Abstract
Proliferative activity of cells is one of the most critical factors in breast cancer diagnosis. It is used to evaluate tumor cell progression and to predict treatment responses in chemotherapy. Ki-67 is a nuclear biomarker commonly used to measure cellular proliferation rate. The ratio between the number of Ki-67 positive tumor nuclei and all tumor nuclei defines Ki-67 index. However, manual cell counting is tedious and time consuming because hundreds of nuclei must be labeled. To speed up the analysis process, nuclei can be segmented automatically and then classified based on staining color. Unfortunately, segmentation of individual nuclei is a big challenge because they often create complex clusters comprised of many touching and overlapping nuclei. To deal with complexities and ambiguities of cytological material we propose a generative model which approximates nuclei using ellipses. We assume that the process of generating a cytological sample has stochastic nature. Therefore it is possible to reconstruct this process using marked point process tuned according to observed cytological sample. To verify the potential of the proposed method, we applied it to determine Ki-67 index in breast cancer immunochemistry samples. The results of experiments have shown that Ki-67 indices determined by proposed approach correlate well with those computed manually.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
- Breast cancer
- Ki-67 index
- Nuclei segmentation
- Stochastic process
- Steepest ascent optimization
- Maximum a posteriori estimation
1 Introduction
Biological material necessary for immunochemical examination is taken from affected breast tissue using needle biopsy. The material is then subjected to an immunochemical processing, fixing and staining. Ki-67 positive nuclei are stained by diaminobenzidine (DAB) and have a brown color; negative nuclei appear as blue due to hematoxylin (H) dye. Finally, slide glass with stained material is scanned and evaluated by the pathologist. Ki-67 index is calculated as a ratio between brown-colored nuclei and all nuclei. Individual cases of breast cancer are classified using a fixed cut-off point for the Ki-67 index. During the 2011 St. Gallen International Breast Cancer Conference agreed a cut-off value of 14% based on the study by Cheang et al. [5]. The recommendation had changed in 2013 when most of the panels of St.Gallen conference suggested a threshold of 20%. Finally, the 2015 consensus suggested that cut-off value should be tuned separately for each laboratory based on median value from local results.
The main problem when determining the Ki-67 index is that many pathologists do not have time to visually count the cell nuclei. This process is extremely time-consuming because it requires choosing the most active areas in proliferative rate (hotspots) and then pathologists have to mark manually few hundreds of nuclei. Counting by visual inspection is highly subjective because it is linked to interpretation. Many studies report that inter-observer variability in determining the Ki-67 index is high [16].
Automatic nuclei counting would have allowed avoiding tedious and in fact unnecessary work for pathologist. Also, the process would become free from the subjective assessment of the pathologist. However, we must remember that the accuracy of nuclei segmentation is critical to performance of Computer-Aided Cytology (CAC). Unfortunately, immunochemical images of cytological material are rather challenging for existing object detection methods. In hotspots, they usually create complex, random and heterogeneous structures like clumps and nests. Nuclei are often overlapping and touching. A number of scientific centers conduct an intensive research to develop efficient algorithms for object segmentation. The most common approaches are based on the image thresholding, color deconvolution, data clustering, watershed, active contours and deep learning [4, 8]. One of the well-known systems for determining Ki-67 index is ImmunoRatio [15]. This system estimates the Ki-67 index as the ratio of the brown-colored area of the image to the brown- and blue-colored area of the image. Staining components (DAB and H) are separated using H+DAB color deconvolution technique and intensity thresholding [12]. For the ImmunoRatio to be effective, it must be appropriately calibrated for data from a specific laboratory. There are a lot of reports in the literature about the high accuracy of this system [18]. Saha et al. proposed deep learning framework for hotspots detection and proliferation scoring [13]. They achieved 93% precision, 0.88% recall and 0.91% F-score value. In addition, in their work, we can find a comprehensive list of methods used for estimating the Ki-67 index. In [1] a classifier based algorithm was proposed for Ki-67 scoring in BC tissue microarray images. Proposed approach reached 90% classification accuracy with 0.64 kappa concordance. Xing et al. applied boundary delineation algorithm to localize tumor and nontumor cells for automatic Ki-67 counting [17]. Their algorithm shows promising performance measure in comparison with other popular Ki-67 scoring techniques. Here, we can also find an overview of the other methods used to estimate Ki-67 index.
