Abstract
With the explosive growth of high-dimensional data, feature selection has become a crucial step of machine learning tasks. Though most of the available works focus on devising selection strategies that are effective in identifying small subsets of predictive features, recent research has also highlighted the importance of investigating the robustness of the selection process with respect to sample variation. In presence of a high number of features, indeed, the selection outcome can be very sensitive to any perturbations in the set of training records, which limits the interpretability of the results and their subsequent exploitation in real-world applications. This study aims to provide more insight about this critical issue by analysing the robustness of some state-of-the-art selection methods, for different levels of data perturbation and different cardinalities of the selected feature subsets. Furthermore, we explore the extent to which the adoption of an ensemble selection strategy can make these algorithms more robust, without compromising their predictive performance. The results on five high-dimensional datasets, which are representatives of different domains, are presented and discussed.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In the context of high-dimensional data analysis, feature selection aims at reducing the number of attributes (features) of the problem at hand, by removing irrelevant and redundant information as well as noisy factors, and thus facilitating the extraction of valuable knowledge about the domain of interest. The beneficial impact of feature selection on the performance of learning algorithms is widely discussed in the literature [1] and has been experimentally proven in several application areas such as bio-informatics [2], text categorization [3], intrusion detection [4] or image analysis [5].
There exists currently a large body of feature selection methods, based on distinct heuristics and search strategies, and several works have investigated their strengths and weaknesses on both real [6] and artificial data [7]. Most of the existing studies, however, concentrate on the effectiveness of the available algorithms in selecting small subsets of predictive features, without taking into account other relevant aspects that only recently have gained attention, such as the scalability [8], the costs associated to the features [9] or the robustness (stability) of the selection process with respect to changes in the input data [10]. This last issue has been recognized to be especially important when the high-dimensionality of data is coupled with a comparatively small number of instances: in this setting, actually, even small perturbations in the set of training records may lead to strong differences in the selected feature subsets.
Though the literature on feature selection robustness is still limited, an increasing number of studies recognize that a robust selection outcome is often equally important as good model performance [11, 12]. Indeed, if the outcome of the selection process is too sensitive to variations in the set of training instances, the interpretation (and the subsequent exploitation) of the results can be very difficult, with limited confidence of domain experts and final users. Moreover, as observed in [13], the robustness of feature selection may have practical implications for distributed applications where the algorithm should produce stable results across multiple data sources.
Further research, from both a theoretical and empirical point of view, should be devoted to better characterizing the degree of robustness of state-of-art selection algorithms in multiple settings, in order to achieve a better understanding of their applicability/utility in knowledge discovery tasks. On the other hand, the definition of feature selection protocols which can ensure a better trade-off between robustness and predictive performance is still an open issue, though a number of studies [11, 14] seem to suggest that the adoption of an ensemble selection strategy can be useful in this regard.
To give a contribution to the field, this work presents a case study which aims to provide more insight about the robustness of six popular selection methods across high-dimensional classification tasks from different domains. Specifically, for each method, we evaluate the extent to which the selected feature subsets are sensitive to some amount of perturbation in the training data, for different levels of perturbation and for different cardinalities of the selected subsets.
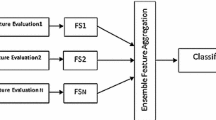
In addition, for each selection algorithm, we implement an “ensemble version” whose output is built by a bagging procedure similar to that adopted in the context of multi-classifier systems [15], i.e. (i) different versions of the training set are created through a re-sampling technique, (ii) the feature selection process is carried out separately on each of these versions and (iii) the resulting outcomes are combined through a suitable aggregation function. The studies so far available on the robustness of this ensemble approach are limited to a single application domain [11, 16], to a single selection method [14] or to a given number of selected features [17], so it is worth providing the interested reader with a more comprehensive evaluation which encompasses different kinds of data, different selection heuristics (both univariate and multivariate) and different subset sizes.
The results of our experiments clearly show that, when comparing the overall performance of the considered selection methods, the differences in robustness can be significant, while the corresponding differences in accuracy (or other metrics, such as the AUC) are often null or negligible. In the choice of the best selector for a given task, hence, the degree of robustness of the selection outcome can be a discriminative criterion. At the same time, our study shows that the least stable methods can benefit, at least to some extent, from the adoption of an ensemble selection strategy.
