Abstract
Unused fertilizers are the single largest source of nitrous oxide greenhouse gas emissions globally, apart from causing N-pollution in ground and surface waters and eutrophication. While short-term improvements in N-use efficiency (NUE) can be made by agronomic practices, long-term crop improvement is only possible through biological interventions. The lack of clearly defined phenotype and genotype has delayed this process till recently, but the advent of omics and reverse genetics is opening up new avenues to improve crop NUE. It is becoming increasingly evident that several genes and pathways contribute to NUE. Many N-responsive regulators of root development, sensing and signaling, transportation, utilization and remobilization have been targeted to improve NUE, such as C-terminally encoded peptides (CEPs), CLAVATA3/endosperm surrounding region-related peptides (CLE), MADS-box transcription factors, and NAC transcription factors. The nitrate transporter of NRT1.1 is proposed to be a sensor or transceptor, which regulates the crop yield by acting as a component in the Ca2+-mediated signaling cascade. Several other signaling pathways involving target of rapamycin (TOR) complex, general amino acid control non-derepressible 2 (GCN2), ionotropic glutamate-like receptor (iGLR), and PII proteins have been found to play a important roles in maintaining proper N balance in plants. In cereals, cytosolic glutamine synthase, glutamate synthetase, and alanine/aspartate aminotransferases are important targets, as they are involved in remobilization of N from senescing leaves during grain filling and in maintaining proper C/N balance. Several post-transcriptional regulators such as non-coding small RNAs and post-translational regulators such as kinases and phosphatases regulate the expression level of genes involved in N-response/NUE and are emerging as novel targets.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Food security is closely linked to nutrient availability for cropping, whereas its sustainability is directly linked to nutrient use efficiency. This is particularly true for nitrogen , which is quantitatively the most important component of all fertilizers. By 2050, there will be 70% increase in the global food demand, as the world population will increase to over 9.7 billion (Yang et al. 2012; York et al. 2016; Chen and Liao 2017). Unfortunately, the average N-use efficiency (NUE) in crops is about 30%, and the unutilized reactive N species that accumulate in the environment cause water and air pollution affecting health, biodiversity, and climate change (Sutton et al. 2013; Zhang et al. 2015). While short-term improvement of NUE at the farm level can be done using better agronomic practices, slow release fertilizers etc., the inherent ability of the crop to take up the available N and use it efficiently for maximal yield and minimal loss has to be tackled biologically.
A major biological challenge is that our idea of yield itself may vary between grain, fruit, seed, flower, leaf, and tuber depending on the crop. Another biological challenge is that out of the several dozens of definitions of NUE, very few are biologically relevant, such as uptake and utilization efficiency (Pathak et al. 2011; Yu et al. 2016). It is also not uncommon for yield -centric researchers to project N responsiveness as NUE. For example, a cultivar that keeps responding to increasing doses of N-fertilizer with slightly higher yield is misinterpreted as N-use efficient, even if its yield differential may keep falling with increasing N, making it actually less efficient. Such approaches also often push biologists to search for NUE within the narrow genetic pool of high-yielding varieties, rather than trying to find the true extent of genetic diversity that exists for NUE.
The identification of the biological avenues for crop improvement toward NUE is hampered by the incomplete characterization of its phenotype and genotype (Pathak et al. 2011; Sinha et al. 2018). This is extremely important to identify contrasting varieties or to rank all the available germplasm in the increasing or decreasing order of NUE, so as to benefit from the fast-growing genomic data for association mapping. Some of the phenotypic traits associated with NUE so far include root length/number/branching/density, (Morita et al. 1988; Yang et al. 2012; Steffens and Rasmussen 2016), dense and erect panicle in rice (Sun et al. 2014), onset of post-anthesis senescence, and plant height in wheat (Gaju et al. 2011). This chapter is primarily focused on the recent advances in the molecular approaches to improve NUE in plants through the identification of the genes involved in N response and NUE and their manipulation by various means.
Molecular Aspects of N Response for NUE
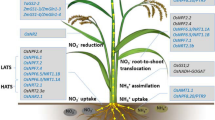
N is present in soil in the form of nitrate (NO3−) or ammonium (NH4+) in aerobic or flooded (anaerobic/acidic) conditions, respectively. A small portion of N can also be absorbed in the form of amino acids or as urea directly by plants with the help of specific transporters. They are mainly absorbed through the roots and translocated throughout the plant through xylem. N-compounds are also recycled and remobilized from internal stores or senescing tissues through the phloem to the sites of demand, such as for grain filling in cereals . The genes involved in all these processes of N uptake, assimilation , and remobilization are important for N-use efficiency, which makes it a complex, quantitative trait. On an organism-wide scale, N response encompasses many more genes/processes that may contribute to NUE, including C metabolism, redox metabolism, and root/shoot development (Fig. 5.1). The molecular biology of N response has been elaborated in several reviews (Pathak et al. 2008; Krapp et al. 2014; Li et al. 2017; Sinha et al. 2018). Therefore, the following sections deal with various molecular targets that have been explored toward improvement of N-use efficiency.
