Abstract
Automatic polyp detection in endoscopy (or colonoscopy) images is challenging because the types of polyp and their appearances are diverse, and the colors and textures of polyps are quite similar to those of normal tissues in many cases. It is thus often very difficult to distinguish polyps from normal tissues using conventional methodology. To effectively resolve these challenges, we propose a framework based on multi-classifier learning and a contour intensity difference (CID) measure. To detect polyps of diverse appearances, we first classify polyps into K types according to their shape via unsupervised learning. We then train K classifiers to detect the K types of polyp. This multi-classifier learning improves the polyp detection rate. However, false positives also increase because colon structures look similar to polyps. To reduce false positives while preserving the high detection rate, we propose a CID measure. Experimental results using public and our own datasets show that the proposed methods are promising for detecting polyps with diverse appearances.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Colon cancer is one of the most common types of cancer and the leading cause of cancer death [1]. One major cause of colon cancer is precancerous polyps such as adenoma, which can develop into cancer if they remain untreated. It is known that the risk of developing colon cancer increases with the number and size of polyps present. Baxter et al. [2] showed that detecting early-stage polyps is highly associated with fewer death from colon cancer; thus, it is very important to detect early-stage polyps through colon examination techniques.
In order to detect potentially cancerous polyps, colonoscopy inspection is commonly used. However, it shows a significant miss rate of around 22% even with high-quality colonoscopes [3]. To improve detection performance, automatic polyp detection methods using computer vision and machine learning techniques have received a lot of research attention. Such methods provide approximate positions and sizes of polyps to doctors during colonoscopy.
Although there has been much progress in automatic polyp detection, it is still a challenging task because of several issues, as illustrated in Fig. 1: (1) high diversity of polyp types and their colors, textures, shapes, and sizes (Fig. 1a); (2) frequent false positives (FPs) due to colon wrinkles and passages, specular highlights, blood vessels, and so on (Fig. 1b); (3) frequent occlusions caused by other colon structures (Fig. 1c); (4) shape variations of polyps caused by unconstrained endoscope motion and position (Fig. 1d); (5) textural and color ambiguities between polyps and normal tissues (Fig. 1e).
In recent years, to handle these difficulties, many studies have focused on developing feature extraction and classifier learning algorithms, which are core parts of object detection schemes. For effectively describing polyps, color- and texture-based features have been proposed or used, such as local binary patterns (LBPs) [4, 5], gray-level co-occurrence matrix (GLCM) [4], and color wavelet covariance (CWC) [5–7]. To train a polyp classifier, learning algorithms based support vector machine (SVM) [4, 8], backpropagation neural networks (BPNNs) [9], and linear discriminative analysis (LDA) [6] have been proposed. However, such features are not discriminative enough to differentiate polyps from normal tissues since their colors and textures are very similar, as illustrated in Fig. 1e. Moreover, the single-classifier-based approach is insufficient to handle various appearance changes of polyps, as shown in Fig. 1a, c, and d. In addition, previous works did not try to reduce FPs caused by factors shown in Fig. 1b.
In this paper, we propose a novel framework for automatic polyp detection, which overcomes the limitations of previous works. The proposed framework consists of feature extraction, multiple-classifier learning, and FP removal. We adopt the histogram of oriented gradients (HOG) [11], which is a shape-based feature extraction method, as the feature descriptor to deal with textural and color ambiguities. We experimentally verify the effectiveness of the HOG feature. In addition, we propose a multi-classifier learning method for building a polyp detector that can handle diverse appearance variations of polyps. The multi-classifier learning method consists of two stages: we first categorize training polyp samples into K subsets using K-means clustering in the HOG feature space, and then generate multiple SVM classifiers with clustered polyp samples and normal tissue samples. When detecting polyps in a test image, we exploit all outputs of the trained multi-classifier by aggregating them using the noisy-OR model [12]. As a result, we can improve the polyp detection rate under the large variation of polyp appearances. Furthermore, based on the observation that FPs are frequently due to colon wrinkles and passages that have similar shapes to polyps, we propose a contour intensity difference (CID) measure for effectively removing FPs while preserving the high detection rate. We extensively evaluate and compare the performance of the proposed method with that of other polyp detection methods using public [13] and our own datasets, which contain many challenging factors.