The rest of this paper is organized as follows. Section 2 summarizes background concepts and related works. Section 3 describes all the materials and methods relevant to our study, i.e. the methodology used for the robustness analysis, the ensemble strategy and the selection algorithms here considered, and the datasets used as benchmarks. The experimental results are presented and discussed in Sect. 4. Finally, Sect. 5 gives the concluding remarks.
2 Background and Related Work
As discussed in [10], the robustness (or stability) of a given selection method is a measure of its sensitivity to changes in the input data: a robust algorithm is capable of providing (almost) the same outcome when the original set of records is perturbed to some extent, e.g. by adding or removing a given fraction of instances.
Recent literature has investigated the potential causes of selection instability [18] and has also focused on suitable methodologies [19] for evaluating the degree of robustness of feature selection algorithms. This evaluation basically involves two aspects: (a) a suitable protocol to generate a number of datasets, different to each other, which overlap to a great (“soft” perturbation) or small (“hard” perturbation) extent with the original set of records; (b) a proper consistency index to measure the degree of similarity among the outputs that are produced (in the form of feature weightings, feature rankings or feature subsets) when a given algorithm is applied to the above datasets. The higher the similarity, the more robust the selection method.
As regards the data perturbation protocols, simple re-sampling procedures are adopted in most cases, though some studies have investigated how to effectively measure and control the variance of the generated sample sets [13]. The influence of the amount of overlap between these sets is discussed by Wang et al. [20], who propose a method for generating two datasets of the same size with a specified degree of overlap.
As regards the similarity measure used to compare the selection outcomes, various approaches have been proposed [10, 21, 22], each expressing a slightly different view of the problem. For example, the Pearson’s correlation coefficient can be used if the output is given as a weighting of the features, the Spearman’s rank correlation coefficient if the output is a ranking of the features, the Tanimoto distance or the Kuncheva index if the output is a feature subset. A good review of stability measures can be found in [18].
From an experimental point of view, a number of studies have compared the robustness of different selection methods on high-dimensional datasets [23,24,25]. This work extends and complements the available studies by encompassing different application domains; besides, stability patterns are derived for feature subsets of different cardinalities and for different levels of data perturbation. As a further contribution, we investigate the impact, in terms of selection robustness, of using an ensemble selection strategy; though presented as a promising approach to achieve more stable results, indeed, it has been so far evaluated in a limited number of settings, particularly with biomedical/genomic data [11, 14, 16, 17].
3 Materials and Methods
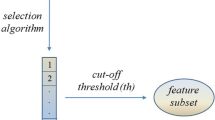
In our study we focus on selection techniques that provide, as output, a feature ranking, i.e. a list (usually referred as ranked list) where the available features appear in descending order of relevance. In turn, the ranked list can be cut at a proper threshold point to obtain a subset of highly predictive features. In the context of high-dimensional problems, indeed, this ranking-based approach is a de facto standard to reduce the dimensionality of the feature space; then, the filtered space can be either refined through more sophisticated (and computationally expensive) techniques or directly used for predictive and knowledge discovery purposes.
The robustness of six popular ranking techniques is here evaluated in a two-fold setting (simple and ensemble ranking), according to the methodology presented in Subsect. 3.1; next, Subsect. 3.2 provides some details on the chosen techniques and describes the datasets used as benchmarks and the specific settings of the experiments.
3.1 Methodology for Robustness Evaluation: Simple vs Ensemble Ranking
Leveraging on best practices from the literature, we evaluate the robustness of the selection process in conjunction with the predictive performance of the selected subsets. Both the aspects, indeed, must be taken into account when assessing the suitability of a given selection approach (actually, stable but not accurate solutions would be not meaningful; on the other hand, accurate but not stable results could have limited utility for domain experts and final users).
In more detail, given the input dataset, we repeatedly perform random sampling (without replacement) to create m different training sets, each containing a fraction f of the original records. For each training set, a test set is also formed using the remaining fraction (1 − f) of the instances. The feature selection process is then carried out in a two-fold way:
-
Simple ranking. A given ranking method is applied separately on each training set to obtain m distinct ranked lists which in turn produce, when cut at a proper threshold (t), m different feature subsets (here referred as simple subsets).
-
Ensemble ranking. An ensemble version of the same ranking method is implemented using a bagging-based approach, i.e. each training set is in turn sampled (with replacement) to construct b samples of the same size (bootstraps). The considered ranking method is then applied to each bootstrap, which results in b distinct ranked lists that are finally combined (through a mean-based aggregation function [26]) into a single ensemble list. In turn, this list is cut at a proper threshold (t) to obtain an ensemble subset of highly discriminative features. Overall, m ensemble subsets are selected, one for each training set.
