Abstract
Experimental chemistry and the younger discipline of computational chemistry have always aspired to increase data volume, velocity, and variety. The recent software developments in machine learning, databases and automation and hardware advances in fast co-processors, networking, and storage have boosted automation and digitization. Computational chemistry is seemingly on the verge of a big-data revolution.
In this chapter, we discuss how many of these data-driven paradigms are part of long-term trend and data have long been at the heart of many chemical problems. Historical repositories of chemical data where the modern cheminformatician can mine high value curated training data are reviewed. Modern automation tools and datasets available for high-data computational chemistry are described. Current applications of computer-driven discovery of molecular materials in optoelectronics (photovoltaics and light-emitting diodes) and electrical energy storage are discussed. Finally, the impact of machine learning approaches to computational chemistry areas of structure-property relationships and chemical space, with an emphasis on generative models, are analyzed.
Access provided by Autonomous University of Puebla. Download reference work entry PDF
Similar content being viewed by others
1 Introduction
Chemistry has been a big-data enterprise for multiple decades. Long before the term was popularized, chemists strived to wield chemical datasets whose volume, variety, and velocity exceeded the ability of tools available to them. For one, chemists have long struggled to address the immensity of chemical space. Since the arrival of computers, chemists have been creators and early adopters of digital technologies for the storage, processing, and retrieval of chemical data. Along those same lines, the advent of the Internet set off a new era of accelerating knowledge creation and sharing in the chemical sciences.
In fields such consumer analytics, advertising, transportation, healthcare or finance, the clearest gains from big-data, and machine learning approaches have come from subareas that combine as many favorable features as possible: data is inexpensive to generate, digital in origin or easy to digitize, highly organized and consistent, and extremely abundant. While abundant, chemical data tends to be expensive to generate, heterogeneous, hand-made, and sparse. Chemical data entry and curation is generally done manually; these tasks require significant expertise because of the heterogeneous sources and reporting criteria, and the technical complexity of chemical reports. In recent years, as chemical instruments, such as microscopes or spectrometers, produce digitized data, very large datasets can be generated by a single device. This very much opens the door to leverage local big data, but one still faces similar barriers when attempting to consolidate results from different groups or apparatuses.
Computational chemistry approaches share a series of features that make them very amenable to this data-driven paradigm and are proving to be the perfect testing ground for realizing the big-data vision in chemistry. The output of computational chemistry software is digitized from beginning to end. Even if it relies on custom-formatted input and output files, automating the creation and processing of these files is a simple task. Computational results are replicable to floating point precision when using the same computer code, and with extreme accuracy even among different programs as long as they use the same underlying algorithms. Because compute is essentially a commodity, results can be generated in arbitrary amounts with linear scaling in cost – or even sublinear, thanks to machine learning approaches. Finally, there is a well-understood tradeoff between computational cost and accuracy that can be leveraged through data-driven techniques.
In this chapter, we will analyze how computational chemistry has embraced big-data for accelerated discovery of much needed new molecules and how these newly generated datasets have allowed to apply, and develop, machine learning techniques that make them even more powerful.
2 Repositories of Chemical Data
The initial development of big-data approaches in chemistry was historically focused on consolidating, curating, and indexing literature and empirical data. Tackling these tasks was one of the first applications of computers in chemistry and remains a key driver in the development of new computational tools for chemistry.
The Chemical Abstracts Service (CAS) was created in 1907 to keep complete records of the chemical literature and reported substances, following the idea of previous abstracts journals in Europe such as Chemisches Zentralblatt and nineteenth-century compilations such as Beilsteins Handbuch der organischen Chemie, Gmelins Handbuch der anorganischen Chemie, or the Merck Index. It contained over 12,000 indices in its first year, its one millionth record in 1937. CAS historically relied on thousands of volunteers to create abstracts and index information for each paper – not that far from today’s Mechanical Turks at Amazon – and slowly phased them out between the late sixties and the nineties. In addition, CAS employed hundreds of staff members by the 1950s, underscoring both the great value of big data to chemistry and the great cost of getting that same data into neatly formatted into useable form.
The adoption of digital computers for chemical applications in the late 1960s initiated the digitization of chemical big data by facilitating the storage, entry, and very importantly querying of chemical databanks. CAS computer-based Chemical Registry System, like the Beilstein Registry Number before it, assigns unique individual arbitrary numbers to chemicals. It debuted internally in 1964 and was progressively updated and expanded, making the basis for increasingly modern querying and retrieval tools, using command-line and graphical interfaces to retrieve data from local or online records. With around three million records and growing at hundreds of thousands per year in the mid-seventies, the registry grew exponentially to its 30 millionth chemical substances in 2007, 50 millionth in 2009, and 100 millionth in 2015. Competing commercial efforts that also build on older academic work are Elservier’s Reaxys (Beilstein and Gmelin) and Thomson-Reuters, now Clarivate.
In 1965, the Kennard group at Cambridge University started collecting published crystallographic data for small molecules. This effort grew into what is known as the Cambridge Structural Database, which now hosts nearly one million curated entries from x-ray and neutron diffraction analyses (Groom et al. 2016). Also in the late 1960s, joint efforts between Brookhaven National Laboratory and Texas A&M University initiated what would become the Protein Data Bank, a repository for three-dimensional structural data of large biological molecules, such as proteins and nucleic acids, and now holds over 120,000 entries (Berman et al. 2000; Bernstein et al. 1978; Meyer 1997).
Other repositories of experimental crystallographic information (Bruno et al. 2017) are the Crystallography Open Database (COD) (Gražulis et al. 2009, 2012), which stores user-submitted data; the Inorganic Crystal Structure Database, produced cooperatively by FIZ Karlsruhe and NIST that stores crystal structures of inorganic solids (Belsky et al. 2002); and the International Centre for Diffraction Data (ICDD) that stores powder diffraction patterns (Faber et al. 2002).
The rise of the Internet sparked the launch of other big-data repositories and the expansion of online availability of existing ones both in chemistry and materials. The Registry of Research Data Repositories aggregates lists of multiple repositories and datasets along many scientific disciplines including chemistry and materials science (Pampel et al. 2013).