2 Literature Review
In general, most previous works on polyp detection have focused on feature extraction and classifier learning methods since they significantly affect the performance of polyp detection. Thus, in this section, we review previous works by categorizing them into feature extraction methods and classifier learning methods.
2.1 Feature Extraction Methods for Automatic Polyp Detection
For extracting features of polyps, several texture- and color-based feature extraction methods have been proposed. Textural cues have been used as discriminative features for polyp detection in many studies. For example, LBPs and GLCM, which represent texture patterns, have been used for polyp detection [4]. The extension series of LBPs, such as curvelet-based LBPs and rotation-invariant uniform LBPs, have been successfully applied for polyp detection [7].
Color-based feature extraction methods have been used for polyp detection as well. Alexandre et al. [8] extracted low-dimensional but abundant features using RGB values and pixel positions within each sub-image. Tjoa and Krishnan [9] designed a color component histogram from the RGB and HSI channels of endoscopic images. For enhancing description power, combined features with color and textural cues have been also presented. Karkanis et al. [6] proposed the CWC feature, which utilizes the covariance of second-order textural measures calculated over the wavelet transform of different color bands. The performance of various texture and color features has been compared [5, 7, 9, 14, 15]. It was found that combined features significantly improve the performance of polyp detection compared to that obtained using only a color or texture feature.
In addition, shape-based polyp detection methods based on sub-region classification have been proposed. In these methods, image sub-regions are first constructed based on an image segmentation algorithm (e.g., watershed [16]). Then, each sub-region is classified as a polyp or non-polyp. Hwang et al. [17] proposed an elliptical shape fitting and polyp candidate selection method. Bernal et al. [13] proposed region classification based on the SA-DOVA shape descriptor. These methods can detect polyps without training data since region classification does not rely on trained classifiers. However, these methods can be dependent on the segmentation results; for example, polyps that are not fully surrounded with strong edges (such as flat polyps) cannot be segmented properly, which leads to polyp detection failure.
2.2 Classifier Learning Methods for Automatic Polyp Detection
In order to train a classifier for polyp detection, SVM has been widely used in many studies [4, 7, 8, 14, 18]. Since SVM can produce an optimal margin for separating different class samples, it has been widely applied to many computer vision applications such as object detection, tracking, and recognition. Tjoa and Krishnan [9] employed a BPNN trained by various training algorithms (e.g., Marquardt, SCG, RPROP) with different numbers of neurons in the hidden layer for analyzing colon status, and Karkanis et al. [6] used LDA, which is a simple model involving a small set of parameters, to train a classifier for polyp detection.
The aforementioned detection methods [4, 6–9, 18] only use a single classifier when discriminating between polyps and normal tissues. However, it is not easy to discriminate polyps with many different appearances from other tissues using only one classifier. Dietterich et al. [10] found that training sub-classifiers for solving sub-problems and voting them for the final decision is efficient. This is because the sub-problems are more tractable for training classifiers with fewer errors and the voting procedure reduces the risk of choosing wrong classifiers. We therefore train multiple classifiers by clustering polyps into several clusters and aggregate all their outputs when detecting polyps.
3 Methods
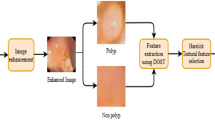
The overall proposed polyp detection framework is shown in Fig. 2. In the training stage, we first extract the HOG features [11] of polyps and categorize the polyps into K classes by performing K-means clustering in the HOG feature space. We then generate K multiple SVM classifiers with the clustered features, as discussed in Sect. 3.1. In the test stage, we obtain polyp hypotheses for an image by combining the outputs of the trained classifiers with the noise-OR model. Among the hypotheses, we remove FPs using the CID measure, as described in Sect. 3.2.
3.1 Multi-Classifier Learning for Polyp Detection
For training a detector that can detect polyps with a variety of appearances, we propose a multi-classifier learning method. The polyp detector is composed of multiple sub-classifiers, each of which is trained independently to specialize in detecting a particular group of polyps. In order to train the sub-classifiers, we first extract the HOG features [11] of polyp sample images. HOG counts occurrences of gradient orientation on a densely sampled grid and makes an orientation histogram. It describes the overall shape of an object.