For both the simple and the ensemble setting, the robustness of the selection process is measured by performing a similarity analysis on the resulting m subsets. Specifically, for each pair of subsets S i and S j (i, j = 1, 2, …, m), we use a proper consistency index [21] to quantify their degree of similarity:
where t is the size of the subsets (corresponding to the cut-off threshold) and n the overall number of features. Basically, the similarity sim ij expresses the degree of overlapping between the subsets, i.e. the fraction of features which are common to them (|S i ∩ S j |/t), with a correction term reflecting the probability that a feature is included in both subsets simply by chance. The need for this correction, which increases as the subset size approaches the total number of features, is experimentally demonstrated for example in [27]. The resulting similarity values are then averaged over all pair-wise comparisons, in order to evaluate the overall degree of similarity among the m subsets and, hence, the robustness of the selection process.
At the same time, in both simple and ensemble settings, a classification model is built on each training set using the selected feature subset, and the model performance is measured (through suitable metrics such as accuracy and AUC) on the corresponding test set. By averaging the accuracy/AUC of the resulting m models, we can obtain an estimate of the effectiveness of the applied selection approach (simple or ensemble) in identifying the most discriminative features. This way, the trade-off between robustness and predictive performance can be evaluated for different values of the cut-off threshold.
3.2 Ranking Techniques, Datasets and Settings
The above methodology can be applied in conjunction with any ranking method. To obtain useful insight on the robustness of different selection approaches, as well as on the extent to which the ensemble implementation affects their outcome, we included in our study six algorithms that are representatives of quite different heuristics. In particular, we considered three univariate methods (Symmetrical Uncertainty, Gain Ratio and OneR), which evaluate each feature independently from the others, and three multivariate methods (ReliefF, SVM-AW and SVM-RFE) which take into account the inter-dependencies among the features. More details on these techniques and their pattern of agreement can be found in [28]. In brief:
-
Symmetrical Uncertainty (SU) and Gain Ratio (GR) both leverage the concept of information gain, that is a measure of the extent to which the class entropy decreases when the value of a given feature is known. The SU and GR definitions differ for the way they try to compensate for the information gain’s bias toward features with more values.
-
OneR (OR) ranks the features based on the accuracy of a rule-based classifier that constructs a simple classification rule for each feature.
-
ReliefF (RF) evaluates the features according to their ability to differentiate between data points that are near to each other in the attribute space.
-
SVM_AW exploits a linear Support Vector Machine (SVM) classifier, which has an embedded capability of assigning a weight to each feature (based on the contribution the feature gives to the decision function induced by the classifier); the absolute value of this weight (AW) is used to rank the features.
-
SVM_RFE, in turn, relies on a linear SVM classifier, but adopts a recursive feature elimination (RFE) strategy that iteratively removes the features with the lowest weights and repeats the overall weighting process on the remaining features (the percentage of features removed at each iteration is 50% in our implementation).
Each of the above methods has been applied, in its simple and ensemble version, on five high-dimensional datasets, chosen to be representatives of different domains. In particular:
-
The Gastrointestinal Lesions dataset [29] contains 1396 features extracted from a database of colonoscopy videos; there are 76 instances of lesions, distinguished in ‘hyperplasic’, ‘adenoma’ and ‘serrated adenoma’.
-
The Voice Rehabilitation dataset [30] contains 310 features resulting from the application of speech processing algorithms to the voices of 126 Parkinson’s disease subjects, who followed a rehabilitative program with ‘acceptable’ or ‘unacceptable’ results.
-
The DLBCL Tumour dataset [31] contains 77 samples, including ‘follicular lymphoma’ and ‘diffuse large b-cell lymphoma’ samples, each described by the expression level of 7129 genes.
-
The Ovarian Cancer dataset [32] contains 15154 features describing proteomic spectra generated by mass spectrometry; the instances are 253, divided in ‘normal’ and ‘cancerous’.
-
The Arcene dataset, in turn, is a binary classification problem where the task is to distinguish ‘cancerous’ versus ‘normal’ patterns from mass spectrometric data. Unlike the Ovarian Cancer dataset, it results from the combination of different data sources; a number of noisy features, having no predictive power, were also added in order to provide a challenging benchmark for the NIPS 2003 feature selection challenge [33]. The overall dimensionality is 10000, while the number of instances is 200.
Note that all the above datasets are characterized by a large number of features and a comparatively small number of records, which makes it difficult to achieve a good trade-off between predictive performance and robustness.
