Abstract
Alzheimer disease (AD) is a common form of dementia affecting people older than the age of 65. Moreover, AD is commonly diagnosed by behavioural paradormants, cognitive tests, and is followed by brain scans. Computer Aided Diagnosis (CAD), applies medical imaging and machine learning algorithms, to aid in the early diagnosis of Alzheimer’s severity and advancement from prodromal stages i.e. Mild Cognitive Impairment (MCI) to diagnosed Alzheimer’s disease. In this work, SVM (support vector machine) is used for dementia stage classification. Anatomical structures of the brain were obtained from FreeSurfer’s processing of structural Magnetic Resonance Imaging (MRI) data and is utilized for as features for SVM. To be more precise, the system is processed using T1-weighted brain MRI datasets consisting of: 150 mild cognitive impairment (MCI) patients, 80 AD patients and 130 normal controls (NC) obtained from Alzheimer Disease Neuroimaging Initiative (ADNI) database. The volumes of brain structures (hippocampus, medial temporal lobe, whole brain, ventricular, cortical grey matter, entorhinal cortex and fusiform) are employed as biomarkers for multi-class classification of AD, MCI, and NC.
Alzheimer’s Disease Neuroimaging Initiative-Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
- Alzheimer disease
- Mild cognitive impairment
- Normal control
- Structural magnetic resonance imaging
- FreeSurfer
- Machine learning
- SVM
1 Introduction
Alzheimer’s disease (AD) is a common form of dementia affecting millions of elderly people above the age of 65 worldwide. Before AD, ailments such as (MCI) serves as an intermediary phase between normal cognitive controls (NC) and AD. Furthermore, this MCI phase has a high conversion rate to AD. As a result, there is a need for the development of a sensitive, precise, and specific atrophy biomarkers for early detection of AD progression [1]. These new methods are needed to help researchers develop new treatments for Alzheimer’s as discussed by Hua et al. [2]. Methods for early detection may further differ by the type of imaging biomarkers that can be applied [3,4,5]. For example, neuroimaging methods such as positron emission tomography (PET), functional magnetic resonance imaging (fMRI) and structural magnetic resonance imaging (MRI) are useful in evaluation of anatomical degradation caused by the disease [6,7,8]. Overtime, structural MRI of the brain has progressively become more employed in identifying structural changes in common aging diseases like Alzheimer’s [9]. Structural brain MRI methods have the ability to utilize biomarkers that are presented in the image. These biomarkers are able to illustrate the structural differences for a healthy and diseased individuals. It is important to note that the methods may vary depending on the nature of the employed imaging biomarkers [10]. In spite of this, due to the ease of availability, non-persistent nature, and a high quality of MR images, they are the most suitable for differentiating changes in the brain anatomy due to disease development and progression.
As a result of the ubiquitous use of MRI in research and medicine, simultaneous advances in neuro-informatics have led to the materialization of many free and commercial image analysis software packages for the last 15 years. This includes but is not restricted to SPM, FSL, FreeSurfer, BrainVisa, Minboggle, NeuroQuant and NeurQlab. Premature diagnosis of AD by structural MRI studies is a challenging task because of its difficulty in quantifying patterns seen in the structural changes during early phases of AD or clinically normal phases [11]. Patients at the early stages of AD are classified as MCI, but not all MCI patients convert to AD. An analysis of research and clinical reports show that 5–10% of MCI patients convert to AD per year [12]. Voxel based (VBM) morphometry from high-resolution T1-weighted brain MRI data has been employed for diagnosis. Furthermore imaging biomarkers were obtained from the processed images such as grey matter concentration maps which are registered to a reference location for facilitating voxel by voxel comparisons across subjects [13]. In this work, we focus on the volumetric measurements of various brain structures as they have an impact on dementia diagnosis. Specifically, MCI is known to be effected by volume loss of brain structures like the hippocampus, MTI, the entorhinal cortex, and the total volume of the brain and is therefore exploited for classification.
