Abstract
We propose a novel local image descriptor called the Extended Multi-resolution Local Patterns, and a discriminative probabilistic framework for learning its parameters together with a multi-class image classifier. Our approach uses training data with image-level labels to learn the features which are discriminative for multi-class colonoscopy image classification. Experiments on a three class (abnormal, normal, uninformative) white-light colonoscopy image dataset with 2800 images show that the proposed feature perform better than popular hand-designed features used in the medical as well as in the computer vision literature for image classification.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
More than one million new Colorectal cancer (CRC) cases are diagnosed yearly worldwide, and CRC remains the third leading cause of cancer death in the world [1]. There is compelling evidence that removing adenomas from the colon substantially reduces the risk of a patient developing CRC [1]. If CRC is diagnosed in its earliest stages, the chance of survival is 90% [1]. Clearly, early identification of colonic abnormalities is crucially important.
Adenoma detection rate (ADR) is a commonly used predictor of the risk of developing CRC after undergoing a colonoscopy screening [2]. Although colonoscopy remains the gold standard for CRC screening, CRC miss rate has been reported as high as \(6\%\) [3], posing risk of developing colon cancer due to a failure to detect treatable lesions in time. It is therefore arguable that a reliable computer-aided detection system specialised for identifying suspicious colonic abnormalities in colonoscopy videos could contribute to improve ADR, e.g. by presenting clinicians with a second opinion obtained by objective and repeatable methods.
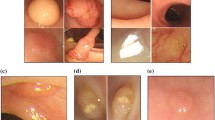
In this paper we propose an automated system to classify colonoscopy images into three classes: abnormal, normal and uninformative. The abnormal images contain various abnormalities such as polyps, cancers, ulcers and bleeding, appearing in a variety of sizes, positions and orientations in the image. The normal images contain none and show a clear healthy colon wall. The uninformative images contain images which are blurred due to out of focus (e.g., camera pushed against the colon wall) or sharp camera movements. Note that we are not specifically interested in detecting uninformative frames as done in the existing approaches (e.g. [4]), but our target is a multi-class colonoscopy image classification (Fig. 1).
Various hand-designed features (e.g. SIFT) have been explored for colonoscopy image classification (discussed in Sect. 2). However, these features may not be optimally discriminative for classifying images from particular domains (e.g. colonoscopy), as not necessarily tuned to the domain’s characteristics. We instead propose a learning approach, which jointly learns discriminative local features together with a multi-class image classifier using training data with image-level labels. Since our features are learned from the data we expect them to be more discriminative than hand-designed ones. Comparative experiments with our colonoscopy dataset show that the learned features perform better than popular features used in the medical as well in the computer vision literature for image classification.
2 Related Work
The approaches proposed for colonoscopy image analysis are mainly focussed on identifying appropriate features; various hand-crafted features such as color wavelet co-variance (CWC) [5], color histograms (CH) [6], gray-level co-occurrence matrices (GLCM) [7], Root-SIFT (rSIFT) [8], Local Binary Patterns (LBP) [8], Local Ternary Patterns (LTP) [8] have been explored. For example, LBP and GLCM for normal/abnormal classification [7, 8], CWC for polyp detection [5], and for classification [7].
Feature learning approaches, e.g. [9,10,11], on the other hand, learn domain-specific discriminative local features and report improved performance compared to hand-crafted features in various applications, e.g. medical image segmentation [9], and natural image retrieval [11, 12]. However, these approaches require a labelled dataset for learning; e.g., Becker et al. [9] uses manual region-level segmentations to learn filters for curvilinear structure segmentation in retinal and microscopy images.
Convolutional neural nets (CNN) have been widely used to jointly learn features and a classifier. Usually CNN requires a large amount of training data [13]; when this is not available, CNN may give worse performance than traditional, hand-crafted features with feature encoding methods such as sparse coding [13]. Recently, transfer learning approaches have been widely used (e.g. [14]) to overcome this, where a CNN model trained on a large dataset (e.g. ImageNet, which contains 1.2 million images with 1000 categories), is used either as an initialization or a fixed feature extractor for the task of interest. CNN is computationally expensive to train, even on the GPU [15].