AtalaAT: A. thaliana Alanine aminotransferase; OsaspAT: O. sativa Aspartate aminotransferase ; ZmGDH: Z. maize NADP-dependent glutamate dehydrogenase; OsCKX2: Cytokinin oxidase; ZmIPT: Cytokinin biosynthesis, OsSGR: Stay green, OsENOD93-1: Mitochondrial membrane protein; AtSTP-13: Hexose transporter, VfAAP1: V. faba Amino acid permease , Rubisco; OsNAP: NAC transcription factor , TaNFYA-B1: T. aestivum CCAAT-binding transcription factors, NtGS1: N. tobacum cytosolic glutamine synthetase , AtGGT1: glutamate: glyoxylate aminotransferase 1; BrUGE: B. rapa UDP-glucose 4-epimerase; OsNADH-GOGAT: NADH- dependent glutamate synthase ; AtTAR2, OsTOND1: Tolerance Of Nitrogen Deficiency 1, OsDRO1: DEEPER ROOTING 1, TaNAC2-5A, OsPTR9: Peptide transporter /nitrate transporter, OsMADS25, GmEXPB2: G. max beta-expansin, ZmFd-NADP+ reductase: Ferredoxin NADP+ reductase, TaEXPB23; AtSWEET1: Sugars Will Eventually Be Exported Transporters; AtGluR2: Glutamate receptor; AtGLB1: PII regulatory protein; OsDEP1: (Dense and erect panicle 1) G protein γ subunit; SlSnRK: S. lycopersicum sucrose non-fermenting-1-related protein kinase 1; AtMKK9-MPK6: Mitogen-activated protein kinase; Hap2-3-5-Gln3: Hap2-3-5 binding domain and Gln3 activation domain; AtNADK2: NAD kinase (Wang et al. 2012; Klemens et al. 2013; Alvarez et al. 2014; Rothstein et al. 2014; Dellero et al. 2015; Wada et al. 2015; Abdula et al. 2016; Li et al. 2016; Chen and Liao 2017; Wan et al. 2017)
Genes/ QTLs Identified for NUE
Marker-assisted genetic mapping has helped identify many genes/QTLs for plant height, panicle weight, and panicle number such as GS1, DEP1, NADH-GOGAT to improve NUE. At the same time, many other genes involved in N transportation, assimilation , signaling, and regulation have been successfully used to improve NUE in rice and other plants (Fig. 5.1). Over-expression of OsNRT2.3b improved nitrate-uptake capacity, C metabolism, grain yield and thereby NUE by 40%. In addition, it also enhanced the uptake capacity of P and Fe by maintaining pH homeostasis (Fan et al. 2016). Therefore, modulation of the expression of N transporters has a beneficial impact on the overall plant NUE. However, it has also been reported that an increased N input may delay flowering time and consequent yield losses especially in high-latitude regions where late-season temperatures hamper grain filling (Li et al. 2017). Transgenic approaches using many other genes involved in N metabolism have also been implicated to improve NUE such as glutamate dehydrogenase (GDH), aspartate aminotransferase (AspAT) , and asparagine synthetase (AS). NLP proteins have been recently reported to increase crop yield by improving plant biomass under both N-rich and poor soil conditions (Xu et al. 2016).
Molecular Manipulation of Root System Architecture for NUE
In the past two decades, scientific community has established a strong basis to target root system architecture as an approach to improve NUE (Forde 2014; Fan et al. 2017; Li et al. 2017). Root system is comprised of embryonic (primary and seminal roots in Arabidopsis and cereals , respectively) and post-embryonic roots (lateral roots in Arabidopsis; lateral, brace, and crown roots in cereals ). Studies carried out in maize helped us to understand the advantages of “steep, cheap, and deep” root morphology to absorb water and nutrients from soil (Lynch 2013). Long and thick primary roots help plants to acquire N from the deeper horizon, while fewer and longer lateral roots with steep root growth angles not only decreases the metabolic cost but also help in exploring greater volume of soil.
Numerous signaling mechanisms are involved in the adjustment of root development to heterogeneous N environments . Studies on the molecular control of N-responsive root development have been mainly carried out in Arabidopsis, though various homologs of the genes involved have also been found in rice and other plants (Forde 2014; Shahzad and Amtmann 2017). A summary of N-responsive regulators of root system architecture is provided in Table 5.1. Arabidopsis shows root adjustment towards different levels and forms of N in the surrounding rhizosphere with the help of various signaling molecules. This includes regulation of lateral root initiation in the xylem pole pericycle cells by CEP5 (C-terminally encoded peptide) in an auxin and N-dependent manner (Roberts et al. 2016); inhibition of lateral root emergence during systemic low N signal by CLE (CLAVATA3/ESR-related) gene family which binds to CLAVATA1 (CLV1 leucine-rich repeat receptor-like kinases (Araya et al. 2014, 2016; Okamoto et al. 2015); inhibition of primary root growth by AFB3 (auxin receptors which are a part of the SCFTIR1/AFB E3 ubiquitin ligase complex) in the presence of nitrate and promotion of lateral root growth by AFB3/NAC4/OBP4-signaling module (Vidal et al. 2010, 2013). Recently, several miRNAs (miR167, miR393, miR160 and miR171) and N-responsive transcription factors have been reported to regulate root system morphology (Table 5.1) under various N conditions in Arabidopsis and rice (Vidal et al. 2010; Yan et al. 2014; Bellegarde et al. 2017; Chien et al. 2017; Gifford et al. 2017; Sun et al. 2017; Undurraga et al. 2017). Generally lateral roots are much more sensitive to the fluctuating nutritional conditions and their response depends on the degree of stress in the surrounding region. Low N deficiency tends to promote lateral root initiation but moderate to severe N deficiency hampers further root emergence and elongation. Root morphology is also determined by the ratio of NO3−:NH4+. High NO3−:NH4+ ratio showed positive effect on the lateral root length, whereas low ratio has a contrary impact (Qin et al. 2017).
N Transporters as Targets to Improve NUE
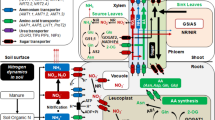
Understanding the molecular mechanism of N uptake and its regulation is of great significance toward the improvement of NUE. Nitrogen is taken up from soil mainly in the form of nitrate (NO3−), ammonium (NH4+), amino acids or peptides, and urea with the help of substrate-specific transporters. Most of these transporters mediate active transport depending on the proton gradient across plasma membrane except few which act as channels and mediate passive transport of solutes. These transporters are classified into low affinity transport systems (LATS) and high affinity transport systems (HATS), as well as in terms of constitutive or inducible. LATS function at a relatively higher concentration of N (>0.5 mM) and have larger Km values (5 mM). On the other hand, HATS mediate transport at low N concentration (0.2–0.5 mM) and have smaller Km values (of about 50 μM). Analysis of tissue-specific expression under varying concentration of N is very important for an efficient N uptake and therefore determining crop yield (Li et al. 2017).