The aim of our research is to develop and test the new method of Ki-67 scoring based on stochastic geometry. Stochastic geometry is a branch of probability theory that deals with the analysis of random spatial patterns [6]. They are successfully applied wherever there are heterogeneous structures whose spatial distribution is random [7, 10]. We can observe such distribution for cytological material in immunochemical images.
The idea of our approach is to transform H+DAB stained image into a model build up with ellipses which vary in the location, size and orientation. The goal is to find such configuration of ellipses that fit the image data best without violating a priori preferences regarding the distribution of nuclei. The next step is to divide ellipses into two classes due to the color of actual nuclei. This step was carried out with the help of color deconvolution, which allowed us to discover DAB staining areas and H staining areas. The ellipses are by default classified to blue-colored (H) objects and then they are changing their assignment if they are touching the brown-colored (DAB) area. Finally, nuclei models are counted in order to determine Ki-67 index.
To verify the effectiveness of proposed approach, it was applied to estimate Ki-67 index for cytological images of breast cancer. The obtained results were compared with the reference results obtained for manual segmentation and with the results coming from ImmunoRatio.
The remainder of this paper is organized as follows. Section 2 gives the description of methods applied to determine Ki-67 index. Results of experiments are presented in Sect. 3. Concluding remarks are given in Sect. 4.
2 Method
2.1 Method Overview
Before it is possible to build immunochemical models of actual images, pre-processing must be applied to these images to separate the areas stained by H and DAB. To achieve this, the RGB image is subjected to H+DAB deconvolution using the procedure implemented in ImageJ plugin [12]. Three separate intensity images are created as a result. The first represents how much of H has been deposited in nuclei, second how much of DAB has been deposited in nuclei, and third residuals (see Fig. 1). For further processing, we are using the image of H density and DAB density. Next step is carried out to determine binary masks of H area and DAB area. Both masks are determined based on intensity thresholding. The H mask is combined with the DAB mask using AND logic operator. As an effect, we are obtaining H+DAB mask used by stochastic based algorithm to detect elliptical objects which resemble the nuclei. The DAB mask alone cannot be used to segment Ki-67 positive nuclei because in some cases only small part of the nucleus is colored by DAB but anyway this means that the nucleus should be counted as Ki-67 positive. Therefore, H+DAB mask is used to segment all nuclei using stochastic geometry approach, and then DAB mask is used to indicate Ki-67 positive nuclei by checking which ellipses overlaps with the area of the DAB mask. The entire procedure is illustrated in Fig. 1.
The crucial step of the proposed strategy is to find a proper configuration of ellipses that fit the H+DAB mask. Stochastic geometry based approach is trying to cover as many as possible pixels of nuclei area on H+DAB mask using a collection of ellipses of different size and orientation [2, 9]. At the same time, the algorithm is trying to avoid to cover background pixels. Of course, there are many different collections of ellipses that fit well to the given H+DAB mask. To choose the best configuration, we need a prior knowledge about preferable configurations of nuclei. For this purpose, we control pairwise interactions between ellipses to limit the number of overlaps between objects. Thus, configurations with a large number of overlapping ellipses are less likely than those that have less overlaps.
2.2 Marked Point Process
In the considered approach nuclei segmentation in H+DAB image boils down to finding the configuration of ellipses varying in size, location and orientation which covers precisely the H+DAB mask and follows the distribution given by aprior model. To find such a configuration of ellipses, we had to assume that the image generation process can be described using marked point process (MPP). So the crucial element necessary to reconstruct the input image is the knowledge of conditional probability mass function (pmf) \(p(\mathbf x |\mathbf y )\) which governs such process:
where likelihood term \(f(\mathbf y |\mathbf x )\) evaluates the consistency of ellipse configuration \(\mathbf x =\{x_1,\ldots ,x_n\}\) respect to H+DAB mask \(\mathbf y \) and a prior term \(p(\mathbf x )\) reflects constraints on pairwise interactions between ellipses within configuration \(\mathbf x \) [2]:
where \(\alpha ,\beta > 0\) are constants, \(n(\mathbf x )\) is the number of disks in configuration \(\mathbf x \), h is the interaction function and \(\sim \) is a symmetric and reflexive relation describing ellipse overlaps.