Since obtaining region-level annotations (to learn features as in [9,10,11]) is a difficult, time-consuming task, we propose a feature learning approach which uses only the image-level labels. Requiring image-labels instead of region-level labels makes annotations less expensive, hence more feasible in practice. Compared to CNN, our approach does not require pre-training on large dataset, or specialized hardware such as GPU for training.
3 Method
First we introduce our notation, and then we define the structure of our feature in Sect. 3.1. Section 3.2 proposes the learning algorithm to learn the parameters of the feature together with a multi-class image classifier. We call the learned feature Extended Multi-Resolution Local Patterns (xMRLP).
We characterize an image \(I_i\) by a set of local features \(\{\mathbf {x}_{ij}\}_{j=1}^{N_i}\), where \(N_i\) is the number of local features in \(I_i\). Let’s consider the general case of labels, whereby an image is associated with an image-level soft label indicating, for e.g., class probabilities. Our goal is to learn the parameters of the xMRLP features as well as a multi-class classifier based on the given training data, which is formed by the set of tuples \(\mathcal {D} = \{(I_i, \tilde{P}_i)\}_{i=1}^M\), where M is the number of images in \(\mathcal {D}\), and \(\tilde{P}_i \in [0,1]^C\) corresponds to a C-dimensional vector of soft labels of the \(i^\text {th}\) training image associated with the C classes. We assume that \(\sum _{c=1}^C \tilde{P}(y_i=c)=1\), where \(\tilde{P}(y_i=c)\) is the latent class assignment of the image \(I_i\) to class c.
3.1 Extended Multi-resolution Local Patterns
Let \(I_{ij}\) be the intensity of the \(j^{\text {th}}\) pixel in the \(i^{\text {th}}\) image. To capture local context and to make the descriptor less sensitive to noise, we use the sampling pattern widely adopted in feature descriptors e.g. [16]. Figure 2 shows a 3-resolution version of the sampling pattern, where the local neighbourhood around the \(j^{th}\) pixel of image \(I_i\) is quantized at three resolution levels. Eight sampling points are considered at each resolution. At each sampling point, a Gaussian filter with standard deviation proportional to the size of the support region (circle around each sampling point in Fig. 2) is applied to collect information from that region.
Let \(I_{ij}^s, \, s=1, \dots , d\), represents the intensity value at the s-th sampling point in the pattern around the \(j^{th}\) pixel of image \(I_i\) (e.g. \(d=24\) in Fig. 2). We define \(\mathbf {x}_{ij}\in \mathbb {R}^d\) as the xMRLP descriptor vector at pixel j in image \(I_i\) using the multi-resolution sampling pattern with d sampling points:
where \(\varvec{a} = \left[ a_1, \dots , a_d\right] \) defines the weights for different neighbourhood regions.
Note that, xMRLP is an improved version of the Multi-resolution Local Patterns (MRLP) descriptor proposed in [17, 18] for cell image classification. In MRLP the weights for the local neighborhoods were fixed to 1, i.e. \(a_i=1, \forall i\) (Eq. 1).
3.2 A Discriminative Multi-class Framework for Learning
In this section we propose a discriminative framework based on image-to-class distances (I2CD) [19] to jointly learn the feature parameter (\(\varvec{a}\) in Eq. 1) and an image-level multi-class probabilistic classifier for colonoscopy image classification.
Image to Class Distances. The I2CD was first introduced by Boiman et al. [19] in the NBNN classifier. It requires no training phase, and classifies an image by comparing its distance to different classes. A relaxed version of I2CD was proposed in [20], showing improved performance over the original version for complex datasets. The relaxed version of I2CD is given as:
where \(\mathbf {x}_{ij}^{cp}\) is the \(p^{th}\) nearest neighbour of \(\mathbf {x}_{ij}\) in the \(c^{th}\) class, P is the number of considered neighbours. In all the reported experiments we set \(P=3\).