Five main families of nitrate transporters are present in plants: nitrate transporter 1/peptide transporter/nitrate peptide transporter family ( NRT1/PTR /NPF), NRT2/nitrate nitrite porter (NRT2/NNP), chloride channels (CLCs), slow anion channel-associated 1 homolog 3 (SLAC1/SLAH), and aluminum-activated malate transporters (ALMT) (Li et al. 2017).
Many ammonium transporters (AMTs) have also been targeted to improve NUE by analyzing the phenotypic changes of specific overexpressing or mutant lines. AMTs belong to the AMT/MEP/Rhesus transporter family, which are highly conserved in bacteria, fungi, and plants with more than 700 homologs in bacteria and plants. In Arabidopsis, there are 6 AMTs and rice genome has 12 AMTs which have been classified into two subfamilies: OsAMT1 and OsAMT2 (Li et al. 2017; Xuan et al. 2017). The activities of these transporters are also controlled by phosphorylation, thereby preventing the accumulation of NH4+ to toxic levels within the plant system.
Urea uptake and metabolism within the plant and its evaluation as a target for NUE has not received requisite attention, despite the fact that most of Asian agriculture depends on urea fertilization. Urea transport occurs in plants through five different types of urea transporters, out of which DUR3 type have high affinity and others have low affinity. DUR3 is a 1 urea /1H+ symporter, whereas the low affinity urea transporters (tonoplast intrinsic protein, TIP) act as channels and are pH independent (Reddy and Ulaganathan 2015). Gene expression of DUR3-type transporters is controlled by ammonia, nitrate, and urea .
Nitrate and ammonium transporters are also sensitive to the pH changes of the rhizosphere, the apoplast, or the cytoplasm, as exemplified by modulation in the activity of AtNPF6.3/AtCHL1/AtNRT1.1 and OsNRT2.3b. Similarly, water also affects N uptake and under the condition of drought stress, plants activate specific signaling pathways to overcome reduction in the N uptake. N starvation-induced basic leucine zipper (bZIP) transcription factor gene, AtTGA4, cytokinin synthesis gene isopentenyltransferase (IPT), and nodule inception-like 7 protein (NLP7) along with NITRATE REGULATORY GENE2 (NRG2) are reported to regulate the process of N uptake under these conditions.
Other regulators of the N transporters include transcription factors (TF) such as MADS-box TF ANR1, LOB Domain-Containing proteins (LBD37/38/39), Nin like proteins (NLP6, NLP7), Hypersensitivity to Low Pi-Elicited Primary Root Shortening 1 (HRS1), TGACG Sequence-specific Binding Protein 1 (TGA1/4), Squamosa Promoter Binding Protein-Like 9 (SPL9), Auxin Signaling F-Box 3 (AFB3), Nitrate Regulatory Gene (NRG2), Teosinte Branched 1/Cycloidea/Proliferating Cell Factor 20 (TCP20), GATA transcription factor , High Nitrogen Insensitive 9 (HNI9), shoot-derived peptide signals such as bZIPTF, HY5, root-derived peptide signals such as CEP and CLE, and miRNAs such as miR393 and miR169a (Marchive et al. 2013; Chien et al. 2017; Xuan et al. 2017).
Apart from these transporters, roots also release exudates in the form of ions, organic compounds, and enzymes to improve nutrient acquisition efficiency (Chen and Liao 2017). Symbiotic association with arbuscular mycorrhizal fungi (AMF) also enables plants such as rice, maize, wheat , and soybean to acquire diffusible nutrients and fixed carbon beyond the rhizosphere and at the same time also reduces the inefficient use of applied N to the soil. It improves the N availability in the rhizosphere through varying the composition of rhizobial microbial community. Recently, Verzeaux et al. (2017) reported improved NUE in wheat by AMF-assisted increased N uptake and accumulation .
Components of N Sensing and Signaling as Targets to Improve NUE
Transcriptomic studies carried out in Arabidopsis, rice, maize, and several other plant species have provided ample support to the fact that N in the form of either nitrate, ammonium , nitric oxide or nitrogen metabolites (L-Glutamate) plays pivotal role in controlling many biological processes in plants, such as root development , crop yield , seed dormancy, flowering time, and leaf development (Wang et al. 2004; Forde et al. 2013; Sun et al. 2016; O’Brien et al. 2016; Noguero and Lacombe 2016). During this, N mainly acts as a signaling molecule to regulate the expression of genes involved in nutrient transport, metabolism, glycolysis, gluconeogenesis, hormonal activities, etc., in both roots and shoots (Chakraborty and Raghuram 2011). Genes involved in these processes include transcription factors (MADS-Box Transcription Factor ), phosphoenolpyruvate carboxylase, Gln synthetase, Asn synthetase, tryptophan amino transferase, ribosomal proteins, initiation factors and many more (Calatrava et al. 2017; Okumoto and Versaw 2017; Liu et al. 2017; Undurraga et al. 2017). Therefore, the knowledge of sensing and signaling components will further enhance our ability to develop improved crop variety (Table 5.2). For example, NRT1.1 and NRT2.1 sense changes in N concentration occurring in the external medium and initiate Ca2+-mediated signaling cascade involving phospholipase C (PLC). CHL1/NPF6.3/NRT1.1 acts as a dual affinity nitrate transceptor and therefore have the ability to sense both high and low concentrations of N. This property is dependent on the phosphorylation status which is under the tight control of CBL-interacting protein kinase23 (CIPK23) (Ho et al. 2009; Bouguyon et al. 2015; Riveras et al. 2015; Undurraga et al. 2017).