Kloppel et al. applied support vector machine (SVM) to classify grey matter segments in T1-weighted MR scans obtained from diagnosed AD patients and the NCs obtained from two centers with dissimilar scanning equipment in order to generalize across different medical centers [14]. Magnin et al. proposed a new classification method of whole-brain (1.5-T) MRI to discriminate AD patients from NC subjects based on SVMs [15]. Here brain is divided into five regions of interest by using a previously developed anatomically labelled template for the brain and created a mask to exclude voxels of the skull. Vemuri et al. [16] developed a tool for Alzheimer’s diagnosis through structural classification of MRI using SVM. As the dimension of these brain structures is collinear, it is essential to know which of them is more likely linked to severity of illness; the amount of atrophy in the other explains further variation in overall symptom severity. The studies in this field typically evaluate the diagnostic accuracy of AD and MCI patients with healthy control subjects. This study proposes volumetric measurement of hippocampus, medial temporal lobe, ventricles, amygdala, whole brain volume, cortical grey matter and entorhinal cortex and fusiform structures used as MRI biomarkers to predict different forms of dementia including the AD and the MCI. The MRI database scan for the proposed work has been taken from the AD Neuroimaging Initiative (ADNI) [17]. FreeSurfer software is employed to obtain hippocampal, MTL, and whole brain volumes, as well as ventricles, amygdala and cortical grey matter by cortical and sub-cortical segmentation. Furthermore, the SVM classification from LibSVM package is utilized for multi-class classification of AD, MCI and NC.
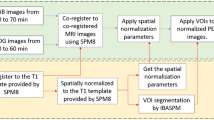
The organization of this paper is as follows. In Sect. 2, possible volume biomarkers of AD are discussed. In the first part of Sect. 3, the dataset and the FreeSurfer tool are briefly presented, followed by explaining the inner workings of SVM for Alzheimer’s classifications. The whole process of classification is given in Fig. 1. Section 4 is devoted for discussing the performance of the presented method. Finally, in Sect. 5 the conclusion and the future work are communicated.
2 Volume Biomarkers of AD
Manual volumetric measurements of brain structures is regarded as “the gold standard” for detecting symptoms of AD. However, it is time consuming and has an operator bias. In comparison, automatic measuring methods such as voxel-based morphometry (VBM) are fast and are extensively employed in the field [23,24,25]. However, this method is not to define every gyrus in the brain and is criticized by some to have confounding issues [26]. Lies et al. has addressed some of these issues where it is found that a VBM method is measuring the same effects as “the gold standard” concerning to the subcortical brain structures [27]. Overall, major structures in the brain like hippocampus, medical temporal lobe, ventricles, amygdala, cortical grey matter, entorhinal cortex and the whole brain volume are investigated for indications of atrophy that lead to AD.
2.1 Hippocampus
The hippocampus creates the majority of the temporal lobe and is commonly used for AD diagnosis. Moreover, hippocampal atrophy is a well-known cause of dementia [9]. Specifically, hippocampal atrophy differentiates the three main disease stages of AD, MCI and NC [21]. It is also speculated that a low hippocampus volume can be utilized as a new diagnostic criterion for MCI patients with high risks of AD conversion [11].
2.2 Medial Temporal Lobe
The medial temporal lobe (MTL) region contains structures that are key in long-term memory. As a result, a structural MRI of the MTL’s atrophy is an effective indicator for the initial diagnosis of AD. Visser et al. reported these results in 1999 among 45 patients in their study [17].
2.3 Ventricles
Ventricles are cavities in the cerebral hemispheres filled with cerebrospinal fluid. Furthermore, their volume variations indicate the existence of AD. These cavities are found to expand in size steadily in AD patients [20]. In particular, Apostolova et al. has reported that the use of cerebral ventricular volume for measurement of AD development. They claimed that the hemispheric atrophy rate calculated by ventricular enlargement correlates strongly with changes on cognitive tests and are able to capture significant variations among levels the stages of Alzheimer’s [18].
2.4 Amygdala
Amygdala is a primary limbic structure anatomically interconnected with the neocortex. In particular, the amygdala serves as a structure for how emotions are processed. In cases of AD, neural lost and alterations in glial cell population have been reported. In support, Poulin et al. reported the magnitude of amygdala atrophy is considerable in AD stages [19].
2.5 Whole Brain
Volumetric MRI studies have found relationships between increasing age and decreasing brain volumes. In particular, there is an age-correlated decrease in hippocampal, temporal, frontal lobe structure volumes, and an increase in cerebrospinal spaces [20]. Moreover, there are more sensitive predictors of AD and MCI are achievable by exploiting the whole brain’s atrophy rate along with the hippocampal volume [21].
2.6 Cortical Grey Matter
MRI measurements of cortical grey matter and abnormal white matter are independently connected with dementia severity. Both biomarkers have their own contributions to the performance in MCI domains as well. For example, quantitative MRI provides a strong conformation that cortical grey matter volume are related to atrophy and abnormal white matter volume are separately related to the dementia severity in AD subjects [22].