The National Institute of Standards and Technology has hosted the NIST Chemistry WebBook since the 1996 (Linstrom and Mallard 2001), a compendium of mostly spectroscopic and thermodynamic data originally compiled in handbooks and tables. In addition, the nearly 200 Standard Reference Data sets include other data of interest to chemists. NIST 101 (III 1999) in particular contains calculated data quantum chemical data for about 1800 gas-phase atoms and small molecules and tools for comparing experimental and computational ideal-gas thermochemical properties.
Since patents and patent applications are open, they are a big-data, information-rich asset. Patent documents, however, tend to be only text-based or image-based, and their digitization into a format that is useable for data-driven approaches is a standing challenge. The European, United States, Japanese Patent Offices, and the World Intellectual Property Organization can be accessed and queried online, but they are not systematically annotated. The US Patent and Trademark Office, through their Complex Work Unit program, made available digital representation of molecules in patent applications, easing the data-processing pipelines.
In the open-data arena, NextMoveSoftware released close to million chemical reactions, extracted by means of automated text mining of the relevant experimental sections reported in patents, covering the period between 1976 and 2013. SCRIPDB contains curated syntheses, chemicals, and reactions from the patent literature, collected from CWU files coming from granted US patents (Heifets and Jurisica 2012).
SureChem is another molecule database, open-sourced in 2012 to SureChemBL. It used by IBM’s Strategic IP Insight Platform and initially released more than two million chemical structures extracted from about 4.7 million patents (1976–2000, only text) and subsequently extended to include patents published up to the end of 2010 and chemical structures were additionally derived from US CWUs and images (2001–2010) (Papadatos et al. 2016). UniChem attempts to unify and cross reference these multiple databases (Chambers et al. 2013).
ChemSpider is a database of chemicals with more than 60 million entries from over 480 data sources. Originally a private enterprise, it was acquired by the Royal Society of Chemistry (RSC) in 2009. It includes a crowd-sourced component, but only limited downloads are available (Pence and Williams 2010).
Because of the large size of the set of possible small organic molecules with biological activity, the large number and diversity of biological targets, the high dimension of many biological assays and measurements and the high value of healthcare applications, cheminformatics, biological chemistry, and drug discovery applications are key target for big data applications in chemistry. Toxicological information of drugs and chemicals are also interesting biological interactions that are recorded in several publicly available databases.
The National Center for Biotechnology Information (NCBI), part of the United States National Library of Medicine (NLM) from the National Institutes of Health (NIH), hosts multiple datasets and informatics tools in biotechnology. Of particular interest in chemistry is PubChem, a database of molecules and their activities against biological assays. Initiated in 2004, it now aggregates over 540 sources, over 90 million compounds, and 233 million bioactivity results for nearly 2.5 million of those. As a federally sponsored service, PubChem has been seen to be in conflict with for-profit repositories such as CAS (Kaiser 2005).
ChemBL is an open database that contains binding, functional, and ADMET (absorption, distribution, metabolism, excretion) measurements for drug-like biologically active compounds, with nearly 15 million bioactivity measurements for more than 1 million compounds and 11,500 protein targets (Gaulton et al. 2012). DrugBank is a web-accessible cheminformatics database and service combining structural and biological target data for drug molecules. The database contains in excess of 9000 small molecule drugs, 3000 FDA-approved drugs, and data for nearly 17,000 drug-target associations (Wishart et al. 2006, 2018). The database for Chemical Entities of Biological Interest follows a similar focus as well (Degtyarenko et al. 2008).
The PDBbind database matches published affinity constants from the literature for the ligand-protein systems whose 3D structures are stored in PDB, undergoing periodic updates (Liu et al. 2015; Wang et al. 2004). BindingDB (Chen et al. 2001; Gilson et al. 2016) and AffinDB (Block et al. 2006) also aim to fulfill a similar task. The comparative Toxicogenomics Database is a volunteer-based genomic resource devoted to toxicologically relevant genes and proteins and their interactions with chemicals and toxins (Davis et al. 2017; Mattingly et al. 2003). MACiE contains enzyme reaction mechanisms focused on the evolution of enzyme catalytic mechanisms and the classification with respect to chemical mechanism (Holliday et al. 2005).
These services are big-data pioneers in chemistry and highlight both the early understanding of how chemistry is a high-dimensional, sparse-data arena where big-data approaches can create great value, as well as the high cost of gathering and curating chemical data. For machine learning applications, these repositories provide extremely valuable labeled data.
3 Big Computational Chemistry
The field of computational chemistry quickly followed the deployment of the first digital computers and has grown at a fast pace, matching developments in algorithms and hardware and also contributing its own. Oftentimes, the chemical calculations carried out will push the hardware available to run them: running in parallel over many cores in supercomputers, using large amounts of memory or storage.
In that sense, judging by the volume and velocity of the output data they produce, and by the strain on hardware requirements, computational chemistry calculations have been big-data all along, essentially as big as the available resources allowed and have been drivers of big-data technologies.
In this chapter, we will focus on applications of computational chemistry that combine volume and velocity with also high variety because of a large degree of granularity. These are the cases of (i) distributive computing, where small calculation payloads are distributed to a large grid of small computers, and after computation, the results are consolidated and processed, and (ii) high-throughput virtual screening, where many thousands of candidate molecules or materials are calculated individually in an automated fashion and a data-driven search for the most performant candidate materials is carried out.
3.1 Automation and Databases in Computational Chemistry
Many tools have been developed to automate the creation, submission, transferring, processing, parsing, storage, and querying of computational chemistry data. One of the most venerable web-based examples is the Basis Set Exchange, originally assembled at the Environmental Molecular Sciences Laboratory, where a myriad of curated basis sets for most of the periodic table are available for download in multiple formats (Feller 1996; Schuchardt et al. 2007).
Close to half dozen platforms exist with a similar philosophy towards achieving some or all the following: automation of materials science and solid-state electronic structure calculations; data processing and analysis of those calculations; and centralized, web-accessible repositories of the output of these calculations for virtual discovery and machine learning purposes. The multiple solutions offer somewhat overlapping functionality, generally in the materials space, and have been reviewed recently (Lin 2015).