After extracting HOG features, we cluster the polyps according to their HOG features by performing K-means clustering in the HOG feature space, as shown in Fig. 3. As a result, we obtain K subsets with different tendencies of shape (Sets 1 and 2 in Fig. 3) and edge intensities (Sets 2 and 3 in Fig. 3). Subsequently, linear SVM classifiers are trained using the K subsets (i.e., positive samples) and non-polyp image patches (i.e., negative samples) randomly extracted from training images.Footnote 1 Finally, we have K sub-classifiers expressed as:
where x is an input HOG feature vector, and W k and b k are the weight and bias of each classifier, respectively.
Using the standard logistic sigmoid function, the probability of the input x (i.e., HOG feature) being positive can be defined with the output of a sub-classifier as:
P k (x) lies in [0, 1]. When aggregating the outputs P k (x) of the trained K classifiers, we aim at maximizing the detection rate rather than reducing the FP rate, since the former is more important in polyp detection. To achieve this, we employ the noisy-OR model [12], defined as:
As a result, we can classify an input vector as a positive response (i.e., high detection probability P(x)) when any output P k (x) of the classifiers is high. We only classify an input vector as a negative response when all outputs of the classifiers are low. P(x) also lies in [0, 1].
Figure 4 illustrates the aggregation process. A true polyp location is marked with a black dashed circle, and the probability maps P 1(x), P2(x), and P 3(x) of the three classifiers H 1(x), H 2(x), and H 3(x), respectively, are drawn, where x = {x 1, x 2, …x N } and N is the total number of HOG features obtained by the sliding window search. Probability maps P 2(x) and P 3(x) show low probability responses in the actual polyp region. However, the final map after aggregation shows high responses in the polyp region due to map P 1(x) of classifier H 1(x), which is trained for detecting polyps whose appearance is similar to that of a given polyp in Fig. 4. This strategy greatly improves the detection rate, and handles polyp appearance variations by using multiple classifiers trained for detecting polyps with different appearances.
3.2 Contour Intensity Difference Measure for Removing False Positives
Even though we can detect polyps with various appearances using the multi-classifier detector, FPs are frequently caused by colon wrinkles and passages with appearances similar to those of polyps. For discriminating them, we propose a CID measure by considering intensity variations around edges of polyps and other tissues. In order to analyze the intensity variations, we consider both an illumination model and the structural difference between polyps and other tissues (i.e., colon wrinkles and passages).
3.3 Observations: Illumination and Structural Models
For the illumination model, we assume that the reflection on the colon can be described by Phong’s illumination model [19], defined as:
where K a is the ambient reflection constant, I a is the ambient intensity, C p is the reflection coefficient of the object at point p, K d is the diffuse reflection coefficient, θ is the angle between the surface normal and the illumination source (i.e., incident angle of the ray), W(θ) is a function of the specular reflected light, i is the angle between the direction of the reflected light and the camera, and n is a power of the specular reflected light for each material. In our polyp detection system, an endoscope is equipped with a camera and an illumination source is placed in almost the same position and direction as those of the camera. In this case, θ is the incident angle of the camera, and i is equal to 2θ; therefore the incident angle of the camera (θ) affects the illumination of the colon, as shown in Fig. 5a. For the structural model of a polyp, we observed that polyps usually have a roughly semi-spherical shape and protrude from the flat colon surface, as shown in Fig. 3.
Based on the illumination model and the structural model of a polyp, we can simulate intensity variations around contours of a polyp under various camera angles (θ = 0°, 55°, 95°). As shown in Fig. 5b, intensities I in of the inside region of the polyp contour tend to be higher than intensities I out of the outside region in all cases. The colon wrinkles and passages have a tube shape and their walls are uneven, as shown in Fig. 5c. As a result, I in is lower than I out for colon wrinkles and passages, as depicted in Fig. 5d, since most of the light cannot reach behind wrinkles and passages. Although our illumination and structural models are simple and imperfect, observations based on them are very helpful for removing FPs.
3.4 Contour Intensity Difference Measure
Based on the observations described in Sect. 3.2.1, we define the CID measure as:
where N is the number of total sampling points around a contour, and I in (j) and I out (j) are the intensity values of the inside and outside regions of the contour around the j-th sampling point, respectively. CID reflects the average intensity difference between the inside and outside regions of the contour.