Plastid localized PII proteins in plants interact with N-acetyl-L-glutamate kinase (NAGK) and acetyl-CoA carboxylase to promote arginine synthesis and fatty acid synthesis, respectively. Glutamine binds to the C-terminal extension of PII proteins to enhance its ability to form complex with NAGK (Gent and Forde 2017).
Through the work carried out in yeast, target of rapamycin (TOR) was identified. In budding yeast, it is found to participate in signaling pathway including nutrient and hormonal signaling and then passing the information to downstream effectors. Plant genomes also have homologs of mammalian or yeast TORC1 complex. Activity of TOR and sucrose non-fermenting 1 (Snf1) kinase (SnRK1 in plants) complement with one another to maintain C/N homeostasis under different environmental conditions by regulating several biologically important processes such as photosynthesis, tricarboxylic-acid cycle, and N assimilation by mainly controlling protein synthesis (Dobrenel et al. 2016; Sesma et al. 2017). Similarly, general amino acid control non-derepressible 2 (GCN2) kinases also plays a very important role in controlling protein synthesis by causing phosphorylation of eIF2α initiation factor under N starvation. In Arabidopsis, there are 20 ionotropic glutamate-like receptors (iGLR) and 24 in rice which have important functional role in stomatal closure, root branching, and maintenance of primary root meristem (Weiland et al. 2014; Gent and Forde 2017).
Nitrogen requirements of crops are fulfilled by the legumes by the process of nodulation by symbiotic relationship with N-fixing bacteria. The availability of genetic mutants has enabled to carry out transcriptomic studies to find out the factors controlling nodulation. Generally, nodulation is promoted under low N supply and excess of N supply has a negative impact on the number of nodules formed. A number of mobile signaling molecules such as CLE peptides, TOO MUCH LOVE (TML), receptor-like kinases, CORYNE and CLAVATA2, CEPs, COMPACT ROOT ARCHITECTURE2 (CRA2) , nodule inception protein (NIN), NIN-like proteins (NLP), and miR172-EARLY NODULIN40 (ENOD40) module regulate nodulation (Murray et al. 2016).
In plants, nitric oxide plays a very important role in regulating many biological processes including seed germination, root development , senescence, plant immunity, and abiotic stress by controlling the expression of many regulatory components (Calatrava et al. 2017). Ammonium is also found to induce the expression of genes involved in N metabolism (PEPC, Gln synthetase, and Asn synthetase) and transport (Amino Acid Permease, AAP1) (Wang et al. 2004). It also affects ammonium uptake, assimilation , hormonal balance, and root system architecture by altering cytosolic pH and post-translational modification of proteins involved in these processes (Liu and Wirén 2017). In Arabidopsis, ammonia is sensed by an ammonium transporter (AtAMT1;1) whose activity is modulated by the calcineurin-B-like-interacting protein kinases (CIPK) proteins by phosphorylation (Xuan et al. 2017).
Molecular Targets Among the Genes of N Assimilation and Remobilization
Nitrate taken up inside the root cells is first reduced by nitrate reductase (NIA) to nitrite and then to ammonium by nitrite reductase (NiR). Two NIA genes exist in Arabidopsis and three in rice. Subsequently, nitrite moves into the plastid and is then metabolized into ammonium by the glutamine synthetase /glutamate synthase (GS/GOGAT) cycle . Ammonium is further incorporated into amino acids . This process of amino acid formation depends on the availability of photosynthates. GS is a very important enzyme for N assimilation and remobilization and there are two isoforms of the enzyme: GS1 that carries out primary ammonium assimilation in roots or re-assimilation of ammonium in leaves and GS2 that carries out assimilation of ammonium in chloroplast. Three-to-five members of GS have been found in different plant species; for example, there are three in rice. Depending on the electron donor specificity, there are two types of GOGAT, viz. ferredoxin-dependent (Fd-GOGAT) and NADH-dependent (NADH-GOGAT). GLU1 and GLU2 are two Fd-GOGATs and GLT is the only NADH-GOGAT gene present in the genome of Arabidopsis. Similarly, rice genome encodes one Fd-GOGAT and two NADH-GOGAT.
Single gene transgenics overexpressing the genes of primary N assimilation (NR, NiR and plastidic GS, GOGAT) did not radically improve NUE (Pathak et al. 2008, 2011; Krapp et al. 2014; Sinha et al. 2018). This was expected in a quantative, multigenic trait like NUE, which involves the coordinated expression of several genes including, but not limited to N-assimilation. This made regulatory targets more attractive than metabolic targets, but the inability to find specific nitrate response elements common to all N-responsive genes has delayed progress in this direction (Das et al. 2007; Pathak et al. 2009). Circadian clock master regulator, CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) also controls the expression of genes involved in N assimilation and thereby establishes a link between N metabolism and circadian clock (Gutiérrez et al. 2008). Kinases and phosphatases are also involved in the regulation of expression of genes coding for N assimilatory enzymes such as NR , NiR, GS2, and Fd-GOGAT (Undurraga et al. 2017). Another level of control of metabolism is carried out by transcription factors (Dof, NLP7, GATA), N metabolites (glutamine and glutamate), and miRNAs (Chien et al. 2017; Zuluaga et al. 2017). miR5640 targets phosphoenolpyruvate carboxylase (PEPC) which plays a very important role in maintaining C/N balance. The expression of PEPC and several other enzymes of tricarboxylic-acid cycle are also under the control of Dof1 (DNA BINDING WITH ONE FINGER) TF (He et al. 2015). Castaings et al. (2009) reported the role NLP7 protein in N assimilation and sensing. All NLP proteins can bind nitrate-responsive cis-element NRE and mediate nitrate-dependent gene expression (Marchive et al. 2013; Xu et al. 2016; Yu et al. 2016) and improve C/N balance under both N-sufficient and N-deficient conditions. On the one hand, proper N assimilation is required for chloroplast development, synthesis of chlorophyll, and proteins such as Rubisco ((ribulose-1,5-bisphosphate carboxylase/oxygenase and PEPC), whereas on the other hand, C assimilation provides energy source for N metabolism in the form of reducing equivalents (Ferredoxin and NADH) and C skeleton for synthesis of amino acids .