The Electronic Structure Project (Klintenberg et al. 2002; Ortiz et al. 2009) utilized the structural data from the ICSD to screen for novel inorganic materials.
The Computational Materials Repository (Landis et al. 2012) proposes an integrated software solution for computer-driven materials design. It is part of the ecosystem of the Quantum Materials Informatics Project that also includes the Atomic Simulation Environment, a python library for working on atomistic simulations (Hjorth Larsen et al. 2017).
AFLOW is an automatic framework for high-throughput materials discovery (Calderon et al. 2015; Curtarolo et al. 2012a), and the matching repository Aflowlib.org hosts and serves the results of those calculations to the public (Curtarolo et al. 2012b). It now contains 1,748,704 material compounds – with over 173,121,696 calculated properties.
The Python Materials Genomics (pymatgen) is an open-source python library for materials for the analysis of solid-state DFT calculations (Ong et al. 2013) which also has a matching Materials Application Programming Interface (Ong et al. 2015) to interact with the Materials Project, a large-scale database of materials calculations (Jain et al. 2013). Other computational tools in this ecosystem include tools such as Custodian for error handling, Fireworks for workflow management. The Materials Project tools and data have been used in over 100 published papers, and nearly 200 by its creators.
The Open Quantum Materials Database (OQMD) is a fully open project that hosts over 400,000 DFT energy calculations of compounds from the ICSD and also for hypothetical compounds, potentially uncovering valid, but yet to synthesize chemistries (Kirklin et al. 2015).
AiiDA (Pizzi et al. 2016) is a flexible and scalable informatics’ infrastructure for simulations, data, and workflows with a heavy focus on plane-wave DFT calculations of materials and much attention to data provenance (Merkys et al. 2017).
The Novel Materials Discovery repository was established to host, organize, and share materials data in a pipeline-agnostic way (Goldsmith 2016) and hosts over 44 million open access user-submitted total-energy calculations from a variety of computer codes.
The ioChem-BD Platform provides a similar solution with an emphasis on molecular data (Álvarez-Moreno et al. 2015). Several toolkits address the simulation and role of defects, such as MAST (Mayeshiba et al. 2017), PyDII (Ding et al. 2015), and others (Goyal et al. 2017). PyChemia is a python library for automatize atomistic simulations, with a focus on materials and interfaces to some DFT codes and data mining functionality.
Multiples datasets of computational chemistry results aimed purely at generating diverse training data exist. QM9 contains B3LYP/6–31 G(2df,p) results for 134k stable small organic molecules made up of CHONF, including harmonic frequencies (Ramakrishnan et al. 2014). PubChemQC is a recent attempt to create training data for machine learning approaches that calculated the ground-state electronic structures of three million molecules based on density functional theory (DFT) at the B3LYP/6-31G* level and 10 lowest excited states of over two million molecules at TD-DFT/B3LYP/6–31 + G* level of theory (Nakata and Shimazaki 2017). ANI-1 contains energies and DFT-level properties for 20 million conformations for over 50,000 small organic molecules distorted along normal modes (Smith et al. 2017). Ab initio molecular dynamics are also available: 5,000 frames at 500 K at the PBE level of theory for 113 structural isomers of C7O2H10 and hundreds of thousands frames for 8 small organic molecules (Chmiela et al. 2017; Schütt et al. 2017).
A large number of internal tools for data processing have grown into open libraries that are available for data analysis of calculation outputs: the python-based cclib for parsing and interpreting the results of computational chemistry packages (O’boyle et al. 2008); ESTEST, a free framework for the comparison, validation and sharing of quantum chemical calculation outputs (Yuan and Gygi 2010); ORBKIT, also python-based library for postprocessing of quantum chemical wavefunction outputs from multiple codes; (Hermann et al. 2016); and PyGlobal, spreadsheet-oriented output postprocessing tool for DFT calculations (Nath et al. 2016). Other tools are also available for input generation and pipelining calculations, such as JACOB (Waller et al. 2013) a framework for computational chemistry aimed at enterprise application, PyADF for scripting multiscale quantum chemistry using the ADF package (Jacob et al. 2011).
This ample landscape suggests that increasing both the velocity and volume of quantum chemical calculations is of great interest. These increases, however, come with a tradeoff in the variety of the applications. Because of the added complexity, most of these rarely see adoption outside the groups or consortia that created them (Thygesen and Jacobsen 2016).
3.2 High-Throughput Virtual Screening
As computational methods become more accurate and computing hardware more affordable, the possibility of automatically prescreening compounds virtually before synthesis grows more promising. Various teams have used HTVS for discovery of many inorganic materials. These are treated in depth in other chapters of this book. Here, the focus will be on organic molecular materials in the domains of organic optoelectronics for light-energy interconversion (photovoltaics and light emitting diodes) and for electrical energy storage (Pyzer-Knapp et al. 2015).
3.2.1 Optoelectronics
Merging concepts from both the volunteer, distributive computing efforts and the HTVS vision, lies the Clear Energy Project (CEP). Since its inception around 2006 and throughout two phases, this project ran on IBM World Community Grid, where volunteers donated computer time to virtually screen p-type, and later n-type, organic photovoltaic oligomers.
CEP tackled many of the challenges for automated virtual testing of chemical compounds: programmatic generation of candidate molecules, automation of the quantum chemical calculation, data storage, and analysis.
Combining a pool of 20 fragments through covalent linking at active sites and also by forming fused rings adjacent to these labeled reactive atoms, the CEP molecular generation processed went up to tetramers and produced over two million candidates that were screened exhaustively. The quantum chemical calculations carried out included multiple DFT functionals and amount to the largest computational chemistry project to that date. The candidate tetramers were assessed using the Scharber model for photovoltaic efficiency, assuming fullerenes as electron acceptors and both raw and empirically calibrated donor energy levels. The statistical analysis of the large dataset produced afforded correlations for the most and least promising fragments and fragment combinations (Hachmann et al. 2011, 2014; Olivares-Amaya et al. 2011).
The CEP, because of the large theoretical dataset, and the existence of numerous independent experimental results in the area of OPV has proven a testing ground for novel approaches to materials discovery. A probabilistic kernel-based calibration scheme to improve theoretical gas-phase results and to capture the bulk effects using a collection of experimental results from the literature (Lopez et al. 2016) improved the predictive performance of DFT calculations (Pyzer-Knapp et al. 2016).