2.7 Entorhinal Cortex
Entorhinal cortex is a key pre-processor that stimulates the nearby hippocampus. It serves as an area for memory and navigation. Examinations have confirmed this assumption; also, few observations illustrate that entorhinal cortex is the primary part which is affected in MCI cases even earlier than hippocampus.
3 Materials and Methods
3.1 Data
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). The ADNI was collectively launched by six non-profit organizations in 2003: the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and available at adni.loni.usc.edu. It aims to assess whether structural MRI, positron emission tomography (PET), biomarkers, as well as clinical and neuropsychological assessments can be collectively measure the progression of MCI and early AD. The dataset is divided into categories of AD, MCI and NC, where MCI consists of EMCI and LMCI as shown in Table 1.
3.2 FreeSurfer Processing
FreeSurfer is one of the most widely used software today for volumetric analysis of the brain. It is indeed a set of tools for cortical analysis and visualization and sub-cortical segmentation of MRI data [28]. Accurate and reliable segmentation is a necessity for volumetric analysis of dementia disease. Here, sub-cortical and cortical volumetric measurements were computed by FreeSurfer (version 5.3.0) using atlas based labelling of region of interest (ROI) [29]. Statistical output files generated during FreeSurfer processing stream was used to obtain hippocampus volume and intra-cranial volume (ICV). The volumes of medial temporal lobe, ventricles, amygdala, CGM, entorhinal cortex, fusiform, and the whole brain was computed using anatomical ROI segmentation analysis of their given file: aparc.a2009s+aseg.mgz. The volume of each structure is found by counting the voxels of each of these coloured and labelled structure using an .mgz image that FreeSurfer outputs by using MATLAB. Each volume calculated was then normalized by dividing them with the intra-cranial volume (ICV). The ICV was found from surfer.nmr.mgh.harvard.edu with three aseg.stat files available at 7 head-sized corrections to reduce inter-individual variation. FreeSurfer processing is computationally expensive and takes several hours to process a single image. Therefore, in order to reduce computational time, eight images are processed in parallel using GNU Parallel on an 8 core machine.
Support Vector Machine. Support vector machine is a machine learning method that classifies binary classes by finding a class boundary. This boundary, the hyper plane, is used to find the maximum margin in the given training data. The training data samples along the hyper planes near the class boundary are called support vectors and the margin is the distance between the support vectors and the class boundary hyperplanes. The SVM classifier is based on the concept of decision planes that define decision boundaries. A decision plane is one that separates between assets of objects having different class memberships. Furthermore, a classification task usually involves training and testing data, which consists of some data instances. Where each instance in the training set contains one target value (class labels) and several attributes (features). SVMs have an advantage that its objective function is convex; however, it can only guarantee to converge to a local minimum. Moreover, it is fundamentally a two-class classifier. One commonly used approach to tackle problems involving more than two classes is the one-versus-the-rest approach and is as followed:
Given a training data set with labels \(\{ (x_1,y_1),...(x_n,y_n) \}\) where \(x_i \in R^n\) and \(y_i \in \{+1,-1\}\) and a non-linear map \(\phi ()\), that maps to a higher dimensional space, \(R^n\) \(R^H\) the SVM technique solves:
Subject to the constraints:
specifically w and b define linear classifiers in a feature space. According to Cover’s theorem, a non-linear mapping function \(\phi \) is performed allowing transformed samples to be more likely linearly separable [30]. A regularizer parameter C allows control over penalty assignment to errors model. Slack variable \(\xi \) are introduced to account for non-separable data involved with permitted errors
Owing to the higher dimensionality of vector variable w, the primal function in Eq. (1) is solved by its Lagrangian dual problem which consists of maximizing:
subject to constraints
where \(\alpha _i\) are Lagrange multipliers corresponding to Eq. (2). It can be noted that all \(\phi \) mappings used in the SVM learning occur in the form of inner products. Furthermore, Boster et al. proposed a way to model more complicated relationships by replacing the inner product with a kernel function (such as a Gaussian radial basis function, polynomial kernel or a linear kernel) [31]. This allows us to define a kernel function K where the inner products in the original space (\(x_i,x_j\)) replaced with inner products in the transformed space [\(\phi (x_i).\phi (x_j)\)]:
This kind of kernel function allows us to simplify the solution of the dual problem considerably. This is because it avoids the computation of the inner products in the transformed space \([\phi (x_i ).\phi (x_j )]\). Though \(\phi \) mapping can be explicitly expressed for a linear or polynomial kernel, there is no explicit form of \(\phi \) mapping corresponding to the Gaussian kernel. Moreover, it can be demonstrated that the expansion is an infinite-dimensional functional [32]. Mercer’s theorem avoids to explicitly calculate \(\phi \) in these cases, and then, by introducing (7) into (4), the dual problem can be finally stated as [33]:
After the dual problem is solved, \(w=\sum _{i=1}^n \alpha _i y_i \phi (x_i)\) and express the final result as a decision f(x). Where any test data x is in the original (lower) dimensional feature space:
Furthermore, b can be easily computed from the \(\alpha _i\) that are neither zero nor C.