Remobilization of nitrate from source (leaves) to sink (developing parts) is also a significant determinant of NUE as it recycles organic N to the seeds during the grain-filling stage and therefore determines the crop yield . Leaf senescence is the underlying phenomenon of nutrient remobilization which facilitates the recycling of photosynthates to the developing seeds. Autophagy promotes senescence of aging plant parts. Several senescence-associated genes (ATG and metacaspases) are expressed at different stages of plant senescence (Havé et al. 2016). This process involves the participation of tissue-specific transporters which replenishes the N requirement during reproductive stage of plant development. Several reports suggest the regulators of this process, such as nitrogen limitation adaptation (NLA), which control the expression of AtNRT1.7 by protein ubiquitination pathway. However, NLA is itself under the control of miRNA827 (Liu et al. 2016). Analysis of rice GOGAT mutant leads to the identification of another protein, viz. ferredoxin-dependent glutamate synthase (OsFd-GOGAT), to play a role in this process (Zeng et al. 2016). Fd-GOGAT plays a role in ammonium recycling by photorespiration .
Various Approaches to Identify More QTLs Associated with NUE
Modern technologies have improved our ability to study the regulation at the level of gene expression. These techniques include TARGET (Transient Transformation System for Genome-Wide Transcription Factor Target Discovery) and ChIP-Seq (Bargmann et al. 2013; Marchive et al. 2013) . A major challenge in crop improvement for nitrogen use efficiency (NUE) is that neither the phenotypic traits nor the genes/alleles determining NUE are clearly defined. Therefore, in this scenario it is very necessary to work with chemist and use analogs of different N sources and then carry out the phenotypic screening for NUE-related genes/loci. For example, by using chlorate, the toxic analog of nitrate, OsNRT1.1B, was identified as a critical QTL contributing to NUE divergence between rice subspecies (Hu et al. 2015). Similar strategy had earlier lead to the identification of several regulators of N assimilation in fungi and algae such as NIT2, NIT4, AREA, NIRA, and NIT2 (Castaings et al. 2009). Application of the new high-throughput measuring techniques such as genome-wide association studies (GWAS) also enables us to identify genes/QTLs regulating NUE. For example, Gifford et al. (2013) grew 96 Arabidopsis accessions under two N regimes and studied root phenotypic traits and identified JASMONATE RESPONSIVE 1 (JR1) as one of the candidate genes. Similarly, CALCIUM SENSOR RECEPTOR, PhzC, ROOT SYSTEM ARCHITECTURE 1, and PHOSPHATE 1 were discovered by high-throughput automated root image analysis (Gifford et al. 2013; Rosas et al. 2013; Slovak et al. 2014). This technique helps us to study natural variations among different genotypes of a plant species and understand the complex regulatory mechanism behind NUE. Another such technique is semiautomated confocal microscopy with the help of which KURZ UND KLEIN (F-box family gene), was identified to play a significant role in root development (Li et al. 2017). Systems biology has also enabled us to identify novel interacting partners and further provides the missing knowledge about the components of signal transduction pathway of N sensing, signaling and metabolism (Gutiérrez et al. 2008; Vidal et al. 2013).
Conclusions
The last decade has witnessed tremendous progress in finding several molecular targets towards the improvement of N-use efficiency of plants. Several genes belonging to various processes have been identified including root development , N uptake, assimilation , and remobilization . In addition, genes involved in N sensing, signaling, and the regulation of the above processes have also emerged, including epigenetic regulation involving miRNA. While phenotype development has not kept pace with these developments, functional genomics and reverse genetics are opening newer opportunities for identification and validation of newer molecular targets. These developments strengthen the hope that improved crop varieties for NUE will become increasingly available for sustainable agriculture in the near future.
References
Abdula SE, Lee HJ, Kim J, Niño MC, Jung Y-J, Cho Y-C abd Cho Y-G (2016) BrUGE1transgenic rice showed improved growth performance with enhanced drought tolerance. Breed Sci 66(2), 226–233
Alvarez JM, Riveras E, Vidal EA, Gras DE, Contreras‐López O, Tamayo KP, Aceituno F, Gómez I, Ruffel S, Lejay L, Jordana X (2014) Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. The Plant J 80(1):1–3
Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wirén N, Takahashi H (2014) CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. P Natl A Sci 111(5):2029–2034
Araya T, von Wirén N, Takahashi H (2016) CLE peptide signaling and nitrogen interactions in plant root development. Plant Mol Biol 91(6):607–615
Bargmann BOR, Marshall-Colon A, Efroni I, Ruffel S, Birnbaum KD, Coruzzi GM, Krouk G (2013) TARGET: A Transient Transformation System for Genome-Wide Transcription Factor Target Discovery. Mol Pla 6(3):978–980
Bellegarde F, Gojon A, Martin A (2017) Signals and players in the transcriptional regulation of root responses by local and systemic N signaling in Arabidopsis thaliana. J Exp Bot 68(10):2553–2565