A later subproject has focused on the virtual screening of over 50,000 nonfullerene electron acceptors from the combination of over 100 common organic moieties. Time-dependent density functional theory calculations were also carried out for elected lead compounds. Diketopyrrolopyrroles and quinoidal thiophene derivatives showed good promise and were proposed for additional study (Lopez et al. 2017).
At a less gargantuan scale, other works also addressed screening over bulk heterojunction solar cell components, such as combinatorial band-gap design strategy over 780 different copolymer donor materials (Shin et al. 2014).
Some works have addressed virtual discovery of molecules for other classes of solar cells, such as TiO2-based dye-sensitized solar cells, optimizing over common dyes (Martsinovich and Troisi 2011) and also over porphyrins (Ørnsø et al. 2014).
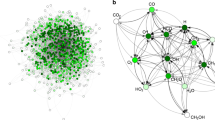
A successful example of blending computational chemistry with deep learning for applied materials discovery has been reported in the area of thermally activated delayed fluorescence (TADF) organic light-emitting diodes (Gómez-Bombarelli et al. 2016). Using custom software that mimics cross-coupling reactions on existing staring materials, a database spanning nearly two million feasible compounds was created from over two hundred starting donor and acceptor fragments. The TADF character of the compounds, as well as their color, was estimated using accurate, empirically calibrated TD-DFT calculations. For accelerated results, the candidates were screened through a neural network, using topological fingerprints as features. The leading compounds with the most promising predicted chemical properties were assessed by a team of experts who synthesized and tested the consensus champion compounds. These were then tested in optoelectronic devices, where they matched the performance of human-generated champion compounds (Fig. 1).
Overview of TADF OLED chemical space. Red dots represent overlap with previous experimental reports in the literature, orange triangles correspond to theoretical leads, cyan squares represent experimentally confirmed theoretical leads. The log-density of molecular candidates as a function of singlet-triplet gap and oscillator strength of shown. The dashed lines follow iso-contours of estimated rate of delayed fluorescence
3.2.2 Electrolytes and Energy Storage
Energy storage is one of the most active areas of materials science and engineering, given the strong demand in both lightweight, high-energy density applications such as mobile phones or transportation and static, low-cost, grid-scale storage.
Flow batteries are large, static batteries where liquid electrolytes are stored in tanks and circulated across an electrochemical cell when charging or discharging. Although they show somewhat inferior energy densities compared to solid-state batteries such as lithium-ion, flow batteries are potentially a much better solution to grid-scale electrical storage because of the lower cost and use of earth-abundant and cheap electrolyte materials. The independent scaling of power (depending on effects such as the kinetics of the electrochemical reaction and the electrode surface area) and energy (related to the total size of the tanks, the concentration of the electrolyte in solution, and the degree of ionization of the electrolyte) allows more flexible engineering for flow-battery solutions. Using water as solvent further reduces the installation costs and increases the operational safety. Because they can be sources from oil and from sustainable biomatter, the great design flexibility of organic molecules, and their promising performance, organic electrolytes for flow batteries are heavily considered contestants (Kowalski et al. 2016; Leung et al. 2017; Wei et al. 2017).
High-throughput simulation, particularly when coupled with experimental follow-on, has become a powerful tool in the discovery of electrolyte materials for organic flow batteries. The report of an efficient, metal-free, low-cost flow battery design using acidic aqueous solutions of anthraquinone disulfonate and bromine/hydrobromic acid leveraged thousands of DFT calculations of potential quinone molecules to optimize reduction potential of the negolyte and its aqueous solubility (Huskinson et al. 2014). The careful analysis of the theoretical predictions allowed to identify useful design rules and structure-property relationships over the domain of benzo-, naphto-, and anthraquinones bearing any of 13 functional groups (Er et al. 2015). Other high-volume calculations of bio-inspired thiophenoquinone derivatives also identified potentially useful electrolytes for flow batteries (Pineda Flores et al. 2015).
The Electrolyte Genome project used the Materials Project backbone to perform high-throughput calculations on a set of nearly 5000 molecules derived mostly from quinoxaline and anthraquinone, thiane, thiophene, and bipyridine derivatives (Qu et al. 2015). Special attention was paid to error handling and estimation of redox potentials, ion pair dissociation, and complex salt formation. A detailed analysis of nearly 1400 quinoxalines for nonaqueous flow batteries estimating redox potentials, solvation energies, and structural changes was also reported (Cheng et al. 2015), as well as over 4000 five- or six-membered rings with one or two functional groups attached (Pelzer et al. 2017).
Somewhat smaller scale virtual searches have also been reported. Starting from experimentally reported molecules available in PubChem, and down-selecting manually to 315 after deploying a neural network, improved experimental performance was observed for a quinoxaline derivative (Park et al. 2015, 2016).
The chemical stability of organic molecules for electrochemical applications, which impact battery shelf-life and long-term cyclability, is one of the standing issues of virtual discovery applications. For this purpose, the first and second reduction and oxidation of organic solvents for lithium-ion batteries have been analyzed using DFT (Borodin et al. 2015). Similarly, bio-inspired alloxazine electrolytes have been shown experimentally to provide excellent stability in alkaline aqueous flow batteries, by including stability criteria in the virtual search, in addition to redox and solubility requirements (Lin et al. 2016).
Smaller approaches with less focus on automation and extreme throughput have been reported, focusing on aspects such as low Li+ binding affinity over 32 organic molecules (Park et al. 2011), 15 counterions for magnesium electrolytes (Qu et al. 2017), redox-switchable polymer-based membranes (Ward et al. 2017), or tuning the reduction potential of organic molecules to optimize the combination of small redox active molecules with conducting polymers (Araujo et al. 2017).
HTVS has also been applied for materials related to other electrochemical applications such as double layer capacitors (Schütter et al. 2016) or other green-chemistry applications such as switchable-hydrophilicity solvents (R. Vanderveen et al. 2015).