The shape of the discriminant function depends on the kind of kernel functions adopted. A common kernel type that fulfills Mercer’s condition is the Gaussian radial basis function where \(\gamma \) controls the shape of the peaks and the data points are transformed to a higher dimension:
where \(\gamma \) is a free parameter inversely proportional to the width of the Gaussian kernel.
A small \(\gamma \) means a Gaussian with a large variance resulting a stronger influence of \(x_j\). In other words, if \(x_j\) is a support vector, a small \(\gamma \) implies the class of this support vector will have influence that has a high bias on deciding the class of the vector \(x_i\) even if the distance between them is large. If \(\gamma \) is large, then variance is small implying that the support vector does not have a wide-spread influence (a low bias). A low bias is utilized because the cost of misclassification is penalized heavily. However, a large \(\gamma \) leads to a high bias and low variance models and vice versa.
The FreeSurfer tool is used to take volume of different brain regions such as medial temporal lobe, ventricles, amygdala, cortical grey matter (CGM), entorhinal cortex, and fusiform in each subject. In the training data, each row is a sample, and the columns consists the above stated feature and labels for each sample. For example, hippocampus training data for AD vs. NC classification consists 400 rows and each row represents a sample/subject; one column consists the feature for each sample; and one more column with labels: here +1 for AD and \(-1\) for NC. All training data is prepared in a similar manner for all the aforementioned brain regions.
The data is scaled before SVM is applied [34], The main advantage of scaling is to avoid attributes in greater numeric ranges dominating those in smaller numeric ranges. Another advantage is to avoid numerical difficulties during the calculation. In order to develop an SVM, penalization parameter C; and kernel parameter \(\gamma \) must be tuned. The best C and \(\gamma \) hyper-parameters are found using Grid-Search. Grid search is when given a set of models (which differ from each other in their parameter values, which lie on a grid), train each of the models and evaluate it using 5 - fold cross-validation. Then select the one that performed best. The best C value is 512 and \(\gamma \) is 0.03125. Finally, from 700 subjects’ data, 75% training and 25% testing data are taken randomly, and used for training and then to evaluate the model’s performance respectively. Here we have implemented SVM using the libSVM [35] software package.
4 Results and Discussions
The simulated results presented are obtained using an 8 Core machine with 8 Giga Bytes of random access memory and 3 Mega Bytes of cache.
The area under the curve (AUC) of a two-class classification of combinations of the prodromal stages of dementia are shown in Table 2. AUC analysis, a commonly chosen metric, is chosen to compare the performance of classification models. The predominate reason for using AUC as an alternative to accuracy is that it is not as sensitive to differences between the class distribution within the training and test samples [36, 37]. To be precise, an AUC driven analysis helps in deciding a correct model when one may have been trained on a skewed data set.
From Table 2, features from the hippocampus are shown to act as better discriminators for most stages of dementia except for LMCI/AD and EMCI/NC. This is in support of the argument that, the hippocampus acts as a sensitive biomarker for earlier stages of dementia. The second highest performing biomarker is utilizing the medial temporal lobe (MTL). Though MTL as a biomarker does not perform as the best discriminator for any individual combination, it performs the best on average. Ventricles and entorhinal cortex structures are shown to be below average discriminators, as they do not even discriminate one combination of dementia stage. Moreover, despite combining all the biomarkers, it does not perform as the best discriminators overall and only excels at EMCI/NC classification. The CGM biomarker performs well for AD/MCI and LMCI/AD. The whole brain performs well for AD/LMCI and EMCI/AD, and EMCI/NC. The Fusiform performs best for LMCI from NC. The combined features perform well for MCI, EMCI discrimination from NC. The performance curve for AD/MCI, AD/NC and MCI/NC using the hippocampus features are shown in Figs. 2, 3, 4, 5, 6 and 7.