Discriminative Probabilistic Softmax Classifier. Equation (3) below defines a discriminative probabilistic classifier. This classifier outputs the posterior probability of an image \(I_i\) belonging to a class c based on the I2CD.
The class c maximising the probability above is the one associated with the smallest I2CD over all classes. In Eq. (3) \(\{\gamma _l\}_{l=1}^C\) are the classifier parameters.
The Objective Function. Equation (4) defines the objective function to learn the feature parameter \(\varvec{a}\) and the classifier parameters \(\{\gamma _l\}_{l=1}^C\).

where, the first term maximizes the target posterior probabilities of the images in the training set and second term is a regularisation term, prevents the parameters \(\varvec{a}\) from becoming arbitrary high and makes their values close to \(-1\) (as in MRLP). We set \(\beta =1\) for all the reported experiments.
We use a coordinate descent method to optimize Eq. (4), where we learn one parameter at a time while keeping the others constant.
Note that, learning the feature parameters is similar to metric learning approaches. For example in [21], class-specific distance metrices were learned to compare images with different classes, and the class which gives the smallest I2CD was considered as the target class for that image. However, in Sect. 4.2 we show that the learned features when they are combined with the traditional feature encoding methods such as sparse coding and a SVM classifier performs better than directly using them (as in [21]).
4 Experiments
This section reports our comparative experiments and the results based on the xMRLP descriptor and other features such as LBP, LTP, rSIFT, RP.
Materials: We collected 82 white-light colonoscopy video segments from the Internet. K-means clustering was applied to select a representative subset of images from each video segment based on color statistics (mean, std, skewness and entropy in RGB color chennels) and texture features (LBP histograms). From each video one frame per cluster was randomly selected and annotated by a clinical expert who provided image-level annotations. It is observed that the movement of the colonoscope is fast in normal videos compared to the abnormal ones as the corresponding colon segments do not need a careful inspection of the colonic walls. Therefore the number of clusters for a video \(v_i\) was experimentally set to \(\frac{V}{7}\) for normal and \(\frac{V}{10}\) for abnormal videos, where V is the total number of images in \(v_i\). The final dataset contains 1000 abnormal, 900 normal and 900 uninformative images. All images in the final dataset are rescaled preserving the aspect ratio so that the maximum dimension (row or column) of each image is 300 pixels.
Experimental setup and evaluation criteria: All the local features are extracted from RGB color patches of size \(3\times 16\times 16\) with an overlap of Q pixels in vertical and horizontal directions. The sampling pattern shown in Fig. 2 (3 resolution, 8 sampling points in each resolution) is used for xMRLP, LBP and LTP features. We rescale the sampling pattern such that all the sampling points lie inside the \(16\times 16\) image patches.
The classification performance is measured as the average of the per-class accuracies (mean-class accuracies, MCA) measured on the test test. All the experiments were repeated 10 times and the MCA averaged over these iterations are reported. In each run we randomly selected 300 images from each class for training and use the rest for testing.
4.1 Effect of Feature Learning
This section compares the xMRLP feature with baseline features rSIFT, RP and MRLP.
For each feature the representation of a patch was obtained by concatenating the features extracted from each of the color channels of the RGB color space. This led to a dimensionality of 72 (\(3 \text { colors } \times 3 \text { resolutions }\times 8\text { sampling points}\)) for MRLP and xMRLP, and \(3\times 128\) for rSIFT. Each of the vectorized color patch of dimension \(3 \times 16 \times 16\) is projected to a compressed space of dimension 200 using a random projection matrix [22] to get a RP feature.
Example of wrongly classified images (abnormal - first two columns, normal - next two columns, uninformative - last two columns) and their confidence values using the xMRLP features. The values in the brackets are correspond to \(P(y=\text {abnormal})\), \(P(y=\text {normal})\) and \(P(y=\text {uninfomative})\) respectively.