Bouguyon E, Brun F, Meynard D, Kubeš M, Pervent M, Leran S, Lacombe B, Krouk G, Guiderdoni E, Zažímalová E, Hoyerová K (2015) Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat Plants 1(3):15015
Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet‐Mercey S, Taconnat L, Renou J, Daniel‐Vedele F, Fernandez E, Meyer C, Krapp, A (2009) The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. The Plant J 57(3):426–435
Calatrava V, Chamizo-Ampudia A, Sanz-Luque E, Ocaña-Calahorro F, Llamas A, Fernandez E, Galvan A (2017) How Chlamydomonas handles nitrate and the nitric oxide cycle. J Exp Bot 68(10):2593–2602
Chakraborty N. and Raghuram N. (2011) Nitrate sensing and signaling in genomewide plant N response. In Nitrogen Use Efficiency in Plants, V. Jain, P. Anandakumar (eds) New India Publishing Agency, New Delhi. pp. 45–62
Chen L, Liao H (2017) Engineering crop nutrient efficiency for sustainable agriculture. J Integr Plant Biol 59(10):710–735
Chien P-S, Chiang C-B, Wang Z, Chiou T-J (2017) MicroRNA-mediated signaling and regulation of nutrient transport and utilization. Curr Opinion in Plant Biol 39:73–79
Das SK, Pathak RR, Choudhury D, Raghuram N (2007) Genomewide computational analysis of nitrate response elements in rice and Arabidopsis. Mol Genet Genomics 278(5):519–525
Dellero Y, Lamothe-Sibold M, Jossier M, Hodges M (2015) Arabidopsis thaliana ggt1 photorespiratory mutants maintain leaf carbon: nitrogen balance by reducing RuBisCO content and plant growth. The Plant J 83(6):1005–1018
Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C (2016) TOR signaling and nutrient sensing. Ann Rev Plant Biol 67(1):261–285
Fan X, Tang Z, Tan Y, Zhang Y, Luo B, Yang M, Lian X, Shen Q, Miller AJ, Xu G (2016) Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. P Natl A Sci 113(26):7118–7123
Fan X, Naz M, Fan X, Xuan W, Miller AJ, Xu G (2017) Plant nitrate transporters: from gene function to application. J Exp Bot 68(10):2463–2475
Forde BG (2014). Nitrogen signalling pathways shaping root system architecture: an update. Curr Opin Plant Biol 21:30–36
Forde BG, Cutler SR, Zaman N, Krysan PJ (2013) Glutamate signalling via a MEKK1 kinase-dependent pathway induces changes in Arabidopsis root architecture. The Plant J 75(1):1–10
Gaju O, Allard V, Martre P, Snape JW, Heumez E, LeGouis J, Moreau D, Bogard M, Griffiths S, Orford S, Hubbart S (2011) Identification of traits to improve the nitrogen-use efficiency of wheat genotypes. Field Crops Res 123(2):139–152
Gan Y, Bernreiter A, Filleur S, Abram B, Forde BG (2012) Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol 53(6):1003–1016
Gent L, Forde BG (2017) How do plants sense their nitrogen status? J Exp Bot 68(10):2531–2539
Gifford ML, Banta JA, Katari MS, Hulsmans J, Chen L, Ristova D, Tranchina D, Purugganan MD, Coruzzi GM, Birnbaum KD (2013) Plasticity regulators modulate specific root traits in discrete nitrogen environments. PLoS Genet 9(9):e1003760
Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD (2017) Cell-specific nitrogen responses mediate developmental plasticity. P Natl A Sci 105(2):803–808
Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, Coruzzi GM (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. P Natl A Sci 105(12):4939–4944
He X, Qu B, Li W, Zhao X, Teng W, Ma W, Ren Y, Li B, Li Z,Tong Y (2015) The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol 169(3):1991–2005
Havé M, Marmagne A, Chardon F, Masclaux-Daubresse C (2016) Nitrogen remobilisation during leaf senescence: lessons from Arabidopsis to crops. J Exp Bot 68(10):2513–2529
Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138(6):1184–1194
Hu B, Wang W, Ou S, Tang J, Li H, Che R, Zhang Z, Chai X, Wang H, Wang Y, Liang C, Liu L, Piao Z, Deng Q, Deng K, Xu C, Liang Y, Zhang L, Li L, Chu C (2015) Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nature Genetics 47(7):834–838
Huang S, Chen S, Liang Z, Zhang C, Yan M, Chen J, Xu G, Fan X, Zhang Y (2015) Knockdown of the partner protein OsNAR2.1 for high-affinity nitrate transport represses lateral root formation in a nitrate-dependent manner. Sci Rep 5:18192
Klemens PAW, Patzke K, Deitmer J, Spinner L, Le Hir R, Bellini C, Bedu M, Chardon F, Krapp A, Neuhaus HE (2013) Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol 163(3):1338–1352
Krapp A, David LC, Chardin C, Girin T, Marmagne A, Leprince A-S, Chaillou S, Ferrario-Méry S, Meyer C, Daniel-Vedele F (2014) Nitrate transport and signaling in Arabidopsis. J Exp Bot 65(3):789–798
Li H, Hu B, Chu C (2017) Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J Exp Bot 68(10):2477–2488
Li X, Zeng R, Liao H (2016) Improving crop nutrient efficiency through root architecture modifications. J Integr Plant Biol 58(3):193–202
Liu KH, Niu Y, Konishi M, Wu Y, Du H, Chung HS, Li L, Boudsocq M, McCormack M, Maekawa S, Ishida T (2017) Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 545(7654):311–316
Liu W, Han X, Zhan G, Zhao Z, Feng Y, Wu C (2015) A novel sucrose-regulatory MADS-box transcription factor GmNMHC5 promotes root development and nodulation in soybean (Glycine max [L.] Merr.). Int J Mol Sci 16(9):20657–20673
Liu W, Sun Q, Wang K, Du Q, Li W-X (2016) Nitrogen limitation adaptation (NLA) is involved in source-to-sink remobilization of nitrate by mediating the degradation of NRT1.7 in Arabidopsis. New Phytol 214(2):734–744
Liu Y, von Wirén N (2017) Ammonium as a signal for physiological and morphological responses in plants. J Exp Bot 68(10):2581–2592
