Abstract
Manual estimation of Ki67 Proliferation Index (PI) in breast carcinoma classification is labor intensive and prone to intra- and interobserver variation. Standard Digital Image Analysis (DIA) has limitations due to issues with tumor cell identification. Recently, a computer algorithm, DIA based on Virtual Double Staining (VDS), segmenting Ki67-positive and -negative tumor cells using digitally fused parallel cytokeratin (CK) and Ki67-stained slides has been introduced. In this study, we compare VDS with manual stereological counting of Ki67-positive and -negative cells and examine the impact of the physical distance of the parallel slides on the alignment of slides. TMAs, containing 140 cores of consecutively obtained breast carcinomas, were stained for CK and Ki67 using optimized staining protocols. By means of stereological principles, Ki67-positive and -negative cell profiles were counted in sampled areas and used for the estimation of PIs of the whole tissue core. The VDS principle was applied to both the same sampled areas and the whole tissue core. Additionally, five neighboring slides were stained for CK in order to examine the alignment algorithm. Correlation between manual counting and VDS in both sampled areas and whole core was almost perfect (correlation coefficients above 0.97). Bland–Altman plots did not reveal any skewness in any data ranges. There was a good agreement in alignment (>85 %) in neighboring slides, whereas agreement decreased in non-neighboring slides. VDS gave similar results compared with manual counting using stereological principles. Introduction of this method in clinical and research practice may improve accuracy and reproducibility of Ki67 PI.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ki67 is a nuclear protein expressed in all active phases (G1, S, G2, and M) of the cell cycle [1]. This makes Ki67 an excellent marker for cellular proliferation. Ki67 can be visualized immunohistochemically, which makes it possible to calculate a Proliferation Index (PI) by determining the fraction of Ki67-stained tumor cells. High Ki67 PI is in many tumor types associated with malignancy, and in many cancer types such as breast carcinoma [2] and endocrine neoplasms [3, 4] associated with a higher probability of relapse and decreased survival.
In breast carcinoma, Ki67 PI has been suggested as a surrogate marker to discriminate the molecular subtype groups of luminal type A and type B. The St Gallen international expert consensus report from 2011 suggested a cut-point of 14 % based on a gene expression study, although this was later changed to a more vague formulation of “Ki67 low or high” [5, 6]. According to the WHO classification, Ki67 PI can be used in neuroendocrine neoplasia as an alternative to counting mitoses. The cut-points of 3 and 20 % are used to classify grade 1 to 3 neuroendocrine neoplasias [3]. No matter the exact level, cut-points challenge clinical practitioners of surgical pathology to deliver precise Ki67 counts. Several studies document this challenge both in regard to intra- and interobserver variation when using “eye ball” estimation [7–9]. Standardized counting methods are more precise but time consuming [10, 11].
Both commercial and public Digital Image Analysis (DIA) systems offer quantitative analysis of Ki67. As an example, the freely available ImmunoRatio can calculate a PI from an uploaded image albeit of limited size [12]. Several authors have examined computer-assisted scoring of proliferation. One group found correlation between human and computer-assisted assessment of Ki67-based PI in breast carcinomas [13]. Another group found that digital assessed Ki67 in breast carcinomas added prognostic value to the traditional mitotic count [14].
One of the barriers for DIA is the current requirement for detailed supervision by a pathologist in order to ensure that the analysis is performed on the correct subset of cells. Tumor tissue contains non-neoplastic cells that will influence the calculated PI, either by underestimation (due to e.g., non-proliferating stromal cells) or overestimation (due to e.g., proliferating lymphocytes). One group addressed this issue in DIA of malignant melanoma by applying a physical double stain that allowed identification and analyses of the melanocytic compartment of the tissue [15]. However, overlapping chromogens may impair the analysis.
In order to circumvent the issues of physical double stains a new method, Virtual Double Staining (VDS), has been developed, which is able to digitally delimitate tumor areas using one section stained with a tumor marker (such as Pan Cytokeratin (PCK) in epithelial tumors) and transfer these areas to a parallel slide stained for another marker, such as Ki67. During the process, the algorithm digitally fuses parallel slides and performs local rotation and distortion of the image in order to correct small procedure-related differences between the two neighboring slides.
The purpose of this study was to compare the VDS-based DIA (abbreviated VDS) of Ki67 expression with the manually obtained PIs by Systematic Uniform Random Sampling (SURS). This was achieved by examining Ki67 PIs in tissue microarrays (TMAs) containing cores of breast carcinomas. Additionally the digital alignment performed by the algorithm was examined by comparing the overlap of PCK-stained tumor tissue in five parallel sections.