4 Applied Machine Learning for Accelerated Discovery
As is the case in many other areas, machine learning applications to chemistry are not a novel enterprise (Pierce and Hohne 1986). As is also the case in many other applications, the combination of deep learning algorithms with larger datasets and specialized computing hardware has resulted in many effective applications of machine learning for chemistry.
4.1 Quantitative Structure-Property Relationships
Most applications of machine learning to computational chemistry applications are related to building fast statistical proxies to expensive calculations on three-dimensional atomic arrangements or chemical graphs. This is, in a sense, building quantitative structure-property relationships trained either on molecular structures or on 3D geometries, often generated on-the-fly using molecular mechanics.
Other chapters in this same publication give a thorough review of many recent contributions to this area, so only a brief overview will be provided. Readers are encouraged to explore further in this same book.
The idea of using neural network models to interpolate over potential energy surfaces goes back to the 1990s (Blank et al. 1995). Very early contemporary works applying machine learning to computational chemistry are those by Behler where neural networks were used for molecular dynamics simulations and other PES exploration techniques (Behler 2011b, a; Behler et al. 2007; Behler and Parrinello 2007). These and many other applications of neural-networks, mostly to bulk and heterogenous systems, have been reviewed recently (Behler 2017).
Another line of application of machine learning techniques to computational chemistry, with a heavier focus on molecules, rather than bulk materials initially leveraged kernel methods, with emphasis on mapping 3D coordinates (Rupp et al. 2012; Schütt et al. 2017) or electronic densities (Brockherde et al. 2017; Snyder et al. 2012) to properties. These approaches are well covered in other chapters of this publication.
One of the most important issues in predictions based on molecular structures is the choice of features to represent molecules. Chemists tend to think of molecules as undirected graphs, with atoms in the nodes and bonds in the edges, and some additional considerations for stereochemistry. In quantum chemistry, molecules are three-dimensional arrangements of atoms that correspond to a local minimum in the potential energy hypersurface. For multiple decades, cheminformatics applications, mostly in pharma and drug discovery, have created descriptive, rich features for machine learning over both graphs: pharmacophore fingerprints, topological fingerprints, and also 3D conformations.
Recent applications of machine learning to molecules in chemistry have focused on QSPR over quantum-chemical properties and placed their focus on more information-rich features. The Coulomb matrix, for instance, is a permutation-dependent symmetry invariant modified distance matrix. Contemporary highly powerful neural network approaches also leverage 3D information (Gilmer et al. 2017; Lubbers et al. 2017; Schütt et al. 2017). In the case of molecules whose 3D structure is unknown, which is generally the case when carrying out predictions, these features are calculated from guess geometries calculated on the fly at the molecular mechanics level of theory. Molecular mechanics methods, through the underlying force fields, are heavily parametrized, generally on quantum chemical calculations but eventually on the experimental data used to parametrize those too. Therefore, the machine learning methods applied over these features are heavily supervised, with their inputs essentially embedding quantitatively a large amount of prior chemical knowledge and hand-tuning.
Learning on graphs presents a very unique set of challenges, because of graph-isomorphism. Machine learning approaches over graph structures have much interest beyond chemistry, such as in networks of any kind in transportation, databases, telecommunications. Whereas extended connectivity circular fingerprints address graph isomorphism, the process of encoding the graph, hashing, muddles chemical information and can lead to different chemical substructures activating the same feature. Neural fingerprints, a differentiable deep-learning extension of ECFP, have recently been proposed as a more flexible and learnable alternative (Duvenaud et al. 2015). These continuous counterparts of topological fingerprints and derivative graph convolutions for chemistry (Kearnes et al. 2016) have proven to outperform some traditional cheminformatics descriptors from QSPR, particularly for larger datasets, and open a new avenue for molecular screening.
4.2 Searching and Optimizing in Chemical Space
An interesting subset of applications of machine learning to the computational chemistry/cheminformatics community is moving in the discrete, high-dimensional space spanned by all possible molecules, or by a relevant subset, such as the small organic molecules with aromatic rings, or similar subsets of interest.
Chemical space is extremely large, with estimates ranging by many orders or magnitude, from 1023 to 1060. Rule-based efforts to enumerate molecules, exhaustively for small compounds and culling nonpractical but formally valid and even potentially stable molecules, have been reported as follows: GDB-11 lists 26.4 million small organic molecules of up to 11 atoms of C, N, O, and F (Fink and Reymond 2007); GDB-13 enumerates 978 billion molecules with up to up to 13 atoms of C, N, O, S, and Cl (Blum and Reymond 2009); and GDB-17 contains 166.4 billion molecules of up to 17 atoms of C, N, O, S, and halogens (Ruddigkeit et al. 2012). Extrapolating from those sets, the size of drug-like chemical space has been extrapolated to 1033 (Polishchuk et al. 2013).
In addition to the size of the space, molecules are discrete graphs and rules exist regarding the types and degrees of connectivity that are allowed. Even further, molecules that are formally valid may still be chemically unstable at the temperatures, pressures, and timescales of interest. Hence, performing local, or even more challenging global, optimization is of big interest in materials and drug design and also a great challenge. The Chemical Space Project leverages the GDB databases to develop visualization and exploration tools, with a focus on drug candidate molecules (Reymond 2015).
Custom molecular libraries have proven a very effective way of navigating chemical space, as the human-driven design allows to embed rules and chemical requirements that are hard to capture without strong supervision, particularly regarding synthetic accessibility of molecules. This success is reflected in the ease of experimental applicability of the examples in the previous section.
Multiple machine-learning approaches to this area suggest that the large existing datasets of chemical reactions from the patent literature (Lowe 2012) or from commercial databases can be leveraged to automatically construct reactivity prediction tools for organic synthesis reactions and retrosynthetic analysis software. Recent examples include prediction the outcome of chemical reaction from fingerprints of the reactants after training on rule-generated examples (Wei et al. 2016) and selection of the major product by ranking a self-generated list of candidates (Coley et al. 2017a; Jin et al. 2017). Sequence-to-sequence models following approaches to machine translation (Fooshee et al. 2018; Schwaller et al. 2018) or similarity searches (Coley et al. 2017b) have proven effective at automatically predicting organic synthesis reactions in an automatic, data-driven way. Even more promising results have been obtained from larger, better-curated datasets (Segler et al. 2017; Segler and Waller 2017a, b).