Lynch JP (2013) Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann Bot 112(2):347–357
Ma W, Li J, Qu B, He X, Zhao X, Li B, Fu X, Tong Y (2014) Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. The Plant J 78(1):70–79
Marchive C, Roudier F, Castaings L, Bréhaut V, Blondet E, Colot V, Meyer C, Krapp A (2013) Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun 4:1713
Młodzińska E, Kłobus G, Christensen MD, Fuglsang AT (2015) The plasma membrane H(+)-ATPase AHA2 contributes to the root architecture in response to different nitrogen supply. Physiol Plant 154(2):270–282
Mochizuki S, Jikumaru Y, Nakamura H, Koiwai H, Sasaki K, Kamiya Y, Kamiya Y, Ichikawa H, Minami E, Nishizawa Y (2014) Ubiquitin ligase EL5 maintains the viability of root meristems by influencing cytokinin-mediated nitrogen effects in rice. J Exp Bot 65(9):2307–2318
Morita S, Suga T, Yamazaki K (1988) The relationship between root length density and yield in rice plants. Jpn J Crop Sci 57(3):438–443
Murray JD, Liu C-W, Chen Y, Miller AJ (2016) Nitrogen sensing in legumes. J Exp Bot 68(8):1919–1926
Nishizawa Y, Mochizuki S, Koiwai H, Kondo K, Kishimoto K, Katoh E, Minami E (2015) Rice ubiquitin ligase EL5 prevents root meristematic cell death under high nitrogen conditions and interacts with a cytosolic GAPDH. Plant Signal Behav 10(3):e990801
Noguero M, Lacombe B (2016) Transporters involved in root nitrate uptake and sensing by Arabidopsis. Front Plant Sci (7):1391
O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA (2016) Nitrate transport, sensing, and responses in plants. Mol Plant 9(6):837–856
Ohyama K, Ogawa M, Matsubayashi Y (2008) Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. The Plant J 55(1):152–160
Okamoto S, Suzuki T, Kawaguchi M, Higashiyama T, Matsubayashi Y (2015) A comprehensive strategy for identifying long-distance mobile peptides in xylem sap. The Plant J 84(3):611–620
Okumoto S, Versaw W (2017) Genetically encoded sensors for monitoring the transport and concentration of nitrogen-containing and phosphorus-containing molecules in plants. Curr Opin Plant Biol 39:129–135
Ondzighi-Assoume CA, Chakraborty S, Harris JM (2016) Environmental nitrate stimulates abscisic acid accumulation in Arabidopsis root tips by releasing it from inactive stores. Plant Cell 28(3):729–745
Pathak RR, Ahmad A, Lochab S, Raghuram N (2008) Molecular physiology of plant N-use efficiency and biotechnological options for its enhancement. Curr Sci 94(11):1394–1403
Pathak RR, Das SK, Choudhury D, Raghuram N (2009) Genomewide bioinformatic analysis negates any specific role for Dof, GATA and Ag/cTCA motifs in nitrate responsive gene expression in Arabidopsis. Physiol Mol Biol Pla 15(2):145–150
Pathak RR, Lochab S, Raghuram N (2011) Improving nitrogen use efficiency. In Compr Biotechnol, vol 4, 2nd edn. Elsevier, Oxford, pp 209–218
Qin S, Sun X, Hu C, Tan Q, Zhao X, Xin J, Wen X (2017) Effect of NO3−:NH4+ ratios on growth, root morphology and leaf metabolism of oilseed rape (Brassica napus L.) seedlings. Acta Physiol Plant 39(9):198
Qu B, He X, Wang J, Zhao Y, Teng W, Shao A, Zhao X, Ma W, Wang J, Li B, Li Z (2015) A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol 167(2):411–423
Reddy MM, Ulaganathan K (2015) Nitrogen nutrition, its regulation and biotechnological approaches to improve crop productivity. Am J Plant Sci 6(18):2745–2798
Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A (2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA 103(50):19206–19211
Riveras E, Alvarez JM, Vidal EA, Oses C, Vega A, Gutiérrez RA (2015) The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis 1. Plant Physiol 177(1):00961
Roberts I, Smith S, Stes E, De Rybel B, Staes A, Van De Cotte B, Njo MF, Dedeyne L, Demol H, Lavenus J, Audenaert D (2016) CEP5 and XIP1/CEPR1 regulate lateral root initiation in Arabidopsis. J Exp Bot 67(16):4889–4899
Rosas U, Cibrian-Jaramillo A, Ristova D, Banta JA, Gifford ML, Fan AH, Zhou RW, Kim GJ, Krouk G, Birnbaum KD, Purugganan MD (2013) Integration of responses within and across Arabidopsis natural accessions uncovers loci controlling root systems architecture. P Natl A Sci 110(37):15133–15138
Rothstein SJ, Bi Y-M, Coneva V, Han M, Good A (2014) The challenges of commercializing second-generation transgenic crop traits necessitate the development of international public sector research infrastructure. J Exp Bot 65(19):5673–5682
Sesma A, Castresana C, Castellano MM (2017) Regulation of translation by TOR, eIF4E and eIF2α in plants: current knowledge, challenges and future perspectives. Front Plant Sci 8:644
Shahzad Z, Amtmann A (2017) Food for thought: how nutrients regulate root system architecture. Curr Opin Plant Biol 39:80–87
Sinha VB, Jangam AP, Raghuram N (2018) Biological determinants of crop N use efficiency and biotechnological avenues for improvement. In: Masso C, Bleeker A, Raghuram N, Bekunda M, Sutton M (eds) Proceedings of the N2013. Springer
Slovak R, Göschl C, Su X, Shimotani K, Shiina T, Busch W (2014) A scalable open-source pipeline for large-scale root phenotyping of Arabidopsis. Plant Cell 26(6):2390–2403
Steffens B, Rasmussen A (2016) The physiology of adventitious roots. Plant Physiol 170(2):603–617
Sun C-H, Yu J-Q, Hu D-G (2017) Nitrate: a crucial signal during lateral roots development. Front Plant Sci 8:485
Sun H, Qian Q, Wu K, Luo J, Wang S, Zhang C, Ma Y, Liu Q, Huang X, Yuan Q, Han R, Zhao M, Dong G, Guo L, Zhu X, Gou Z, Wang W, Wu Y, Lin H, Fu X (2014) Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat Genet 46(6):652–656