Materials and methods
Tissue
Tissue samples from 140 resection specimens of breast carcinomas, consecutively obtained from the archives of the Institute of Pathology, Aalborg University Hospital, Denmark. All tissues had been routinely fixed for 24–48 h in 10 % neutral-buffered formalin and subsequently paraffin embedded. Areas containing tumor tissue, but not carcinoma in situ (CIS) and normal glandular epithelium, were marked in a hematoxylin and eosin-stained slide and cores with diameter of 2 mm were drilled out using an automated microarray device (3DHISTECH, Hungary). These cores were transferred to three recipient paraffin blocks and fused in an oven for 20 min at 37 °C and for 10 min at 60 °C. Ki67 PI assessed by an experienced breast pathologist at the primary diagnostic examination of the whole tumor varied between 2 and 70 %.
Immunohistochemistry
Sets of two neighboring 3-μm-thick slides were cut from each of the TMAs and mounted on positively charged slides (SuperFrost + , Menzel Gläser, Germany), dried at room temperature and baked for 1 h at 60 °C. For each set, slides were stained for either Ki67 or PCK using staining protocols optimized according to guidelines of the NordiQC organization (www.nordiqc.org). In short, deparaffinization, rehydration, and heat-induced epitope retrieval were performed on the Ventana Benchmark Ultra. Epitope retrieval was performed in Cell Conditioning 1 (cat. 950-124; Ventana, USA), pH 8.5 at 99 °C for 64 min. Endogenous peroxidase was blocked with 3 % hydrogen peroxide (ultraView DAB, cat. 760–500; Ventana). Primary antibodies for Ki67, clone MIB-1 (Dako, Denmark) and PCK, clone AE1/AE3 (Dako) were diluted (Ki67: 1:200, PCK: 1:100), and incubated 20 min at 36 °C. Diaminobenzidine (ultraView DAB, cat. 760–500; Ventana) staining was developed using a multimer-based visualization system (ultraView DAB, cat. 760-500; Ventana). For the second part of the experiment analysis, the consistency of the digital alignment algorithm performed as part of the VDS, five serial sections were cut from the same TMA and stained for PCK using the above-mentioned protocol settings.
Scanning and image analysis platform
All slides were scanned using a Hamamatsu NanoZoomer XR (Hamamatsu Photonics, Japan) applying the ×40 scanning resolution mode (equivalent of 400 times magnification) in a single focus plane automatically identified by the machine. The images of the scanned slides were transferred to an analysis platform with VIS (Visiopharm Integrator System version 4.6.1.630, Visiopharm, Denmark) Image Analysis Software installed.
Manual counting
Every core of breast carcinoma was manually assessed for the Ki67 PI calculated as the number of Ki67-positive tumor cells divided by the total number of tumor cells. Since manual counting of all tumor cells would be too time consuming, areas were selected according to the SURS principle in order to calculate an unbiased estimate of the Ki67 PI. This was done using the stereological newCast function of the VIS program. First, the computer randomly placed a grid covering a known percentage of the total core area. The grid was designed to avoid borders in order to eliminate border confounding and dilution. The area percentage used was calculated from an initial estimation of the number of tumor cells in each slide to include no less than 200 tumor cells. Stereological counting frames [16] covering the estimated area percentage were used to avoid overestimation of cell profiles. All tumor cell profiles within the counting frame were counted as follows: Profiles completely within the counting frame, as well as profiles touching the upper or right border of the frame were included, whereas profiles touching the lower or left border were excluded. A specifically trained pathologist (RR) marked all tumor cells as either Ki67-positive or Ki67-negative. All nuclei, stained with an at least weak but distinctive brown color, were considered positive. Only tumor cells were marked. Complicated cases were discussed with another experienced pathologist (MV). Cores estimated to contain less than 20 % tumor cells were excluded from the analysis. Because the counted areas were selected according to the SURS principle, manually counted PI could be extrapolated to an estimate of the whole core.
Virtual double staining, pan cytokeratin and Ki67 analysis
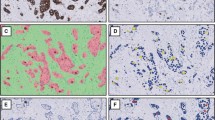
The main principle of VDS consists of digitally aligning scanned images of two parallel slides and fusion to one image (illustrated in Fig. 1). First, all scanned slides containing pairs of parallel scanned slides (one stained for PCK and one for Ki67) were loaded into the ‘TMA Workflow’ module of VIS. The software then created separate images of each core for further analysis. As a final step, the software aligned each paired core using rotation and local deformation.