In order to reliably measure CID, it is important to consider only object-level contours (OLCs) that can cause FPs. In general, OLCs are strong and circular contours similar to those of polyps. Before detecting OLCs, we first detect strong contours using an edge detection method such as the Canny edge detector [20] or a learning-based edge detector [21]. Then we have strong contours e i ∊ E, where E is the initially detected contours, and each contour is denoted by e i = (l i , r i ), where l i is the length of the contour, r i is the radius of circle fitted to the contour. r i represents a curvature of a contour; for example, a large radius implies small curvature. We obtained r i using the least squares circle fitting method [22].
However, as shown in Fig. 6a (Initial), the initially detected contours E contain several outliers. To filter out the outliers, we adopt two-step contour filtering. First, we remove short and noisy contours by checking their length since short contours are caused by clutters in general. To this end, we filter out the noise contours as:
where Wπ is the maximum length of the circle enveloped with W × W (pixels) input image, and α is a parameter determining the allowable length of the contour. We hereby adaptively filter out a number of noisy contours depending on the length of the input image. For instance, Fig. 6a (Step 1) shows the removed noisy contours marked as red and we have OLC length (e i ) = {e 1, e 3}. Next, we further check the curvature of each contour in order to remove remaining outliers (e.g., strong and long enough contours but not circular as a polyp, as shown in Fig. 6a (Step 2)) as follows:
where β defines the allowable curvature range (or radius range) of the contour. In our experiment, we empirically set α = 8, β = 2, and consequently, we have the OLCs (e.g., OLC(e i ) = {e 1}) shown in Fig. 6a (Result). In practice, it is possible that OLC(e i ) contains multiple contours. When OLC(e i ) is obtained, we calculate the normal and inverse normal vectors of each contour point. In order to figure out whether the normal vectors point towardinside or outside of the contour, we check the directions of normal vectors and the position of the fitted circle center. We assume that the vector heading for the circle center (marked as blue arrows in Fig. 6b) is heading for inside of the contour and the vector heading for the opposite direction of the circle center (marked as red arrows in Fig. 6b) is heading for outside of the contour. Likewise, we extract I in (j) and I out (j) for measuring CID, as shown in Fig. 6b. If the value of CID is lower than σ, the detection result is regarded as an FP and ignored. Examples of CID measure values are shown in Fig. 6c.
In order to select a reasonable decision threshold σ, we analyzed CID distributions of polyps and non-polypscausing FPs (i.e., colon wrinkles and passages).Footnote 2 Figure 7 shows both distributions. To separate the two distributions with minimum classification error, the decision threshold is decided as σ opt = − 9. Although σ opt has the minimum classification error (17.3%), polyps will be regarded as FPs with a 8.05% chance. For the polyp detection system, it is more important to maintain a high detection rate than reduce FPs; thus polyps should not be rejected by CID. In that respect, we adjusted the decision threshold to compromise between maintaining the detection rate and rejecting FPs. In this paper, we set the decision threshold σ opt = −25, as shown in Fig. 7 (magenta line). This threshold should keep 97.1% of detected polyps and reject 72.4% of FPs caused by passages or wrinkles.
4 Experimental Design
4.1 Database
In our experiments, we utilized two databases: the CVC colon database (CVC [13]) and our colon databases (ODB and ODB seq ). CVC contains 300 colonoscopy images captured from 15 sequences. Each image contains at least one polyp and the type of polyp is annotated (flat polyp or peduncular polyp). However, CVC has a small number of images of only 15 distinct polyps, which is insufficient to cover challenging situations of polyp detection (e.g., diverse appearances of polyps). To validate the proposed method with diverse appearances of polyps, we built our colon database ODB, which contains 1432 colonoscopic images with 1098 distinct polyps captured from respective examinations. In addition, we built colon sequence database ODB seq , which contains 87 images from three endoscopy sequences. The polyps in ODB and ODB seq were annotated by experts as follows: larger than 5 mm, smaller than 5 mm, laterally spreading tumors, submucosal tumor, and colon cancer. Table 1 shows a summary of the databases.
4.2 Evaluation Settings
In the polyp image datasets, positive samples (i.e., polyp image patches) were manually extracted. Negative samples (i.e., non-polyp image patches), which did not overlap with the positive samples, were automatically and randomly extracted from the images. For the image patches, we extracted the HOG features and used them to train multiple SVM classifiers using a linear kernel. To detect various sizes of polyps, we generated a seven-level image pyramid for each test image with scaling values [0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1]. For example, a 520 × 450 pixel test image with a scaling value of 0.4 will be a 208 × 180 pixel image.