If we assume that variables representing mask values \(y_{t} \in \{0,1\}\) are conditionally independent given configuration \(\mathbf x \) then \(f(\mathbf y |\mathbf x )\) takes the following form:
where \(b(y_t;p_{N})\) and \(b(y_t;p_{B})\) are Bernoulli pmf’s:
They are used to evaluate the likelihood of pixels on H+DAB mask \(\mathbf y \) within nuclei region S(x):
and background region \(S \setminus S(x)\) respectively, where S is a pixel lattice of mask H+DAB and \(S(x_{i})\) is the silhouette of the ellipse \(x_{i}\). Parameter \(p_{N}\) describe a probability of occurring actual nuclei pixel within nuclei region S(x) on H+DAB mask \(\mathbf y \), and \(p_{B}\) is a probability of occurring actual background pixel within background \(S\setminus S(x)\). Both parameters were chosen arbitrarily and are shown in Fig. 2.
Finally, we choose Strauss process to implement interaction model [3, 14]:
where \(\alpha ,\beta ,\gamma > 0\) are constants, n(x) is the number of ellipses in configuration and \(r(\mathbf x )\) is the number of pairwise overlaps in configuration \(\mathbf x \). For \(0<\gamma <1\), model exhibits repulsive forces between ellipses and this prevent an excessive number of overlaps in ellipse configurations.
2.3 Optimization
Finding proper configuration of ellipses boils down to optimization problem where \(\varOmega \) is a set of all possible configurations \(\mathbf x \). To solve the problem of nuclei segmentation, we must find in \(\varOmega \) a configuration \(\mathbf x \) that fit the image best without contravene a prior interaction constraints. In Bayesian framework this problem can be viewed as a maximum a posterior estimation problem:
Unfortunately, direct sampling from \(f(\mathbf y |\mathbf x )\) and \(p(\mathbf x )\) is not straightforward. But, the problem becomes much more tractable if we deal with the following proportion [2, 9]:
where \(\mathbf x _{k}\) is the current configuration, \(\mathbf x _{k+1}\) is the new prospective configuration and \(S_{N} = \big (S(\mathbf x _{k+1})\cup S(\mathbf x _{k})\big ) \setminus \big (S(\mathbf x _{k+1})\cap S(\mathbf x _{k})\big )\). If we limit the ways the new configurations \(\mathbf x _{k+1}\) can emerge by allowing only to add single ellipse u or delete single ellipse u from the current configuration \(\mathbf x _{k}\) then it becomes possible to apply steepest ascent procedure to find the local maximum. Algorithm is always choosing new configuration \(\mathbf x _{k+1}\) to maximize proportion w. Therefore, probability never decreases at any stage and eventual convergence is guaranteed. However, algorithm usually stuck in nearest local maxima. The pseudocode of this procedure is presented in Algorithm 1.

2.4 Ki-67 Scoring
Ki-67 index is computed as the ratio between brown-colored nuclei and all nuclei. Steepest ascent procedure segments nuclei and returns the configuration of ellipses that approximates actual nuclei but it is not able to distinguish Ki-67 positive nuclei form Ki-67 negative nuclei. To tackle this problem, we are marking all ellipses which are within the DAB mask as Ki-67 positive (see Fig. 1). Finally, we can quickly count all nuclei by checking how many nuclei are in the found configuration \(\hat{\mathbf{x }}\). The estimate of Ki-67 index is computed as the number of positive ellipses to the number of all ellipses.