The VDS principle. Left tile: Cytokeratin-stained slide where the automated image analysis detects positive areas (tumor cells) and mark these (green line) as the region of interest. Right tile: Ki67-stained slide with cells profiles classified as either positive (red) or negative (green) in the regions of interest. Scattered weakly stained nuclei are unclassified. Stromal areas (in gray) are excluded from the analysis
The next part of the image analysis was performed in the ‘Image Analysis’ module of VIS. Each of the aligned images was reviewed manually. Cores missing one of the stained slides were excluded from further analysis. Areas containing large tissue artifacts (e.g., tissue folds and cracks) were either excluded from analysis or marked as ‘Region of no interest’. After this, all cores were analyzed using a batch process where the two analysis protocols were run in succession. In brief, PCK-positive areas were enhanced using filtering of the RGB pixel values and segmented in ‘Tumor cell area’ and ‘Stroma’ using a Bayesian classifier based on in-program stored predefined values. These marked areas were then transferred to the image of the Ki67-stained slide for further analysis, where nuclear profiles were detected based on form and size, and segmented as either Ki67-positive or -negative based on pixel-color intensity cut-points. Before analysis, several test sets of breast carcinomas had been examined by manual visual inspection in order to select the best cut-point between Ki67-positive and -negative cells. Finally, the numbers of positive and negative cells, and calculated PI were exported from the program. For all cores, digital PIs were calculated for both the whole core and the SURS areas used for manual counting.
Pan cytokeratin overlap
In order to quantify the potential source of error introduced by small spatial differences between the two slides used in the VDS, and to examine the proficiency of the alignment algorithm, a second experiment measuring the agreement in overlap between slides was setup. Five serial slides were cut from one of the TMA blocks and stained for PCK. To examine the importance of increasing spatial distance on the degree of overlap, the VDS algorithm was applied to different slide pairs: Neighboring (1–2, 2–3, 3–4, and 4–5) and non-neighboring (1–3, 1–4, and 1–5). After this, an adapted version of the PCK algorithm segmented each core in four areas: ‘PCK positive areas both slides’ (+/+), ‘PCK negative areas both slides’ (−/−), and ‘PCK positive areas in one slide’ (±) and (∓) in another, see Fig. 2. Agreement percentages were calculated as the double positive and double negative marked PCK areas divided by the total core area.
Statistics
Statistical analysis and data preparation was performed with the RStudio software (version 0.98.1091) using reproducible scripts (available on request). Figures were produced using the Grammar of Graphics implementation ‘ggplot2′-package. Intra Class Coefficients were calculated using the psych package. P-values lower than 0.05 were considered statistically significant.
Results
Material
Of the 140 cores of breast carcinoma contained in the three TMAs, 103 were analyzed further. 37 cores were excluded due to one of the following reasons: Missing cores in either the PCK- or Ki67-stained slide (N = 12), no or very small amounts of tumor tissue (n = 9) or large areas with folding (N = 16). Of the 103 cases, 83 consisted of ductal carcinomas (NOS), 13 of lobular carcinomas, and seven of other subtypes (apocrine, medullary, basal, tubular, and combined ductal and lobular carcinoma). Clinical summary of the cores are shown in Table 1.
Manual counting in sampled areas
Sampled areas of the tumor cores varied between 4 and 20 % of the total core area. Table 2 lists summary statistics for manual and digital cell profile count and calculated PIs. The number of tumor cell profiles counted inside the frames in each core varied between 202 and 622 (median 295). The derived PIs varied between 0.0 and 73.0 % (median 11.5 %).
VDS in sampled areas
The VDS algorithm was applied to the same sampled areas as used for the manual counting. In these areas, the algorithm identified a median of 258 cell profiles (range 37–420). Overall (74 % of cases), the algorithm identified 14.1 % fewer cell profiles than the human observers. This percentage did vary somewhat (SD = 24.5 %) and had four outliers (> ±50 % of the manual count). All these cases were diffusely infiltrating tumors. The calculated PI varied between 0.0 and 86.0 % (median 10.6 %).