Sun J, Ye M, Peng S, Li Y (2016) Nitrogen can improve the rapid response of photosynthesis to changing irradiance in rice (Oryza sativa L.) plants. Sci Rep 6(1):31305
Sutton MA, Bleeker A, Howard CM, Bekunda M, Grizzetti B, de Vries W, van Grinsven HJM, Abrol YP, Adhya TK, Billen G,. Davidson EA, Datta A, Diaz R, Erisman JW, Liu XJ, Oenema O, Palm C, Raghuram N, Reis S, Scholz RW, Sims T, Westhoek H, Zhang FS, with contributions from Ayyappan S, Bouwman AF, Bustamante M, Fowler D, Galloway JN, Gavito ME, Garnier J, Greenwood S, Hellums DT, Holland M, Hoysall C, Jaramillo VJ, Klimont Z, Ometto JP, Pathak H, Plocq Fichelet V, Powlson D, Ramakrishna K, Roy A, Sanders K, Sharma C, Singh B, Singh U, Yan XY, Zhang Y (2013) Our nutrient world: the challenge to produce more food and energy with less pollution. Global Overview of Nutrient Management, Centre for Ecology and Hydrology, Edinburgh on behalf of the Global Partnership on Nutrient Management and the International Nitrogen Initiative
Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y (2014) Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science (New York, N.Y.) 346(6207):343–346
Undurraga SF, Ibarra-Henríquez C, Fredes I, Álvarez JM, Gutiérrez RA (2017) Nitrate signaling and early responses in Arabidopsis roots. J Exp Bot 68(10):2541–2551
Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA (2013) Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. P Natl A Sci 110(31):12840–12845
Vidal EA, Viviana A, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis Thaliana. P Natl A Sci 107(9):4477–4482
Verzeaux J, Hirel B, Dubois F, Lea PJ, Tétu T (2017) Agricultural practices to improve nitrogen use efficiency through the use of arbuscular mycorrhizae: basic and agronomic aspects. Plant Sci 264:48–56
Wada S, Hayashida Y, Izumi M, Kurusu T, Hanamata S, Kanno K, Kojima S, Yamaya T, Kuchitsu K, Makino A, Ishida H (2015) Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol 168(1):60–73
Wan TE, Xue HE, TONG YP (2017) Transgenic approaches for improving use efficiency of nitrogen, phosphorus and potassium in crops. J Integr Agri 16 (12):60345–60347
Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136(1):2512–2522
Wang X, Peng F, Li M, Yang L, Li G (2012) Expression of a heterologous SnRK1 in tomato increases carbon assimilation, nitrogen uptake and modifies fruit development. J Plant Physiol 169(12):1173–1182
Weiland M, Mancuso S, Baluska F (2014) Signalling via glutamate and GLRs in Arabidopsis thaliana. Funct Plant Biol 43(1):1–25
Xuan W, Beeckman T, Xu G (2017) Plant nitrogen nutrition: sensing and signaling. Curr Opin Plant Biol 39:57–65
Xu N, Wang R, Zhao L, Zhang C, Li Z, Lei Z, Liu F, Guan P, Chu Z, Crawford NM,Wang Y (2016) The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. The Plant Cell 28(2):485–504
York LM, Silberbush M, Lynch JP (2016) Spatiotemporal variation of nitrate uptake kinetics within the maize (Zea mays L.) root system is associated with greater nitrate uptake and interactions with architectural phenes. J Exp Bot 67(12):3763–3775
Yang JC, Zhang H, Zhang JH (2012) Root morphology and physiology in relation to the yield formation of rice. J Integr Agri 11(6):920–926
Yan Y, Wang H, Hamera S, Chen X, Fang R (2014) MiR444a has multiple functions in the rice nitrate-signaling pathway. The Plant J 78(1):44–55
Yu C, Liu Y, Zhang A, Su S, Yan A, Huang L, Ali I, Liu Y, Forde BG, Gan Y (2015) MADS-box transcription factor OsMADS25 regulates root development through affection of nitrate accumulation in rice. PLoS One 10(8):e0135196.
Yu LH, Miao ZQ, Qi GF, Wu J, Cai XT, Mao JL, Xiang CB (2014) MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol Plant 7:1653–1669
Yu LH, Wu J, Tang H, Yuan Y, Wang S-M, Wang Y-P, Zhu QS, Li SG, Xiang C-B (2016) Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Scientific reports 6:27795
Zeng D-D, Qin R, Li M, Alamin M, Jin X-L, Liu Y, Shi C-H (2016) The ferredoxin-dependent glutamate synthase (OsFd-GOGAT) participates in leaf senescence and the nitrogen remobilization in rice. Mol Genet Genomics 292(2):385–395
Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science (New York, N.Y.) 279(5349):407–409
Zhang X, Davidson EA, Mauzerall DL, Searchinger TD, Dumas P, Shen Y (2015) Managing nitrogen for sustainable development. Nature 528(7580):51
Zuluaga DL, De Paola D, Janni M, Curci PL, Sonnante G (2017) Durum wheat miRNAs in response to nitrogen starvation at the grain filling stage. PLoS One 12(8):e0183253
Acknowledgements
This work was supported in part by research grant to NR and fellowship to NS from the Department of Biotechnology, Govt. of India under the Indo-UK Virtual Nitrogen Centre on Nitrogen Efficiency of Whole cropping Systems (NEWS) BT/IN/UK-VNC/44/NR/2015-16. VM is a recipient of DBT fellowship (BCIL/HRD/DBT-JRF/FLSP).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Mandal, V.K., Sharma, N., Raghuram, N. (2018). Molecular Targets for Improvement of Crop Nitrogen Use Efficiency: Current and Emerging Options. In: Shrawat, A., Zayed, A., Lightfoot, D. (eds) Engineering Nitrogen Utilization in Crop Plants. Springer, Cham. https://doi.org/10.1007/978-3-319-92958-3_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-92958-3_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-92957-6
Online ISBN: 978-3-319-92958-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)