In the feature learning stage of xMRLP we use only 50 images from each of the 3 classes, since the I2CD calculations are computationally expensive due to nearest neighbour search. In the classification stage we randomly sample 50, 000 local features from each class of the training images and calculate the I2CD between a test image and the training set to do the classification. In both cases features are extracted densely without overlap (\(Q=0\)).
Table 1 compares the performance of different features; xMRLP improves the performance of MRLP by about \(7\%\), suggesting that learning can capture discriminative information. xMRLP also outperforms rSIFT and RP with low dimensional representation, makes the I2CD classifier computationally efficient.
Since the proposed framework can also provide probabilistic outputs for the test images, Figs. 3 and 4 show example of the wrongly and correctly classified test images and their confidence values based on the probabilistic soft-max classifier given in Eq. (3). As can be seen from Fig. 3 the probability outputs and the wrong classification results are reasonable, as it is hard to assign the ambiguous images (i.e. images with ambiguous appearance) to a single class with high confidence.
4.2 XMRLP with Feature Encoding and SVM Classifier
The softmax classifier used in Sect. 4.1 is computationally expensive due to the nearest neighbour search involved in the I2CD calculations. Feature encoding methods (e.g. [23]) with SVM classifier, on the other hand, are widely used in medical image analysis [8] and are computationally efficient compared to I2CD calculations. Therefore, in this section we evaluate the performance of the learned xMRLP features (after learning them as explained in Sect. 4.1) using a feature encoding method called Locality Constraint Linear Coding (LLC) [23] and a SVM classifier. We show that xMRLP features with LCC+SVM perform better than other features as well as xMRLP features with the soft-max classifier (Sect. 4.1).
Since feature encoding is computationally efficient, we extracted features more densely, with an overlap of \(Q=12\) pixels. For each feature type we randomly sampled 100,000 local features to learn the dictionary using k-means. We used SVM classifier (LIBSVM [24]) with an exponential \(\chi ^2\) kernel and report the performance in Fig. 5. xMRLP feature outperforms other features even with a smaller dictionary size (500) suggesting that learned features are better than other features considered. When the dictionary size is 4000, xMRLP gives a MCA of \(92.8\%\) which is better than the MCA obtained by rSIFT (\(89.7\%\)) and RP (\(89.1\%\)).
4.3 Comparison with the Features Proposed for Colonoscopy
This section compares the performance of various features proposed for colonoscopy image classification literature such as LBP [8], LTP [8], color histograms (CH) [6], GLCM [25], GLCM on wavelet images (WGLCM) [26], CWC [5] and CWC with higher-order statistics (CWC2) [7].
For LBP and LTP features we use a three resolution version of the sampling patterns as explained in Sect. 4. These features are extracted with an overlap of \(Q=12\) pixels. The LTP parameters were learned from a 5-fold cross validation on the training set. To make a fair comparison we used the same SVM classifier with an exponential \(\chi ^2\) kernel for this experiment.
The results are reported in Table 2. The proposed xMRLP feature outperforms others by a large margin. xMRLP feature takes about 0.3 s to classify an image compared to 1.1 s and 1.3 s by RP and rSIFT features respectively on an Intel Core-i7 machine with 8 GB RAM. These times include the time for feature extraction and encoding with a dictionary of size 1000.
4.4 Comparison with Deep Convolutional Neural Nets
Since CNN was widely applied for bio-medical [27] as well in non-medical [13] applications, the following experiments were done with CNN to evaluate its performance on our colonoscopy dataset.
Training using colon dataset: A shallow network (Fig. 6) was trained (from scratch) using only the images from the colon dataset with data augmentation (mirrored images). This network gives an MCA of \(76.1\pm 0.7\%\), which is \(\sim \)15% less compared to our approach (\(92.8\%\)). This is mainly due to the lack of data used for training. Similar findings were reported in [13] on the Caltech101 datasetFootnote 1; CNN trained on this dataset gives an accuracy of \(46\%\), compared to the accuracy of \(84\%\) obtained by the hand-designed features with feature encoding.