Then, a detector consisting of the multi-classifier densely scans across the image pyramid using the sliding window search, and generates decision probabilities P(x) (Eq. (3)) by combining scores of the multi-classifier at scanning points. From extensive evaluation, we empirically set the window size to 128 × 128 pixels. As mentioned above, with the image pyramid, we can search different sizes of polyps using a fixed window size. After scanning, points with positive decision scores are merged via non-maximal suppression and the remaining points with high scores are considered as locations of bounding boxes BB d . Here, the sizes of boxes are computed by dividing the window size by their scaling values used for image pyramid.
To evaluate detection performance, we employ an intersection of union (IOU) measurement [23], defined as:
We consider two boxes as matched if the ratio of an overlap area over a union area between a detection box BB d and a ground truth box BB gt exceeds 0.5. Matched BB d and BB gt are counted as true positives (TPs). On the other hand, unmatched BB d and BB gt are counted as FPs and false negatives, respectively. Using this matching result, we plot detection error tradeoff (DET) curves, which represent the miss rate versus false positives per image in log–log scale. Our method was tested with various test scenarios for unbiased evaluations, as summarized in Table 2 and compared with several existing methods, as summarized in Table 3.
a Performance of multi-classifiers with various numbers of classifiers, b performance of each sub-classifiers when K = 4, c performance comparison with other detection methods (ROC curves) under test scenario 1, and performance comparisons (DET curves) under d test scenario 1, e test scenario 2, and f test scenario 3 (best viewed in color)
Detection results of polyp video sequences (1) detection results under illumination changes, (2) detection results of textureless polyp, (3) detection results of blurred images (our results: yellow box; Ameling’s [4] results green box)
5 Results
In order to evaluate the methods as fairly as possible, we take into account various test scenarios, as described in Table 2. Under the test scenarios, we first evaluated the performance of several single classifiers with various types of kernel [linear, polynomial, radial basis function (RBF)] and multi-classifiers with various numbers of classifiers, as shown in Fig. 8a. The classifier using the linear kernel outperformed single classifiers using non-linear kernels. This means that a non-linear classifier does not always lead to better performance. The proposed multi-classifiers outperformed all single classifiers. This implies that the proposed multi-classifier learning method achieves good generalization for the various appearances of polyps. Among the multi-classifiers, the multi-classifier with K = 4 showed the best performance; therefore, we selected it for subsequent experiments.
Figure 8b shows the performance (for all test images) of a single classifier (SCL), four different sub-classifiers, and a multi-classifier containing the four sub-classifiers (MCL). Overall, all sub-classifiers yield low performance since they are trained for detecting a specific appearance. On the other hand, the performance of the multi-classifier shows good performanceby combining the sub-classifiers. Interestingly, the single classifier that used all training samples performs worse than the second sub-classifier that used training image set 2 in Fig. 3. This result indicates that it is difficult to cover various appearances of polyps using only a single classifier, and that training a classifier with too many diverse samples can degrade performance.
Figure 8c, d show the performances of detectors with different features and learning methods. We implemented other polyp detection methods using LBP, GLCM16 [4], and CWC [5] within our framework for a fair comparison. Since other studies [4, 5] evaluated their performance using the receiver operating characteristic (ROC) curve based on a per-window measure, we conduct the comparison using the ROC curve with a per-window measure in Fig. 8c and the DET curve with a per-image measure in Fig. 8dFootnote 3 under test scenario 1 (fivefold cross-validation using two datasets). In Fig. 8c, the performance (82.5%) of the SCL using the HOG feature is better than that (77.5–71.6%) of the others [4, 5], proving that the HOG feature is more appropriate for polyp detection. In Fig. 8d, the proposed detector MCL-HOG+CID (blue) shows the best performance, proving the effectiveness of the proposed MCL and CID measure. By comparing the DET curves of SCL-HOG (black), MCL-HOG (red), and MCL-HOG+CID (blue), we can see that the proposed learning and CID reduce FPs while preserving the detection rate. As shown in Fig. 6d, CID can extract OLCs and measure the intensity difference around the contour to remove FPs.