According to the methodology described in Subsect. 3.1, different training/test sets have been built for each dataset; specifically, we set m = 20. As regards the amount of data perturbation, i.e. the fraction of the original instances randomly included in each training set, we explored the values f = 0.70, f = 0.80 and f = 0.90. For the number of bootstraps involved in the construction of the ensemble subsets, we also explored different values, i.e. b = 20, b = 50 and b = 80. Further, for both the simple and the ensemble subsets, different values of the cut-off threshold (i.e. different subset sizes) have been considered, ranging from 0.5% to 7% of the original number of features.
4 Experimental Study: Results and Discussion
In this section, we summarize the main results of our robustness analysis. First, it is interesting to consider the effect of varying the amount of perturbation introduced in the input data, i.e., in our setting, the effect of including in the training sets only a fraction f of the original records. Limited to the simple ranking, Fig. 1 shows the robustness of the six selection methods here considered (SU, GR, OR, RF, SVM-AW, SVM-RFE) on the Gastrointestinal Lesions dataset, for different values of f and different subset sizes.
As we can see, even a small amount of perturbation (f = 0.90) affects the stability of the selection outcome in a significant way, since the average similarity among the 20 feature subsets (selected from the m = 20 training sets built from the original dataset) is far lower than the maximum value of 1. As the amount of perturbation increases, the degree of robustness dramatically falls off, for all the selection methods, though some of them exhibit a somewhat better behaviour. Similar considerations can be made for the other datasets here considered (whose detailed results are omitted for the sake of space), thus confirming that the instability of the selection outcome is a very critical concern when dealing with high-dimensional problems.
A further point to be discussed is the extent to which the adoption of an ensemble strategy improves the robustness of the selection process. Figs. 2, 3, 4, 5 and 6 show, for the five datasets included in our study, the stability of both the simple and the ensemble subsets, with a data perturbation level of f = 0.80. In particular, for each selection method, three ensembles have been implemented with different numbers of bootstraps (b = 20, b = 50, b = 80), but only the results for b = 20 (20b-ensemble) and b = 50 (50b-ensemble) have been reported here, since a higher value of b does not further improve the robustness in an appreciable way.
As we can see, the impact of the ensemble approach is different for the different methods and varies in dependence on the subset size and the specific characteristics of the data at hand. In particular, among the univariate selection methods, SU turns out to be intrinsically more robust, with a further (though limited) stability improvement in the ensemble version. The other univariate approaches, i.e. GR and OR, turn out to be less robust in their simple form and take greater advantage of the ensemble implementation. In turn, in the group of the multivariate approaches, the least stable method, i.e. SVM-RFE, is the one that benefits most from ensemble strategy; this strategy, on the other hand, is not beneficial for the RF method, except that in the Gastrointestinal Lesions and in the Voice Rehabilitation datasets, but only for some percentages of selected features. In all cases, it is not useful to use more than 50 bootstraps in the ensemble implementation.
The above robustness analysis has been complemented, according to the methodology presented in Subsect. 3.1, with a joint analysis of the predictive performance. Specifically, the selected feature subsets have been used to train a Random Forest classifier (parameterized with log2(t) + 1 random features and 100 trees), which has proved to be very effective in several domains [34]. For the sake of space and readability, only the results obtained in the f = 0.80 perturbation setting are here reported; specifically, Table 1 summarizes the AUC performance (averaged over the m = 20 training/test sets) achieved with both the simple and the 50b-ensemble subsets, limited to a threshold t = 5% of the original number of features (but the AUC results obtained with feature subsets of different cardinalities confirm what observed in Table 1).
When comparing the overall performance of the six selection methods, in their simple form, it is clear that the differences in AUC are much smaller (and often negligible) than the corresponding differences in robustness. In cases like these, where the AUC/accuracy is not a discriminative factor, the outcome stability can then be assumed as a decisive criterion for the choice of the best selector.
A further important observation is that no significant difference exists between the AUC performance of the simple and the ensemble version of the considered selection methods. Indeed, irrespective of the application domain, each selection algorithm achieves almost the same AUC outcome in both the implementations. When looking at the trade-off between the predictive performance and the robustness of the selection process, we can then conclude that the adoption of an ensemble strategy can lead to more stable feature subsets without compromising at all the predictive power of these subsets.