3 Results
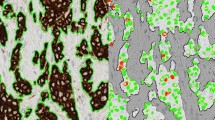
The proposed approach was applied to estimate Ki-67 index in 20 test cases of breast cancer. Immunochemical examinations were obtained for 20 patients from the University Hospital in Zielona Góra, Poland. Immunochemical slides were digitized into virtual slides using the Olympus VS120 Virtual Microscopy System. Selected fragments (size \(500 \times 500\) pixels) of these slides were used in experimental studies (see Fig. 3). Each test image contains from 100 to 400 nuclei. All images were manually marked to get the reference results of Ki-67 scoring. The accuracy of the proposed method was compared with the accuracy of ImmunoRatio system.
To compute Ki-67 index we need segmented nuclei. In our approach stochastic geometry is responsible for extracting nuclei models in the form of ellipses. The method requires an atlas of predefined models of nuclei. In this experiment, we have used atlas comprised of 240 ellipses which vary in sizes and orientations. Each ellipse is described using 3 parameters: major axis length \(r_{M} \in [6,\ldots ,15]\), minor axis length to major axis length ratio \(r_{R} \in [0.5, 0.65, 0.8, 1]\) and orientation \(o \in [0^{\circ }, 30^{\circ }, 60^{\circ }, 90^{\circ }, 120^{\circ }, 150^{\circ }]\). The other parameters that must be defined for stochastic geometry are \(\beta \) and \(\gamma \). As a result of the experiments, it was found that the best results were obtained for \(\ln (\beta ) = -600\), and \(\ln (\gamma ) = -700\).
Segmentation results for all images are presented in Fig. 4. An illustrative example of segmentation results for stochastic geometry, ImmunoRatio, and manual segmentation is presented in Fig. 5. Based on the segmentation results, we were able to estimate the Ki-67 indexes for all test samples. Table 1 summarizes the results obtained for stochastic geometry, ImmunoRatio system and manual segmentation. According to St. Gallen consensus in 2011 and 2013 cut-off points to classify Ki-67 proliferative activity were defined at 14% and 20% levels respectively. We used these cut-off points to classify our results for stochastic geometry, ImmunoRatio, and manual segmentation. Classification results based on automatic segmentation were compared with the reference results using confusion matrices (see Tables 2 and 3). Moreover, we computed Cohen’s kappa coefficient \(\kappa \) to measure agreement between automatic and manual approaches. Stochastic geometry showed high agreement with the reference data because it obtained \(\kappa _{14} = 0.74\) and \(\kappa _{20} = 1\) for the 14% and 20% cut-off points respectively. ImmunoRatio got worse results, \(\kappa _{14} = 0.47\) and \(\kappa _{20} = 0.8\) respectively.
4 Conclusions
We proposed a novel method based on stochastic geometry for estimating Ki-67 index. The preliminary results of the experiments carried out are satisfactory. We showed that this method is more accurate than the approach used in the ImmunoRatio system. Unfortunately, the number of test images is relatively small, thus the statistical significance of the results is not satisfactory. To conduct more reliable tests, a large number of ground truth images must be provided. Unfortunately, manual counting is tedious and requires the involvement of highly qualified medical personnel. For this reason, we decided that the process of building the reference database will be continuous and experimental results will be updated with the expansion of this database.
The segmentation method proposed in the work belongs to a group of methods with high demand for computing power. However, we managed to implement the presented algorithm with intensive use of convolution operation. This allowed us to run our method using parallel computing on GPU. Moreover, we have shown in Eq. 9 that due to iterative nature of the proposed segmentation method only a small part of the prospective ellipses must be evaluated in the consecutive steps of the algorithm. Only in the first step, all prospective ellipses have to be evaluated. Thanks to such manipulations, it was possible to shorten the time needed to process an image containing approximately 250 nuclei up to 2 min.
Future work will concentrate on building more sophisticated models of nuclei based on additional information from input images such as edges and textures [11]. This will allow us to segment the overlapping cell nuclei more accurately.