VDS on whole core
The PCK-stained areas constituted on average 36 % (SD = 20 %) of the total core area. In these areas, a median of 5,262 cell nuclei profiles were counted. Counts varied between 602 and 27,467 cells. PI varied between 0.0 and 83.2 % (median 10.4 %).
Correlation of manual counting and VDS
Correlation of manual estimation of Ki67 proliferation in sampled area, VDS in the same sampled areas, and VDS of the whole core are shown in Fig. 3. One extreme data point (PI >80 %) was removed from the plots, but not from the data analysis, to improve visual interpretation.
There was a good correlation between manual counting and VDS in the same sampled areas. R2 value for linear fit was 0.96. Using Bland–Altman plots, a small tendency to lower PI (mean difference 0.4 %) when using VDS was seen. The plot did not reveal any skewness in specific data ranges. The high degree of correlation was confirmed using Intra Class Coefficient (ICC) calculation where all relevant coefficients, analyzing absolute correlation and consistency, were 0.98 or above (CI: 0.97–0.99).
Since the areas were sampled according to the SURS principle, calculated PI also served as estimates of PI in the whole core. This allowed direct comparison of PI in manual counted sampled areas and VDS of the whole core. The same high level of correlation was observed when comparing manual counting in sampled area to VDS on the whole core. R2 value for linear fit was 0.95. The Bland–Altman plot also showed a tendency to lower PI when using VDS (mean difference was 0.8 %). No skewness was observed across the data. ICC coefficients were all 0.97 or above (CI:0.96–0.99).
Digital estimation of PI was also applied to the whole tissue core without applying the VDS principle (i.e., not excluding the stromal cells) in order to demonstrate unsupervised DIA. This affected the PI significantly (Fig. 4) as the unsupervised PI on average was 4 % (range 1–28 %) lower. ICC comparing DIA and manual counting or VDS varied between 0.78–0.92 and 0.82–0.93, repectively.
There was no significant difference in correlation between VDS and manual counting among the different tumor types (Fig. 5).
PIs were also calculated for stromal tissue (areas identified as PCK-negative but not background or holes). In all tissues, stromal proliferation was lower than 12 %, most being lower than 6 %. There was no statistical significant correlation between PI in stromal and tumor tissue. Cell profile density varied between the two compartments (median ~5,800/mm2 in PCK-positive cells and ~3,200/mm2 among stromal cells).
Overlap experiments
One TMA containing 40 cores of breast carcinoma was examined in five serial sections stained for PCK. Of these cores, 14 were excluded due to missing tissue in one or more of the sections (n = 9), small amounts of tumor tissue (<10 % of total area; n = 2) or large areas with folding tissue (n = 3) impairing correct alignment. Of the remaining 26 cores, mean agreements among neighboring and non-neighboring slides were 91 % (range 82–98 %) and 82 % (range 58–97 %), respectively. Variance in agreement for each core was stable across the neighboring slides with a mean coefficient of variance (CV) of 1.5 %, but varied in most non-neighboring cores significantly with the distance between slides, mean CV 5 % (range 1–11 %).
Comparison of agreement in neighboring and non-neighboring slides can be seen in Fig. 6.
The theoretical impact of suboptimal alignment of slides was calculated using the formula:
Based on the results above, as an example, a tumor with a “true” PI of 20 % would be estimated to 18.5 % in slides with 90 % agreement (assuming 5 % proliferation in stromal cells and same stromal cell density). Figure 7 shows the theoretical impact of agreement on tumors with different PI.
Clinical impact
All examined tumors were classified according to the St. Gallen suggested cut-off point of 20 % as either Luminal type A or B [6]. When manual counting was used, 74 % of cases was classified as Luminal Type A, while this was the case in 75 % of tumors when using VDS in the SURS areas (see Table 3). When VDS was applied on the whole core, all tumors were classified as the manual result. Using DIA on the whole core without applying VDS, a significant higher proportion (85 %) of tumors was classified as Luminal Type A.
Discussion
In this study, we investigated the usability of the VDS principle combined with DIA for the selection of tumor areas in digital slides and calculation of Ki67 PI. Our results showed that PI obtained by the VDS method were in good agreement with the current gold standard of manual counting using stereological principles.
Some differences (Table 2) were observed in the absolute number of cells counted, where the DIA identified fewer nuclei than the human observer. This was due to missed cells with weakly defined nuclei and small differences in the alignment. Despite this, calculated PIs were in good agreement.