5 Conclusions
This work emphasized the importance of evaluating the robustness of the selection process, besides the final predictive performance, when dealing with feature selection from high-dimensional data. The stability of the selection outcome, indeed, is important for practical applications and can be a useful (and objective) criterion to guide the choice of the proper selection method for a given task. Further, the proposed study contributed to demonstrate that the adoption of an ensemble selection strategy can produce better results even in those domains where the selection of robust subsets is intrinsically harder, due to a very low instances-to-features ratio. The beneficial impact of the ensemble approach is more significant for the selection methods that turn out to be less stable in their simple form (e.g., the univariate Gain Ratio and the multivariate SVM-RFE). Actually, the stability gap between the different methods tend to become much smaller (or sometimes null) when they are used in the ensemble version. This is noteworthy for practitioners and final users that, in the ensemble setting, could exploit different, but equally robust, selection methods.
References
Guyon, I., Elisseeff, A.: An introduction to variable and feature selection. J. Mach. Learn. Res. 3, 1157–1182 (2003)
Saeys, Y., Inza, I., Larranaga, P.: A review of feature selection techniques in bioinformatics. Bioinformatics 23(19), 2507–2517 (2007)
Forman, G.: An extensive empirical study of feature selection metrics for text classification. J. Mach. Learn. Res. 3, 1289–1305 (2003)
Bolón-Canedo, V., Sánchez-Maroño, N., Alonso-Betanzos, A.: Feature selection and classification in multiple class datasets: an application to kdd cup 99 dataset. Expert Syst. Appl. 38(5), 5947–5957 (2011)
Staroszczyk, T., Osowski, S., Markiewicz, T.: Comparative analysis of feature selection methods for blood cell recognition in leukemia. In: Proceedings of the 8th International Conference on Machine Learning and Data Mining in Pattern Recognition, pp. 467–481 (2012)
Tang, J., Alelyani, S., Liu, H.: Feature selection for classification: a review. In: Aggarwal, C.C. (ed.) Data Classification: Algorithms and Applications, pp. 37–64. CRC Press, Boca Raton (2014)
Bolón-Canedo, V., Sánchez-Maroño, N., Alonso-Betanzos, A.: A review of feature selection methods on synthetic data. Knowl. Inf. Syst. 34(3), 483–519 (2013)
Bolón-Canedo, V., Rego-Fernández, D., Peteiro-Barral, D., Alonso-Betanzos, A., Guijarro-Berdiñas, B., Sánchez-Maroño, N.: On the scalability of feature selection methods on high-dimensional data. Knowl. Inf. Syst. 1–48 (2018). https://springerlink.bibliotecabuap.elogim.com/article/10.1007/s10115-017-1140-3
Maldonado, S., Pérez, J., Bravo, C.: Cost-based feature selection for support vector machines: an application in credit scoring. Eur. J. Oper. Res. 261(2), 656–665 (2017)
Kalousis, A., Prados, J., Hilario, M.: Stability of feature selection algorithms: a study on high-dimensional spaces. Knowl. Inf. Syst. 12(1), 95–116 (2007)
Saeys, Y., Abeel, T., Van de Peer, Y.: Robust feature selection using ensemble feature selection techniques. In: Daelemans, W., Goethals, B., Morik, K. (eds.) ECML PKDD 2008, Part II. LNCS (LNAI), vol. 5212, pp. 313–325. Springer, Heidelberg (2008). https://doi.org/10.1007/978-3-540-87481-2_21
Pes, B.: Feature selection for high-dimensional data: the issue of stability. In: 26th IEEE International Conference on Enabling Technologies: Infrastructure for Collaborative Enterprises, WETICE 2017, pp. 170–175 (2017)
Alelyani, S., Zhao, Z., Liu, H.: A dilemma in assessing stability of feature selection algorithms. In: IEEE 13th International Conference on High Performance Computing and Communications, pp. 701–707 (2011)
Abeel, T., Helleputte, T., Van de Peer, Y., Dupont, P., Saeys, Y.: Robust biomarker identification for cancer diagnosis with ensemble feature selection methods. Bioinformatics 26(3), 392–398 (2010)
Dietterich, T.: Ensemble methods in machine learning. In: Proceedings of the 1st International Workshop on Multiple Classifier Systems, pp. 1–15 (2000)