5 Conclusions and Future Work
In this study we examined the accuracy and reliability of multi class classification based on ROC using volumetric measurements of different brain structures for an accurate diagnosis of dementia stages. Hippocampal volume measurements are the best discriminate for transitions of: AD from NC, AD from MCI, and NC from MCI. The results obtained are satisfactory and are based on a database of hippocampus features. This database consisted of: 400 images for AD vs. NC, 500 images for AD vs. MCI, and 500 images for NC vs. MCI. Moreover, we were able to achieve an AUC value 95.75%, 79.13% and 64.09% respectively. For future work, will be on the use of raw data to classify stages of dementia using a deep learning approach such as a convolutional neural network. Furthermore, we would like to explore the performance of utilizing combined features of hippocampus, CGM, and volume of the entire brain and how they complement each other on the several stages of dementia classification.
References
Liu, Y., Paajanen, T., Zhang, Y., et al.: Combination analysis of neuropsychological tests and structural MRI measures in differentiating AD, MCI and control groups the AddNeuroMed study. Neurobiol. Aging 32(7), 1198–1206 (2011)
Hua, X., Leow, A., Lee, S., et al.: 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. NeuroImage 41, 19–34 (2008)
Hua, X., Lee, S., Yanovsky, I., et al.: Optimizing power to track brain degeneration in Alzheimer’s disease and mild cognitive impairment with tensor-based morphometry: an ADNI study of 515 subjects. NeuroImage 48, 668–681 (2009)
Markiewicz, P., Matthews, J., Declerck, J., et al.: Robustness of multivariate image analysis assessed by resampling techniques and applied to FDG-PET scans of patients with Alzheimer’s disease. NeuroImage 46, 472–485 (2009)
Walhovd, K., Fjell, A., Amlien, I., et al.: Multimodal imaging in mild cognitive impairment: metabolism, morphometry and diffusion of the temporalparietal memory network. NeuroImage 45, 215–223 (2009)
Tripoliti, E.E., Fotiadis, D.I., Argyropoulou, M., et al.: A six stage approach for the diagnosis of the Alzheimers disease based on fMRI data. J. Biomed. Inform. 43(2), 307–320 (2010)
Shin, J., Lee, S.-Y., Kim, S.J., et al.: Voxel-based analysis of Alzheimer’s disease PET imaging using a triplet of radiotracers: PIB, FDDNP, and FDG. NeuroImage 52, 488–496 (2010)
Frisoni, G.B., Fox, N.C., Jack, C.R., et al.: The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 6, 67–77 (2010)
He, Y., Evans, A.: Magnetic resonance imaging of healthy and diseased brain networks. Front. Hum. Neurosci. 2014(8), 890 (2015). doi:10.3389/fnhum.2014.00890
Johnson, K.A., Fox, N.C., Sperling, R.A., et al.: Brain imaging in Alzheimer Disease. Cold Spring Harbor Perspect. Med. 2, a006213–a006213 (2012)
McGeown, W.J., Shanks, M.F., Forbes-McKay, K.E., et al.: Patterns of brain activity during a semantic task differentiate normal aging from early Alzheimer’s disease. Psychiatry Res. Neuroimag. 173, 218–227 (2009)
Torpy, J.M.: Mild cognitive impairment. JAMA 302, 452 (2009)
Schmitter, D., Roche, A., Marchal, B., et al.: An evaluation of volume-based morphometry for prediction of mild cognitive impairment and Alzheimer’s disease. NeuroImage Clin. 7, 7–17 (2015)
Kloppel, S., Stonnington, C.M., Chu, C., et al.: Automatic classification of MR scans in Alzheimer’s disease. Brain 131, 681–689 (2008)
Magnin, B., Mesrob, L., Kinkingnhun, S., et al.: Support vector machine-based classification of Alzheimers disease from whole-brain anatomical MRI. Neuroradiol. 51, 73–83 (2009). 17 24 P
Vemuri, P., Gunter, J.L., Senjem, M.L., et al.: Alzheimer’s disease diagnosis in individual subjects using structural MR images: validation studies. NeuroImage 39, 1186–1197 (2008)
Visser, P.J., Scheltens, P., Verhey, F.R.J., et al.: Medial temporal lobe atrophy and memory dysfunction as predictors for dementia in subjects with mild cognitive impairment. J. Neurol. 246, 477–485 (1999)
Nestor, S.M., Rupsingh, R., Borrie, M., et al.: Ventricular enlargement as a possible measure of Alzheimer’s disease progression validated using the Alzheimer’s disease neuroimaging initiative database. Brain 131(9), 2443–2454 (2008)