A well-understood approach to chemical optimization are genetic-algorithms, where mutations from a hand-prepared list of chemical transformations and stochastically applied to starting molecules, and the resulting compounds, if the show improvement in a desired property, are kept for further evolution. Because they rely on hand-picked mutations and hyperparameters, and oftentimes also on manually tuned tradeoffs between target properties, these approaches involve a certain degree of chemical intuition. By compounding mutations that may be allowed individually but not in combination, the molecules generated tend to be quite complex. These types of genetic approaches have been used to navigate chemical space (van Deursen and Reymond 2007; Virshup et al. 2013) and to optimize multiple classes of molecules, such as organic light-emitting diodes (Rupakheti et al. 2016; Shu and Levine 2015), organic photovoltaics (Kanal and Hutchison 2017), diamondoids (Teunissen et al. 2017), visible chromophores with high hyperpolarizability (Elward and Rinderspacher 2015), or small molecules with high electrical dipole (Rupakheti et al. 2015).
4.3 Generative Models
Generative models are machine learning models that aim to produce natural-seeming data that capture the intrinsic statistical properties of the training populations. They can be trained in an unsupervised way and thus are not inhered by the need for labeled data. Given the large size of chemical space, and the very abundant number of chemicals known (see Sect. 2) numbering around 100 million known compounds, unsupervised deep learning models based on existing molecules have been assessed recently.
One of the most basic examples of generative models in chemistry has been the use of recurrent neural networks (RNN) to predict the next character of the SMILES representation of molecules. By feeding n characters to the network to predict the n + 1th, RNNs have proven very powerful at generating valid – if nonsensical – text (Karpathy 2015). A simple and comprehensive string representation of molecules that is human readable and can be stored a single string exists: the simplified molecular-input line-entry system (SMILES) (Weininger 1988). SMILES contain the full molecular connectivity using a series of rules and a canonicalization procedure. The same principles shown for text have proven to work well for generating molecules through their string-based SMILES representation. Additional work has been directed towards generating molecules for generating leads that capture the statistical behavior of the training data (Bjerrum 2017; Ertl et al. 2017; Gupta et al. 2018). A further step involves evolutionary refinement of the molecule pool to bias the set towards a given property (Olivecrona et al. 2017).
Autoencoders are deep learning models that are trained to reproduce a high-dimensional input, subjecting it to a low-dimension information bottleneck. Some of the thorniest issues in molecular design emerge from the discrete nature of molecules and the extremely large size of the search domain. Hence, this low-dimension, real-valued, decodable embedding would potentially allow to apply gradient-based optimization algorithms to molecular optimization. A further improvement is the use of a variational autoencoder, where random noise is added to the encoding step, which results in more continuous coverage in the latent space and fewer dead areas that do not correspond to valid decoded points.
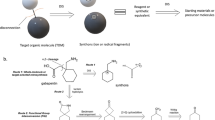
As discussed above, there are available strategies for using chemical graphs as inputs for machine learning systems. However, deep learning tools that efficiently write chemical graphs are still an open problem. Neural networks architectures have been shown to efficiently write text and images, and hence, the first report (Gómez-Bombarelli et al. 2018) of variational autoencoders (VAE) for chemical discovery used a string-based chemical representation. This work showed how a VAE can accurately reconstruct molecules from a continuous real-valued array representation. Even further, it explored how transformations in the latent space, particularly molecular optimization with respect to properties of interest, can be carried out. This application was particularly efficient in the case of jointly trained VAE plus predictor systems, where the deep learning system was simultaneously trained as an unsupervised generator and a supervised predictor. In this case, the latent space is topologically shaped by chemical property (Fig. 2).
Diagram of a Variational Autoencoder with joint property prediction: an encoder neural network takes in the string repetition of a molecule and converts it into a real-valued array that can be decoded back into a string through decoded neural network. By training jointly with a property prediction network, the latent space can be navigated for property optimization
The original VAE model suffers from several flaws, generally related to its ability to write out molecules, particularly the string output is probabilistic. Because of the string representation used, and its need for internal consistency and even arithmetic (opening and closing rings and branches), the VAE models produce many invalid molecules, in more than one sense. On the one hand, syntax errors result in strings that are invalid as SMILES and do not correspond to an actual chemical graph (cycles that open but do not close, parenthesis that open but do not close, representing unfinished branches, etc.), a similar type of failure is to write complete graphs that are not chemically valid graph (generally related to valences and the octet rule: carbon atoms with valence higher than five, oxygen atoms with valence three, etc.). A different type of writeout failure is molecules that are formally and chemically valid, but when re-encoded, do not correspond to the original point in the latent space: the stochastic text generation strays away from the original point.
A number of works are rapidly expanding this area, addressing these performance issues, and exploring further avenues for deep generative models in chemistry (Blaschke et al. 2018; Xu et al. 2017). These include using a series of grammar rules for SMILES as the output of the decoder (Kusner et al. 2017), active learning over the validity of the output (Janz et al. 2017), performing constrained Bayesian optimization to avoid exploring dead areas of the latent space (Griffiths and Hernández-Lobato 2017), performing local optimization near encoded latent points (Mueller et al. 2017), combining an additional RNN to generate higher-quality outputs through reinforcement learning (Jaques et al. 2016).
Further improvements and applications towards generating DNA (Killoran et al. 2017) and protein sequences (Sinai et al. 2017) have been reported, as well as adversarial approaches with the ability to apply bias towards desired properties (Guimaraes et al. 2017; Sanchez-Lengeling et al. 2017).
5 Conclusions
Machine learning and data-driven application have taken computational chemistry by storm. Tasks that have been deemed holy grails for decades seem closer than ever thanks to deep learning approaches: affordable computational predictions that match or surpass experimental accuracy, computerized retrosynthesis that can beat humans, etc.
Chemistry in general and computational chemistry, in particular, have long strived with high-volume, high-velocity, and high-variety data. The pain points are dissimilar however. In computational chemistry, the acquisition velocity is less and less of a bottleneck. Experimentally they are many more constraints on the pace and reproducibility, and they come at a much larger cost. Computational chemistry, on the other hand, has shown difficulty addressing variety: as parametrization increases the transferability and the trust in the predictive power decrease.