Kuncheva, L.I., Smith, C.J., Syed, Y., Phillips, C.O., Lewis, K.E.: Evaluation of feature ranking ensembles for high-dimensional biomedical data: a case study. In: IEEE 12th International Conference on Data Mining Workshops, pp. 49–56. IEEE (2012)
Haury, A.C., Gestraud, P., Vert, J.P.: The influence of feature selection methods on accuracy, stability and interpretability of molecular signatures. PLoS ONE 6(12), e28210 (2011)
Zengyou, H., Weichuan, Y.: Stable feature selection for biomarker discovery. Comput. Biol. Chem. 34, 215–225 (2010)
Awada, W., Khoshgoftaar, T.M., Dittman, D., Wald, R., Napolitano, A.: A review of the stability of feature selection techniques for bioinformatics data. In: IEEE 13th International Conference on Information Reuse and Integration, pp. 356–363. IEEE (2012)
Wang, H., Khoshgoftaar, T.M., Wald, R., Napolitano, A.: A novel dataset-similarity-aware approach for evaluating stability of software metric selection techniques. In: Proceedings of the IEEE International Conference on Information Reuse and Integration, pp. 1–8 (2012)
Kuncheva, L.I.: A stability index for feature selection. In: 25th IASTED International Multi-Conference: Artificial Intelligence and Applications, pp. 390–395. ACTA Press Anaheim (2007)
Somol, P., Novovicova, J.: Evaluating stability and comparing output of feature selectors that optimize feature subset cardinality. IEEE Trans. Pattern Anal. Mach. Intell. 32(11), 1921–1939 (2010)
Dessì, N., Pascariello, E., Pes, B.: A comparative analysis of biomarker selection techniques. BioMed. Res. Int. 2013, Article ID 387673 (2013)
Drotár, P., Gazda, J., Smékal, Z.: An experimental comparison of feature selection methods on two-class biomedical datasets. Comput. Biol. Med. 66, 1–10 (2015)
Wang, H., Khoshgoftaar, T.M., Seliya, N.: On the stability of feature selection methods in software quality prediction: an empirical investigation. Int. J. Soft. Eng. Knowl. Eng. 25, 1467–1490 (2015)
Wald, R., Khoshgoftaar, T.M., Dittman, D.: Mean aggregation versus robust rank aggregation for ensemble gene selection. In: 11th International Conference on Machine Learning and Applications, pp. 63–69 (2012)
Cannas, L.M., Dessì, N., Pes, B.: Assessing similarity of feature selection techniques in high-dimensional domains. Pattern Recogn. Lett. 34(12), 1446–1453 (2013)
Dessì, N., Pes, B.: Similarity of feature selection methods: an empirical study across data intensive classification tasks. Expert Syst. Appl. 42(10), 4632–4642 (2015)
Mesejo, P., Pizarro, D., Abergel, A., Rouquette, O., et al.: Computer-aided classification of gastrointestinal lesions in regular colonoscopy. IEEE Trans. Med. Imaging 35(9), 2051–2063 (2016)
Tsanas, A., Little, M.A., Fox, C., Ramig, L.O.: Objective automatic assessment of rehabilitative speech treatment in Parkinson’s disease. IEEE Trans. Neural Syst. Rehabil. Eng. 22, 181–190 (2014)
Shipp, M.A., Ross, K.N., Tamayo, P., Weng, A.P., et al.: Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat. Med. 8(1), 68–74 (2002)
Petricoin, E.F., Ardekani, A.M., Hitt, B.A., Levine, P.J., et al.: Use of proteomic patterns in serum to identify ovarian cancer. Lancet 359, 572–577 (2002)
Guyon, I., Gunn, S.R., Ben-Hur, A., Dror, G.: Result analysis of the NIPS 2003 feature selection challenge. In: Advances in Neural Information Processing Systems, vol. 17, pp. 545–552. MIT Press (2004)
Rokach, L.: Decision forest: twenty years of research. Inf. Fusion 27, 111–125 (2016)
Acknowledgments
This research was supported by Sardinia Regional Government, within the projects “DomuSafe” (L.R. 7/2007, annualità 2015, CRP 69) and “EmILIE” (L.R. 7/2007, annualità 2016, CUP F72F16003030002).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Pes, B. (2018). Evaluating Feature Selection Robustness on High-Dimensional Data. In: de Cos Juez, F., et al. Hybrid Artificial Intelligent Systems. HAIS 2018. Lecture Notes in Computer Science(), vol 10870. Springer, Cham. https://doi.org/10.1007/978-3-319-92639-1_20
Download citation
DOI: https://doi.org/10.1007/978-3-319-92639-1_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-92638-4
Online ISBN: 978-3-319-92639-1
eBook Packages: Computer ScienceComputer Science (R0)