The VDS principle is based on the digital alignment of two slides cut from the same block. Our investigation also showed that the physical distance between the selected slides, impacted the degree of the alignment (Fig. 6). In solid tumors, agreement of alignment was good even between non-neighboring slides. In contrast, aligning tumors with complex or diffuse growth pattern were affected when the distance between the slides increased. Figure 7 shows the impact of misalignment. In our experiment, agreement in all cores except one was above 85 % when using neighboring slides but significantly lower in some non-neighboring slides. Consequently, only VDS using neighboring slides can be recommended.
The algorithm for alignment was challenged in tumor areas with single cell growth pattern and did in these cases include varying amounts of stromal cells or excluded obvious tumor cells. This could potentially introduce errors, especially in lobular carcinomas. Interestingly, in this study, this did not impact the calculated PIs in these carcinomas significantly, as they were still comparable to that of the human observers. Ki67 PI among the lobular carcinomas in this study varied between 0–45 %, most being below 10 % (median 7 %), similar to other study cohorts [17]. The good agreement despite dilution with stromal tissue may in these cases probably be explained by the somewhat similar Ki67 PI in the neoplastic and stromal tissues.
Normal glandular epithelium and CIS are PCK-positive and will be counted as tumor areas possibly confounding the PI. However, in our study the cores were selected to exclude normal epithelium and CIS. In a clinical setting of full slides, this may prevent a fully unsupervised analysis. One solution could be to expand the analysis with a basal cell marker (e.g., p63 or heavy chain smooth muscle myosin) in virtual triple staining where the cell groups lined with basal markers would be excluded from analysis. Unfortunately, this method was not available to us, but could be interesting to apply in a new study.
Tumors may contain non-neoplastic tumor-infiltrating lymphocytes (TILs) between the neoplastic cells. TILs will be included in the cell counts giving misleading results if their PI deviates from the tumor cell PI. Especially medullary carcinomas contain many TILs [18]. In this experiment, VDS calculated one PI higher and another PI lower than the human observer in the two tumors with medullary features, perhaps indicating that this source of error may be of minor importance. However, more samples are needed to clarify this.
The findings of this study—in terms of both manual counting and VDS—must be interpreted within their limitations: All analyses were carried out on slides stained using in-house optimized protocols selected to secure the highest degree of sensitivity and minimal background staining (best signal to noise ratio). Differences in staining methodology (antibody clone, protocol, and staining platform) among laboratories may challenge computer algorithms due to differences in texture and color nuances (e.g., nuclear morphology or red/brown chromogen). Implementation of standardized immunohistochemical staining protocol parameters across laboratories would reduce this problem significantly. Adding correction factors based on common controls may help overcome day-to-day variations.
Using strict stereological theory, assessing PI in just one non-systematically sampled slide introduces several possible errors; first, the calculated PI may not represent a true estimate of the whole tumor. Secondly, Ki67-positive cells are on average larger than negative cells (in this study with a ratio of 1.09) and thus more likely to be represented in a cut section. Size differences between positive and negative cells may be due to physical differences but could also be related to coating with chromogen. To overcome these problems, one could analyze several systematically randomly selected sections from each tumor implementing the physical dissector principle [19]. This may be suited for future research designs to evaluate the size of the potential bias, but truly randomized sectional planes may interfere with the assessment of resection margins, and the physical dissector may be too laborious for daily diagnostic use.
To the best of our knowledge, this is the first study examining the usability of automated tissue detection and Ki67 calculation using the VDS DIA principle in order to establish a standardized method to obtain objective and reproducible results. Future studies of breast carcinomas will show if this method will improve the prognostic value of the Ki67 assay, potentially identifying the best cut-point in PI between tumor subtypes. The potential of this method ought also to be examined in other tumor types such as neuroendocrine neoplasia across different organs.
In conclusion, the VDS principle for selection of tumor areas, combined with DIA determining a PI of Ki67 in immunohistochemically stained slides, gave similar results compared with the current gold standard of manual counting using stereological principles. Introduction of these methods may improve accuracy and reproducibility, and decrease inter-laboratory variability, thus increasing the usability of the Ki67 marker in research and clinical settings.