The more digitized and automated it is, the more experimental chemistry assimilates to computational chemistry, and hence the easiest to leverage these extremely promising data-driven tools. For a broader impact, and if computational chemistry is truly a sandbox and an accelerator for ideas that will ultimately change experimental chemistry, more focus is needed on addressing heterogeneous, unstructured data. Unsupervised machine learning and transfer learning are promising tools for this task.
References
Álvarez-Moreno M, de Graaf C, López N, Maseras F, Poblet JM, Bo C (2015) Managing the computational chemistry big data problem: the ioChem-BD platform. J Chem Inf Model 55:95
Araujo RB, Banerjee A, Panigrahi P, Yang L, Strømme M, Sjödin M, Araujo CM, Ahuja R (2017) Designing strategies to tune reduction potential of organic molecules for sustainable high capacity battery application. J Mater Chem A 5:4430
Behler J (2011a) Atom-centered symmetry functions for constructing high-dimensional neural network potentials. J Chem Phys 134:74106
Behler J (2011b) Neural network potential-energy surfaces in chemistry: a tool for large-scale simulations. Phys Chem Chem Phys 13:17930
Behler J (2017) First principles neural network potentials for reactive simulations of large molecular and condensed systems. Angew Chemie Int Ed 56:12828
Behler J, Lorenz S, Reuter K (2007) Representing molecule-surface interactions with symmetry-adapted neural networks. J Chem Phys 127:14705
Behler J, Parrinello M (2007) Generalized neural-network representation of high-dimensional potential-energy surfaces. Phys Rev Lett 98:146401
Belsky A, Hellenbrandt M, Karen VL, Luksch P (2002) New developments in the Inorganic Crystal Structure Database (ICSD): accessibility in support of materials research and design. Acta Crystallogr Sect B Struct Sci 58:364
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235
Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M (1978) The protein data bank: a computer-based archival file for macromolecular structures. Arch Biochem Biophys 185:584
Bjerrum EJ (2017) SMILES enumeration as data augmentation for neural Network modeling of molecules arXiv:1703.07076. https://arxiv.org/abs/1703.07076
Blank TB, Brown SD, Calhoun AW, Doren DJ (1995) Neural network models of potential energy surfaces. J Chem Phys 103:4129
Blaschke T, Olivecrona M, Engkvist O, Bajorath J, Chen H (2018) Application of generative autoencoder in de Novo molecular design Mol. Inform 37:1700123
Block P, Sotriffer CA, Dramburg I, Klebe G (2006) AffinDB: a freely accessible database of affinities for protein-ligand complexes from the PDB. Nucleic Acids Res 34:D522
Blum LC, Reymond J-L (2009) 970 million druglike small molecules for virtual screening in the chemical universe database GDB-13. J Am Chem Soc 131:8732
Borodin O, Olguin M, Spear CE, Leiter KW, Knap J (2015) Towards high throughput screening of electrochemical stability of battery electrolytes. Nanotechnology 26:354003
Brockherde F, Vogt L, Li L, Tuckerman ME, Burke K, Müller K-R (2017) Bypassing the Kohn-Sham equations with machine learning. Nat Commun 8:872
Bruno I, Gražulis S, Helliwell JR, Kabekkodu SN, McMahon B, Westbrook J (2017) Crystallography and databases. Data Sci J 16:38
Calderon CE, Plata JJ, Toher C, Oses C, Levy O, Fornari M, Natan A, Mehl MJ, Hart G, Buongiorno Nardelli M, Curtarolo S (2015) The AFLOW standard for high-throughput materials science calculations. Comput Mater Sci 108:233
Chambers J, Davies M, Gaulton A, Hersey A, Velankar S, Petryszak R, Hastings J, Bellis L, McGlinchey S, Overington JP (2013) UniChem: a unified chemical structure cross-referencing and identifier tracking system. J Cheminform 5:3
Chen X, Liu M, Gilson MK (2001) BindingDB: a web-accessible molecular recognition database. Comb Chem High Throughput Screen 4:719
Cheng L, Assary RS, Qu X, Jain A, Ong SP, Rajput NN, Persson K, Curtiss LA (2015) Accelerating electrolyte discovery for energy storage with high-throughput screening. J Phys Chem Lett 6:283
Chmiela S, Tkatchenko A, Sauceda HE, Poltavsky I, Schütt KT, Müller K-R (2017) Machine learning of accurate energy-conserving molecular force fields. Sci Adv 3:e1603015
Coley CW, Barzilay R, Jaakkola TS, Green WH, Jensen KF (2017a) Prediction of organic reaction outcomes using machine learning. ACS Cent Sci 3:434
Coley CW, Rogers L, Green WH, Jensen KF (2017b) Computer-assisted retrosynthesis based on molecular similarity. ACS Cent Sci 3:1237
Curtarolo S, Setyawan W, Hart GLW, Jahnatek M, Chepulskii RV, Taylor RH, Wang S, Xue J, Yang K, Levy O, Mehl MJ, Stokes HT, Demchenko DO, Morgan D (2012a) AFLOW: an automatic framework for high-throughput materials discovery. Comput Mater Sci 58:218
Curtarolo S, Setyawan W, Wang S, Xue J, Yang K, Taylor RH, Nelson LJ, Hart GLW, Sanvito S, Buongiorno-Nardelli M, Mingo N, Levy O (2012b) AFLOWLIB.ORG: a distributed materials properties repository from high-throughput ab initio calculations. Comput Mater Sci 58:227
Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ (2017) The comparative toxicogenomics database: update 2017. Nucleic Acids Res 45:D972
Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M (2008) ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res 36:D344