Transfer learning: In this experiment we fine-tuned the ImageNet (1.2 Million images) trained model “AlexNet” [15] using the colon dataset with data augmentation (mirrored images, and randomly cropped image regions of size \(227\times 227\) from images of size \(256\times 256\)). This fine-tuned net gives a MCA of \(92.9\pm 0.6\%\), which is similar to the MCA obtained by our approach (\(92.8\%\)).
Unlike our approach, CNN is designed to capture features at multiple scales. Therefore, the classification performance of CNN can be expected to be high compared to our approach. However this ImageNet pretrained CNN shows similar performance compared to our approach, as the results on this dataset are saturated at \(\sim \)93%. Although results are similar, our approach does not require a larger dataset for pre-training or higher computational power such as GPU. Our approach takes \(\sim \)1.5 h to train on a CPU with our unoptimized Matlab code on an Intel Core-i7 machine with 8 GB RAM compared to \(\sim \)20 min fine-tuning time required by CNN on NVidia Tesla K40 GPUFootnote 2 with 12 GB RAM.
For the above two experiments we use the following parameters to train the network: learning rate \(10^{-4}\), momentum 0.9, weight decay \(5\times 10^{-4}\). The maximum number of iterations were set to 10000 and 7000 for the first and second experiments respectively. The library Caffe [28] was used in all the CNN-related experiments.
5 Conclusions
We presented a novel discriminative feature learning approach for multi-class colonoscopy image classification, which jointly learns the parameters of the proposed xMRLP features together with an image-level classifier using training data with image-level labels. Various comparative experiments on a colonoscopy dataset with the features proposed in the literature of colonoscopy as well as computer vision show that our learned features outperform others. The proposed approach is not restricted to colonoscopy images, hence our future work will explore applications to other medical image domains.
Notes
- 1.
- 2.
Tesla K40 GPU used for this research was donated by the NVIDIA Corporation.
References
Atkin, W., Cook, C., Cuzick, J., Edwards, R., Northover, J., Wardle, J.: Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: a multicentre randomised controlled trial. Lancet 132, 1624–1633 (2010)
Wallace, M.: Improving colorectal adenoma detection: technology or technique? Gastroenterology 132, 1221–1223 (2007)
Bressler, B., Paszat, L.F., Chen, Z., Rothwell, D.M., Vinden, C., Rabeneck, L.: Rates of new or missed colorectal cancers after colonoscopy and their risk factors: a population-based analysis. Gastroenterology 132(1), 96–102 (2007)
Arnold, M., Ghosh, A., Lacey, G., Patchett, S., Mulcahy, H.: Indistinct frame detection in colonoscopy videos. In: Machine Vision and Image Processing Conference, pp. 47–52(2009)
Karkanis, S.A., Iakovvidis, D.K., Maroulis, D.E., Karras, D.A., Tzivras, M.: Computer aided tumor detection in endoscopic video using color wavelet features. IEEE Trans. IT Biomed. 7, 141–152 (2003)
Khun, P.C., Zhuo, Z., Yang, L.Z., Liyuan, L., Jiang, L.: Feature selection and classification for wireless capsule endoscopic frames. In: Biomedical and Pharmaceutical Engineering, pp. 1–6 (2009)
Lima, C., Barbosa, D., Ramos, A., Tavares, A., Montero, L., Carvalho, L.: Classification of endoscopic capsule images by using color wavelet features, higher order statistics and radial basis functions. In: IEEE Engineering in Medicine and Biology Society, pp. 1242–1245 (2008)
Manivannan, S., Wang, R., Trucco, E., Hood, A.: Automatic normal-abnormal video frame classification for colonoscopy. In: IEEE International Symposium on Biomedical Imaging, pp. 644–647 (2013)
Becker, C.J., Rigamonti, R., Lepetit, V., Fua, P.: Supervised feature learning for curvilinear structure segmentation. In: Medical Image Computing and Computer-Assisted Intervention, pp. 526–533 (2013)