In order to compare the methods more extensively, we provide additional results under test scenarios 2 and 3 (details given in Table 2). Through these test scenarios, we observed the performance of each polyp detector to detect totally unseen polyps. Since the two datasets do not share the same patients, the distributions of images (e.g., identities of polyps, view point, lighting condition, etc.) in CVC and ODB can be different. Figure 8e, f show the results under test scenarios 2 and 3, respectively.
The results of Fig. 8f generally show lower performance than those of Fig. 8e since test scenario 3 is much more challenging.Footnote 4 Nevertheless, our methods still show promising performance under both scenarios. The LBP-based method [4] performed poorlyunder scenario 3, as shown in Fig. 8f. The proposed CID measure under scenario 3 contributed a lot to remove FPs since ODB contains more challenging scenes (e.g., with many colon wrinkles and passages) than those in CVC (Fig. 8f). This result confirms that the CID measure works properly for removing FPs and that its contribution is higher when the colon scenes are complex .
In order to compare our method with more recent work, we also compared the methods using various evaluation metrics, as show in Table 4. F 1 score is a measure of testing accuracy that considers both precision and recall:
where \(\rm precision = \frac{TP}{TP + FP}\) and \(\rm recall = \frac{TP}{TP + FN}\). The results show that our method outperforms SA-DOVA [13] in terms of accuracy (F 1) and speed.
Figure 9 shows the qualitative evaluation results (i.e., detection results) of our methods. Polyps with a roughly semi-spherical shape are detected by both detectors (i.e., SCL and MCL), as shown in Fig. 9a–d. However, the detector based on SCL produces FPs and missed detections due to colon wrinkles, as shown in Fig. 9e–h. Figure 9i–l show that the detector based on MCL successfully detects various shapes of polyps using the multi-classifier and generates few FPs using the CID measure (no polyps are detected by the detector based on SCL in Fig. 9i–l).
As shown in Fig. 8c–f, the LBP-based method [4] shows higher performance than that of other previous methods in the various test scenarios. To show the performance enhancement based on our methods, we provide qualitative evaluation results of our method and Ameling’s method [4] under challenging endoscopic sequence dataset ODB seq , as shown in Fig. 10. Sequence 01 contains severe illumination changes (frames #1–#3), but our polyp detector successfully detects all polyps. In sequence 02, a textureless polyp is not detected by the texture-based detector [4], whereas it is correctly detected by our shape-based detector using the HOG feature. We further show that our detector can produce accurate detections in blurred images caused by severe endoscope motion, as shown in sequence 03 (frames #2–#5). In these sequences, our method achieves high detection accuracy but also reduces FPs and missed detections. These experimental results confirm that our method is far superior to Ameling’s method [4]. They also show that our method is more reliable and appropriate in a clinical environment for polyp detection.
6 Conclusion
In this paper, we proposed a framework for automatic polyp detection. We evaluated several features to discriminate between polyps and normal tissues and adopted the HOG feature, which has larger discriminative power than that of other features. In addition, we proposed a multi-classifier learning method for detecting polyps with significant appearance variations and a CID measure for removing FPs caused by colon winkles and passages. Experimental results using both public and our own datasets verify that our method achieves better performance than other state-of-the-art polyp detection methods.
Notes
We aim to train multiple classifiers that can detect specific shapes of polyps against the background. Therefore, in this paper, we do not cluster the negative samples.
In order to find the CID probability densities, we performed the following process. First, we manually detected all polyps and the FP factors in the datasets. Second, we extracted all CID measures of polyps and FP factors in the test dataset and found the CID probability densities. For example, when we conducted fivefold cross-validation, we have five test datasets, and obtain five CID probability densities of polyps and FP factors, respectively. In practice, we set the CID distributions from the first fold test dataset as the representatives since the five distributions from each fold are very similar to each other.
Test scenario 3 is challenging because ODB contains more diverse polyps and complex scenes than those in CVC and the numbers of training and test samples are not balanced (training: 300 ≪ test: 1432).
References
World cancer report. (2014). IARC Nonserial Publication.
Baxter, N. N., Goldwasser, M. A., Paszat, L. F., Saskin, R., Urbach, D. R., & Rabeneck, L. (2009). Association of colonoscopy and death from colorectal cancer. Annals of Internal Medicine, 150, 1–8.
van Rijn, J. C., Reitsma, J. B., Stoker, J., Bossuyt, P. M., van Deventer, S. J., & Dekker, E. (2006). Polyp miss rate determined by tandem colonoscopy: A systematic review. The American Journal of Gastroenterology, 101, 343–350.