References
Scholzen T, Gerdes J (2000) The Ki-67 protein: from the known and the unknown. J Cell Physiol 182:311–322. doi:10.1002/(SICI)1097-4652(200003)182:3<311:AID-JCP1>3.0.CO;2-9
de Azambuja E, Cardoso F, de Castro G et al (2007) Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. Br J Cancer 96:1504–1513. doi:10.1038/sj.bjc.6603756
Bosman FT, Carneiro F, Hruban R, Theise N (2010) Classification of tumours of the digestive system. WHO, Lyon
Bettini R, Boninsegna L, Mantovani W et al (2008) Prognostic factors at diagnosis and value of WHO classification in a mono-institutional series of 180 non-functioning pancreatic endocrine tumours. Ann Oncol 19:903–908. doi:10.1093/annonc/mdm552
Goldhirsch A, Wood WC, Coates AS (2011) Strategies for subtypes–dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 22:1736–1747. doi:10.1093/annonc/mdr304
Goldhirsch A, Winer EP, Coates AS (2013) Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 24:2206–2223. doi:10.1093/annonc/mdt303
Shui R, Yu B, Bi R et al (2015) An interobserver reproducibility analysis of Ki67 visual assessment in breast cancer. PLoS ONE 10:e0125131. doi:10.1371/journal.pone.0125131
Mikami Y, Ueno T, Yoshimura K et al (2013) Interobserver concordance of Ki67 labeling index in breast cancer: Japan Breast Cancer Research Group Ki67 ring study. Cancer Sci 104:1539–1543. doi:10.1111/cas.12245
Polley M-YC, Leung SCY, McShane LM et al (2013) An international Ki67 reproducibility study. J Natl Cancer Inst 105:1897–1906. doi:10.1093/jnci/djt306
Polley M-YC, Leung SCY, Gao D et al (2015) An international study to increase concordance in Ki67 scoring. Mod Pathol 28:778–786. doi:10.1038/modpathol.2015.38
Laurinavicius A, Plancoulaine B, Laurinaviciene A et al (2014) A methodology to ensure and improve accuracy of Ki67 labelling index estimation by automated digital image analysis in breast cancer tissue. Breast Cancer Res 16:R35. doi:10.1186/bcr3639
Tuominen VJ, Ruotoistenmäki S, Viitanen A et al (2010) Immunoratio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res 12:R56. doi:10.1186/bcr2615
Fasanella S, Leonardi E, Cantaloni C et al (2011) Proliferative activity in human breast cancer: ki-67 automated evaluation and the influence of different Ki-67 equivalent antibodies. Diagn Pathol 6(Suppl 1):S7. doi:10.1186/1746-1596-6-S1-S7
Gudlaugsson E, Klos J, Skaland I et al (2013) Prognostic comparison of the proliferation markers (mitotic activity index, phosphohistone H3, Ki67), steroid receptors, HER2, high molecular weight cytokeratins and classical prognostic factors in T1−2N0M0 breast cancer. Pol J Pathol 64:1–8
Nielsen PS, Riber-Hansen R, Jensen TO et al (2013) Proliferation indices of phosphohistone H3 and Ki67: strong prognostic markers in a consecutive cohort with stage I/II melanoma. Mod Pathol 26:404–413. doi:10.1038/modpathol.2012.188
Gundersen HJ, Bendtsen TF, Korbo L et al (1988) Some new, simple and efficient stereological methods and their use in pathological research and diagnosis. APMIS 96:379–394
Narbe U, Bendahl P-O, Grabau D et al (2014) Invasive lobular carcinoma of the breast: long-term prognostic value of Ki67 and histological grade, alone and in combination with estrogen receptor. Springerplus 3:70. doi:10.1186/2193-1801-3-70
Alvarenga CA, Paravidino PI, Alvarenga M et al (2012) Reappraisal of immunohistochemical profiling of special histological types of breast carcinomas: a study of 121 cases of eight different subtypes. J Clin Pathol 65:1066–1071. doi:10.1136/jclinpath-2012-200885
Sterio DC (1984) The unbiased estimation of number and sizes of arbitrary particles using the disector. J Microsc 134:127–136
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical standards
All experiments in this article were performed in accordance to Danish law.
Conflicts of interest
RR and MV have collaborated with Visiopharm in the development of the software. MV is member of the Visiopharm Scientific Advisory Board. The remaining authors declare that they have no conflict of interest.
Disclosures
None.
Rights and permissions
About this article
Cite this article
Røge, R., Riber-Hansen, R., Nielsen, S. et al. Proliferation assessment in breast carcinomas using digital image analysis based on virtual Ki67/cytokeratin double staining. Breast Cancer Res Treat 158, 11–19 (2016). https://doi.org/10.1007/s10549-016-3852-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-016-3852-6