGcyaDakAdhPdc, pGupCas). While fps1Δgpd2Δ (pGcyaDakAdhPdc, pGupCas) strain consumed almost 17.5 g/L of glycerol, the fps1Δgpd2Δ (pGcyaDak, pGupCas) strain consumed 16.0 g/L of glycerol (Fig. 6). For ethanol production using glycerol, the strain fps1Δgpd2Δ (pGcyaDakAdhPdc, pGupCas) produced 5.4 g/L ethanol, whereas fps1Δgpd2Δ (pGcyaDak, pGupCas) produced 4.0 g/L after 96 h of cultivation. The overall ethanol yield was 1.4-fold higher than what was measured in the fps1Δgpd2Δ (pGcyaDak, pGupCas) strain. The fps1Δgpd2Δ (pGcyaDakAdhPdc, pGupCas) strain produced 5.4 g/L ethanol, compared to 0.69 g/L in the parent strain after 96 h of cultivation. Thus, the ethanol production in fps1Δgpd2Δ (pGcyaDakAdhPdc, pGupCas) strain was 7.8-fold more than in YPH499 (pESC-TRP) (Table 3).
In our previous study, a high rate of glycerol utilization was achieved by simultaneous overexpression of glycerol dehydrogenase (Gcy) and dihydroxyacetone kinase (Dak), and high rates of glycerol uptake by simultaneous overexpression of pGupCas also enhanced ethanol yield by minimizing the synthesis of glycerol. To further increase the ethanol production, an optimized fermentative condition promoting higher rates of growth and glycerol consumption was successfully established for the conversion of pyruvate to ethanol in S. cerevisiae. From the overexpression of the genes involved in the pyruvate to ethanol pathway, the final ethanol yield was significantly increased. Given our success with improving the yield of ethanol production through expression of adh1 and pdc1 with glycerol utilization genes, this study succeeded in optimizing the fermentative conditions required for higher ethanol yield using glycerol.
These results confirmed that through metabolic engineering, we have achieved an improved yield of ethanol from pyruvate under optimal fermentative conditions by overexpression of the native genes involved in conversion of pyruvate to ethanol in S. cerevisiae. Such recombinant hosts are now being constantly improved with the ultimate goal of sustaining chemical production from renewable resources in the near future.
Conclusion
Microbial metabolic engineering presents a unique opportunity to lower the costs associated with the raw materials used in biodiesel production. The purpose of this study was to develop biological processes for the production of ethanol from low-priced glycerol, which is also the main by-product of the chemical transesterification process. Developing organisms to achieve a better product yield and productivity often requires simultaneous manipulation of pathways and optimization of regulatory effects. The final recombinant yeast strain in this study will be very useful in the development of industrial processes for ethanol production via metabolic and process engineering compared to other microbial hosts, as this strain demonstrated good fermentation properties in tandem with its ability to convert cheap substrate, specifically glycerol into valuable biofuel.
References
Choi, W. J. (2008). Recent Patents on Biotechnology, 2, 173–180.
Choi, W. J., Hartono, M. R., Chan, W. H., & Yeo, S. S. (2011). Applied Microbiology and Biotechnology, 89, 1255–1264.
Conway, T., Osman, Y. A., & Ingram, L. O. (1987). Journal of Bacteriology, 169, 2327–2335.
Dinus, R. J. (2001). Applied Biochemistry and Biotechnology, 91–93, 23–34.
Hong, W. K., Kim, C. H., Heo, S. Y., Luo, L. H., Oh, B. R., & Seo, J. W. (2010). Biotechnology Letters, 32, 1077–1082.
Ingram, L. O., Conway, T., Clark, D. P., Sewell, G. W., & Preston, J. F. (1987). Applied and Environmental Microbiology, 53, 2420–2425.
Ishchuk, O. P., Voronovsky, A. Y., Stasyk, O. V., Gayda, G. Z., Gonchar, M. V., Abbas, C. A., & Sibirny, A. A. (2008). FEMS Yeast Research, 8, 1164–1174.
Liu, L. M., Li, Y., & Chen, J. (2005). Wei Sheng Wu Xue Bao, 45, 77–80.
Liu, S., Dien, B. S., & Cotta, M. A. (2005). Current Microbiology, 50, 324–328.
Liu, S., Dien, B. S., Nichols, N. N., Bischoff, K. M., Hughes, S. R., & Cotta, M. A. (2007). FEMS Microbiology Letters, 274, 291–297.
Nikel, P. I., Ramirez, M. C., Pettinari, M. J., Mendez, B. S., & Galvagno, M. A. (2010). Journal of Applied Microbiology, 109, 492–504.
Oh, B. R., Seo, J. W., Heo, S. Y., Hong, W. K., Luo, L. H., Joe, M. H., Park, D. H., & Kim, C. H. (2011). Bioresource Technology, 102, 3918–3922.
Palmer, T. N., Wober, G., & Whelan, W. J. (1973). European Journal of Biochemistry, 39, 601–612.
Robertson, G. H., Doyle, L. R., & Pavlath, A. E. (1983). Biotechnology and Bioengineering, 25, 3133–3148.
Romero, S., Merino, E., Bolivar, F., Gosset, G., & Martinez, A. (2007). Applied and Environmental Microbiology, 73, 5190–5198.
Tokuhiro, K., Ishida, N., Nagamori, E., Saitoh, S., Onishi, T., Kondo, A., & Takahashi, H. (2009). Applied Microbiology and Biotechnology, 82, 883–890.
Wenger, J. I., & Bernofsky, C. (1971). Biochimica et Biophysica Acta, 227, 479–490.
Wiesenfeld, M., Schimpfessel, L., & Crokaert, R. (1975). Biochimica et Biophysica Acta, 405, 500–512.
Yazdani, S. S., & Gonzalez, R. (2007). Current Opinion in Biotechnology, 18, 213–219.
Yu, K. O., Kim, S. W., & Han, S. O. (2010). Bioresource Technology, 101, 4157–4161.
Yu, K. O., Kim, S. W., & Han, S. O. (2010). Journal of Biotechnology, 150, 209–214.
Acknowledgment
This work was supported by the Advanced Biomass R&D Center (ABC) of Korea grant funded by the Ministry of Education, Science and Technology (ABC-2010-0029799) and the Technology Development Program for Agriculture and Forestry, Ministry for Agriculture, Forestry and Fisheries, Republic of Korea (no. 309016–5).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yu, K.O., Jung, J., Ramzi, A.B. et al. Improvement of Ethanol Yield from Glycerol via Conversion of Pyruvate to Ethanol in Metabolically Engineered Saccharomyces cerevisiae . Appl Biochem Biotechnol 166, 856–865 (2012). https://doi.org/10.1007/s12010-011-9475-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-011-9475-9