Brown, M., Hua, G., Winder, S.: Discriminative learning of local image descriptors. IEEE Pattern Anal. Mach. Intell. 33, 43–57 (2011)
Simonyan, K., Vedaldi, A., Zisserman, A.: Learning local feature descriptors using convex optimisation. IEEE Pattern Anal. Mach. Intell. 36, 1573–1585 (2014)
Philbin, J., Isard, M., Sivic, J., Zisserman, A.: Descriptor learning for efficient retrieval. In: European Conference on Computer Vision, pp. 677–691 (2010)
Matthew, Z., Rob, F.: Visualizing and understanding convolutional networks. In: European Conference on Computer Vision, pp. 818–833 (2014)
Oquab, M., Bottou, L., Laptev, I., Sivic, J.: Learning and transferring mid-level image representations using convolutional neural networks. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 1717–1724 (2014)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 25, 1097–1105 (2012)
Winder, S., Hua, G., Brown, M.: Picking the best daisy. In: IEEE Computer Vision and Pattern Recognition, pp. 178–185 (2009)
Manivannan, S., Li, W., Akbar, S., Wang, R., Zhang, J., McKenna, S.J.: An automated pattern recognition system for classifying indirect immunofluorescence images of HEp-2 cells and specimens. Pattern Recogn. 51, 12–26 (2016)
Manivannan, S., Li, W., Akbar, S., Wang, R., Zhang, J., McKenna, S.J.: HEp-2 cell classification using multi-resolution local patterns and ensemble SVMs. In: I3A 1st workshop on Pattern Recognition Techniques for Indirect Immunoflurescence Images, in International Conference on Pattern Recognition (2014)
Boiman, O., Shechtman, E., Irani, M.: In defense of nearest-neighbor based image classification. In: IEEE Computer Vision and Pattern Recognition, pp. 1–8 (2008)
Zhen, X., Shao, L., Zheng, F.: Discriminative embedding via image-to-class distances. In: British Machine Vision Conference (2014)
Wang, Z., Hu, Y., Chia, L.T.: Image-to-class distance metric learning for image classification. In: European Conference on Computer Vision, pp. 706–719 (2010)
Bingham, E., Mannila, H.: Random projection in dimensionality reduction: applications to image and text data. In: ACM Knowledge Discovery and Data Mining, pp. 245–250 (2001)
Wang, J., Yang, J., Yu, K., Lv, F., Huang, T., Gong, Y.: Locality-constrained linear coding for image classification. In: IEEE Computer Vision and Pattern Recognition, pp. 3360–3367 (2010)
Chang, C.C., Lin, C.J.: LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2, 27:1–27:27 (2011)
Engelhardt, S., Ameling, S., Paulus, D., Wirth, S.: Features for classification of polyps in colonoscopy. In: CEUR Workshop Proceedings (2010)
Maroulis, D.E., Iakovidis, D.K., Karkanis, S.A., Karras, D.A.: CoLD: a versatile detection system for colorectal lesions in endoscopy video-frames. Comput. Methods Programs Biomed. 70, 151–166 (2003)
Li, W., Manivannan, S., Zhang, J., Trucco, E., McKenna, S.J.: Gland segmentation in colon histology images using hand-crafted features and convolutional neural networks. In: IEEE International Symposium on Biomedical Imaging (2016)
Jia, Y., Shelhamer, E., Donahue, J., Karayev, S., Long, J., Girshick, R., Guadarrama, S., Darrell, T.: Caffe: convolutional architecture for fast feature embedding. arXiv preprint arXiv:1408.5093 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Manivannan, S., Trucco, E. (2017). Extended Multi-resolution Local Patterns - A Discriminative Feature Learning Approach for Colonoscopy Image Classification. In: Peters, T., et al. Computer-Assisted and Robotic Endoscopy. CARE 2016. Lecture Notes in Computer Science(), vol 10170. Springer, Cham. https://doi.org/10.1007/978-3-319-54057-3_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-54057-3_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-54056-6
Online ISBN: 978-3-319-54057-3
eBook Packages: Computer ScienceComputer Science (R0)