References
Abubakar, M., et al.: High-throughput automated scoring of Ki67 in breast cancer tissue microarrays from the Breast Cancer Association Consortium. J. Pathol: Clin. Res. 2(3), 138–153 (2016)
Baddeley, A.J., van Lieshout, M.N.M.: ICM for object recognition. In: Dodge, Y., Whittaker, J. (eds.) Computational Statistics, vol. 2, pp. 271–286. Physica-Verlag HD, Heidelberg (1992). https://doi.org/10.1007/978-3-642-48678-4_34
Baddeley, A.J., van Lieshout, M.N.M.: Stochastic geometry models in high-level vision. In: Mardia, K.V., Kanji, G.K. (eds.) Advances in Applied Statistics, Statistics and Images, vol. 1, pp. 231–256. Carfax Publishing, Abingdon (1993)
Bembenik, R., Jóźwicki, W., Protaziuk, G.: Methods for mining co-location patterns with extended spatial objects. Int. J. Appl. Math. Comput. Sci. 27(4), 681–695 (2017)
Cheang, M.C., Chia, S.K., Voduc, D., Leung, S., Snider, J.: Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J. Natl. Cancer Inst. 101(10), 736–750 (2009)
Descombes, X.: Stochastic Geometry for Image Analysis. Wiley, Hoboken (2012)
Descombes, X.: Multiple objects detection in biological images using a marked point process framework. Methods 115(Suppl. C), 2–8 (2017)
Kowal, M., Filipczuk, P.: Nuclei segmentation for computer-aided diagnosis of breast cancer. Int. J. Appl. Math. Comput. Sci. 24(1), 19–31 (2014)
van Lieshout, M.N.M.: Markov point processes and their applications in high-level imaging. Bull. Int. Stat. Inst. 56, 559–576 (1995)
Markowsky, P., Reith, S., Zuber, T.E., König, R., Rohr, K., Schnörr, C.: Segmentation of cell structures using model-based set covering with iterative reweighting. In: 2017 IEEE 14th International Symposium Biomedical Imaging (ISBI 2017), pp. 392–396, April 2017
Mazurek, P., Oszutowska-Mazurek, D.: From the slit-island method to the Ising model: analysis of irregular grayscale objects. Int. J. Appl. Math. Comput. Sci. 24(1), 49–63 (2014)
Ruifrok, A.C., Johnston, D.A.: Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 23(4), 291–299 (2001)
Saha, M., Chakraborty, C., Arun, I., Ahmed, R., Chatterjee, S.: An advanced deep learning approach for Ki-67 stained hotspot detection and proliferation rate scoring for prognostic evaluation of breast cancer. Sci. Rep. 7(1), 3213 (2017)
Strauss, D.J.: A model for clustering. Biometrika 62(2), 467–475 (1975)
Tuominen, V.J., Ruotoistenmäki, S., Viitanen, A., Jumppanen, M., Isola, J.: ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 12(4), R56 (2010). https://doi.org/10.1186/bcr2615
Vörös, A., Csörgő, E., Nyári, T., Cserni, G.: An intra- and interobserver reproducibility analysis of the Ki-67 proliferation marker assessment on core biopsies of breast cancer patients and its potential clinical implications. Pathobiology 80, 111–118 (2013). https://doi.org/10.1159/000343795
Xing, F., Su, H., Neltner, J., Yang, L.: Automatic Ki-67 counting using robust cell detection and online dictionary learning. IEEE Trans. Biomed. Eng. 61(3), 859–870 (2014)
Yeo, M.K., Kim, H.E., Kim, S.H., Chae, B.J., Song, B.J., Lee, A.: Clinical usefulness of the free web-based image analysis application ImmunoRatio for assessment of Ki-67 labelling index in breast cancer. J. Clin. Pathol. 70(8), 715–719 (2017). http://jcp.bmj.com/content/70/8/715
Acknowledgments
The research was supported by National Science Centre, Poland (2015/17/B/ST7/03704).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Kowal, M., Skobel, M., Korbicz, J., Monczak, R. (2018). Stochastic Geometry for Automatic Assessment of Ki-67 Index in Breast Cancer Preparations. In: Rojas, I., Ortuño, F. (eds) Bioinformatics and Biomedical Engineering. IWBBIO 2018. Lecture Notes in Computer Science(), vol 10814. Springer, Cham. https://doi.org/10.1007/978-3-319-78759-6_15
Download citation
DOI: https://doi.org/10.1007/978-3-319-78759-6_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-78758-9
Online ISBN: 978-3-319-78759-6
eBook Packages: Computer ScienceComputer Science (R0)