Poulin, S.P., Dautoff, R., Morris, J.C., et al.: Amygdala atrophy is prominent in early Alzheimer’s disease and relates to symptom severity. Psychiatry Res. Neuroimag. 194(1), 7–13 (2011)
Sluimer, J.D., van der Flier, W.M., Karas, G.B., et al.: Whole-brain atrophy rate and cognitive decline: longitudinal MR study of memory clinic patients 1. Radiology 248(2), 590–598 (2008)
Henneman, W.J.P., Sluimer, J.D., Barnes, J., et al.: Hippocampal atrophy rates in Alzheimer disease added value over whole brain volume measures. Neurology 72(11), 999–1007 (2009)
Stout, J.C., Jernigan, T.L., Archibald, S.L., et al.: Association of dementia severity with cortical gray matter and abnormal white matter volumes in dementia of the Alzheimer type. Arch. Neurol. 53(8), 742–749 (1996)
Yokum, S., Stice, E.: Initial body fat gain is related to brain volume changes in adolescents: a repeated-measures voxel-based morphometry study. Obesity 25(2), 401–407 (2017)
Riddle, K., Cascio, C.J., Woodward, N.D.: Brain structure in autism: a voxel-based morphometry analysis of the Autism Brain Imaging Database Exchange (ABIDE). Brain Imaging Behav. 11(2), 541–551 (2017)
Chen, Q., et al.: Brain gray matter atrophy after spinal cord injury: a voxel-based morphometry study. Front. Hum. Neurosci. 11, 211 (2017)
Focke, N.K., Trost, S., Paulus, W., Falkai, P., Gruber, O.: Do manual and voxel-based morphometry measure the same? a proof of concept study. Front. Psychiatry 5, 39 (2014). doi:10.3389/fpsyt.2014.00039
Clerx, L., et al.: Can FreeSurfer compete with manual volumetric measurements in Alzheimer’s Disease? Curr. Alzheimer Res. 12(4), 358–367 (2015)
Fischl, B.: Freesurfer. NeuroImage 62, 774–781 (2012)
Fischl, B., Salat, D.H., Busa, E., et al.: Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33(3), 341–355 (2002)
Cover, T.M.: Geometrical and statistical properties of systems of linear inequalities with application in pattern recognition. IEEE Trans. Electron. Comp. 14, 326–334 (1965). (reprinted. In: Mehra, P., Wah, B. (eds.) Artificial Neural Networks: Concepts and Theory. IEEE Computer Society Press, Los Alamitos, California (1992))
Boser, B.E., Guyon, I.M., Vapnik, V.N.: A training algorithm for optimal margin classifiers. In: Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Pittsburgh, Pennsylvania, USA, 27–29 July 1992, pp. 144–152 (1992)
Schölkopf, B., Smola, A.: Learning with Kernels-Support Vector Machines, Regularisation, Optimization and Beyond. The MIT Press Series, Cambridge (2001)
Mercer, J.: Functions of positive and negative type and their connection with the theory of integral equations. Philos. Trans. Roy. Soc. A 209(441–458), 415–446 (1909)
Sarle, W.S.: Neural network FAQ (1997). ftp://ftp.sas.com/pub/neural/FAQ.html. Periodic posting to the Usenet newsgroup comp.ai.neural-nets
Chang, C.-C., Lin, C.-J.: LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2, 27:1–27:27 (2011)
Hanley, J.A., McNeil, B.J.: The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36 (1982)
Rathore, S., Habes, M.: A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer’s disease and its prodromal stages. NeuroImage (2017). doi:10.1016/j.neuroimage.2017.03.057
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Author information
Authors and Affiliations
Consortia
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG
About this paper
Cite this paper
Kruthika, K.R., Rajeswari, Pai, A., Maheshappa, H.D., Alzheimer’s Disease Neuroimaging Initiative. (2018). Classification of Alzheimer and MCI Phenotypes on MRI Data Using SVM. In: Thampi, S., Krishnan, S., Corchado Rodriguez, J., Das, S., Wozniak, M., Al-Jumeily, D. (eds) Advances in Signal Processing and Intelligent Recognition Systems. SIRS 2017. Advances in Intelligent Systems and Computing, vol 678. Springer, Cham. https://doi.org/10.1007/978-3-319-67934-1_23
Download citation
DOI: https://doi.org/10.1007/978-3-319-67934-1_23
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-67933-4
Online ISBN: 978-3-319-67934-1
eBook Packages: EngineeringEngineering (R0)