Ameling, S., Wirth, S., Paulus, D., Lacey, G., & Vilarino, F. (2009). Texture-based polyp detection in colonoscopy. Bildverarbeitung für die Medizin, 346–350. doi:10.1007/978-3-540-93860-6_70.
Iakovidis, D. K., Maroulis, D. E., & Karkanis, S. A. (2006). An intelligent system for automatic detection of gastrointestinal adenomas in video endoscopy. Computers in Biology and Medicine, 36, 1084–1103.
Karkanis, S. A., Iakovidis, D. K., Maroulis, D. E., Karras, D. A., & Tzivras, M. (2003). Computer-aided tumor detection in endoscopic video using color wavelet features. IEEE Transactions on Information Technology in Biomedicine, 7, 141–152.
Li, B.-P., & Meng, M. Q.-H. (2012). Comparison of several texture features for tumor detection in CE images. Journal of Medical Systems, 36, 2463–2469.
Alexandre, L. A., Casteleiro, J., & Nobreinst, N. (2007). Polyp detection in endoscopic video using SVMs. Knowledge Discovery in Databases, 2007, 358–365.
Tjoa, M. P., & Krishnan, S. M. (2003). Feature extraction for the analysis of colon status from the endoscopic images. BioMedical Engineering OnLine, 2, 1–17.
Dietterich, T. G. (2000). Ensemble methods in machine learning. Multiple Classifier Systems, 1857, 1–15.
Dalal, N., & Triggs, B. (2005). Histograms of oriented gradients for human detection. Computer Vision and Pattern Recognition, 1, 693–886.
Zhang, C., Platt, J. C., & Viola, P. A. (2005). Multiple instance boosting for object detection. Advances in Neural Information Processing Systems, 7, 1417–1424.
Bernal, J., Sánchez, J., & Vilarino, F. (2012). Towards automatic polyp detection with a polyp appearance model. Pattern Recognition, 45, 3166–3182.
Iakovidis, D. K., Maroulis, D. E., Karkanis, S. A., & Brokos, A. (2005). A comparative study of texture features for the discrimination of gastric polyps in endoscopic video, Computer-Based Medical Systems, 575–580.
Li, B., & Meng, M. Q.-H. (2012). Automatic polyp detection for wireless capsule endoscopy images. Expert Systems with Applications, 39, 10952–10958.
Bieniek, A., & Moga, A. (2000). An efficient watershed algorithm based on connected components. Pattern Recognition, 6, 907–916.
Hwang, S., and Oh, J-H., Tavanapong, W., Wong, J., & de Groen, P. C. (2007). Polyp detection in colonoscopy video using elliptical shape feature, International Conference on Image Processing, 2, II-465.
Li, P., Chan, K. L., & Krishnan, S. M. (2005). Learning a multi-size patch-based hybrid kernel machine ensemble for abnormal region detection in colonoscopic images. Computer Vision and Pattern Recognition, 2, 670–675.
Phong, B. T. (1975). Illumination for computer generated pictures. Communications of the ACM, 18, 311–317.
Canny, J. (1986). A computational approach to edge detection. Pattern Analysis and Machine Intelligence, 6, 679–698.
Dollár, P., & Zitnick, C. L. (2013). Structured Forests for Fast edge Detection, International Conference on Computer Vision, 1841–1848.
Coope, I. D. (1993). Circle fitting by linear and nonlinear least squares. Journal of Optimization Theory and Applications, 76, 381–388.
Everingham, M., Zisserman, A., Williams, C. K., Van Gool, L., Allan, M., Bishop, C. M., et al., (2006). The 2005 pascal visual object classes challenge. Machine Learning Challenges. Evaluating Predictive Uncertainty, Visual Object Classification, and Recognising Tectual Entailment, 117–176. doi:10.1007/11736790_8.
Dollár, P., and Wojek, C., Schiele, B., & Perona, P. (2009). Pedestrian detection: A benchmark, Computer Vision and Pattern Recognition, 304–311.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cho, YJ., Bae, SH. & Yoon, KJ. Multi-Classifier-Based Automatic Polyp Detection in Endoscopic Images. J. Med. Biol. Eng. 36, 871–882 (2016). https://doi.org/10.1007/s40846-016-0190-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-016-0190-4