Ding H, Medasani B, Chen W, Persson KA, Haranczyk M, Asta M (2015) PyDII: a python framework for computing equilibrium intrinsic point defect concentrations and extrinsic solute site preferences in intermetallic compounds. Comput Phys Commun 193:118
Duvenaud DK, Maclaurin D, Aguilera-Iparraguirre J, Gómez-Bombarelli R, Hirzel T, Aspuru-Guzik A, Adams RP, Iparraguirre J, Bombarell R, Hirzel T, Aspuru-Guzik A, Adams RP (2015) Adv Neural Inf Process Syst 2:2215–2223
Elward JM, Rinderspacher BC (2015) Smooth heuristic optimization on a complex chemical subspace. Phys Chem Chem Phys 17:24322
Er S, Suh C, Marshak MP, Aspuru-Guzik A (2015) Computational design of molecules for an all-quinone redox flow battery. Chem Sci 6:885
Ertl P, Lewis R, Martin E, Polyakov V (2017) In silico generation of novel, drug-like chemical matter using the LSTM neural network arXiv:1712.07449. https://arxiv.org/abs/1712.07449
Faber J, Fawcett T, IUCr (2002) The powder diffraction file: present and future. Acta Crystallogr Sect B Struct Sci 58:325
Feller D (1996) The role of databases in support of computational chemistry calculations. J Comput Chem 17:1571
Fink T, Reymond JL (2007) Virtual exploration of the chemical universe up to 11 atoms of C, N, O, F: assembly of 26.4 million structures (110.9 million stereoisomers) and analysis for new ring systems, stereochemistry, physicochemical properties, compound classes, and drug discove. J Chem Inf Model 47:342
Fooshee D, Mood A, Gutman E, Tavakoli M, Urban G, Liu F, Huynh N, Van Vranken D, Baldi P (2018) Deep learning for chemical reaction prediction. Mol Syst Des Eng 3:442–452
Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40:D1100
Gilmer J, Schoenholz SS, Riley PF, Vinyals O, Dahl GE (2017) Neural message passing for quantum chemistry. arXiv1704.01212. https://arxiv.org/abs/1704.01212
Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J (2016) BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 44:D1045
Goldsmith B (2016) NoMaD repository. https://nomad-repository.eu
Gómez-Bombarelli R, Aguilera-Iparraguirre J, Hirzel TD, Duvenaud D, Maclaurin D, Blood-Forsythe MA, Chae HS, Einzinger M, Ha D-G, Wu T, Markopoulos G, Jeon S, Kang H, Miyazaki H, Numata M, Kim S, Huang W, Hong SI, Baldo M, Adams RP, Aspuru-Guzik A (2016) Design of efficient molecular organic light-emitting diodes by a high-throughput virtual screening and experimental approach. Nat Mater 15:1120
Gómez-Bombarelli R, Wei JN, Duvenaud D, Hernández-Lobato JM, Sánchez-Lengeling B, Sheberla D, Aguilera-Iparraguirre J, Hirzel TD, Adams RP, Aspuru-Guzik A (2018) Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent Sci 4:268
Goyal A, Gorai P, Peng H, Lany S, Stevanović V (2017) A computational framework for automation of point defect calculations. Comput Mater Sci 130:1
Gražulis S, Chateigner D, Downs RT, Yokochi AFT, Quirós M, Lutterotti L, Manakova E, Butkus J, Moeck P, Le Bail A (2009) Crystallography open database – an open-access collection of crystal structures. J Appl Crystallogr 42:726
Gražulis S, Daškevič A, Merkys A, Chateigner D, Lutterotti L, Quirós M, Serebryanaya NR, Moeck P, Downs RT, Le Bail A (2012) Crystallography Open Database (COD): an open-access collection of crystal structures and platform for world-wide collaboration. Nucleic Acids Res 40:D420
Griffiths R-R, Hernández-Lobato JM (2017) Constrained bayesian optimization for automatic chemical design. ArXiv:1709.05501. https://arxiv.org/abs/1709.05501
Groom CR, Bruno IJ, Lightfoot MP, Ward SC, IUCr (2016) The Cambridge structural database. Acta Crystallogr Sect B Struct Sci Cryst Eng Mater 72:171
Guimaraes GL, Sanchez-Lengeling B, Outeiral C, Farias PLC, Aspuru-Guzik A (2017) Objective-Reinforced Generative Adversarial Networks (ORGAN) for sequence generation models. ArXiv:1705.10843. https://arxiv.org/abs/1705.10843
Gupta A, Müller AT, Huisman BJH, Fuchs JA, Schneider P, Schneider G (2018) Generative recurrent networks for De Novo drug design. Mol Inf 37:1700111
Hachmann J, Olivares-Amaya R, Atahan-Evrenk S, Amador-Bedolla C, Sanchez-Carrera RS, Gold-Parker A, Vogt L, Brockway AM, Aspuru-Guzik A (2011) The Harvard clean energy project: large-scale computational screening and design of organic photovoltaics on the world community grid. J Phys Chem Lett 2:2241
Hachmann J, Olivares-Amaya R, Jinich A, Appleton AL, Blood-Forsythe MA, Seress LR, Roman-Salgado C, Trepte K, Atahan-Evrenk S, Er S, Shrestha S, Mondal R, Sokolov A, Bao Z, Aspuru-Guzik A (2014) Lead candidates for high-performance organic photovoltaics from high-throughput quantum chemistry – the Harvard Clean Energy Project. Energy Environ Sci 7:698
Heifets A, Jurisica I (2012) SCRIPDB: a portal for easy access to syntheses, chemicals and reactions in patents. Nucleic Acids Res 40:D428
Hermann G, Pohl V, Tremblay JC, Paulus B, Hege H-C, Schild A (2016) ORBKIT: a modular python toolbox for cross-platform postprocessing of quantum chemical wavefunction data. J Comput Chem 37:1511
Hjorth Larsen A, Jørgen Mortensen J, Blomqvist J, Castelli IE, Christensen R, Dułak M, Friis J, Groves MN, Hammer B, Hargus C, Hermes ED, Jennings PC, Bjerre Jensen P, Kermode J, Kitchin JR, Leonhard Kolsbjerg E, Kubal J, Kaasbjerg K, Lysgaard S, Bergmann Maronsson J, Maxson T, Olsen T, Pastewka L, Peterson A, Rostgaard C, Schiøtz J, Schütt O, Strange M, Thygesen KS, Vegge T, Vilhelmsen L, Walter M, Zeng Z, Jacobsen KW (2017) The atomic simulation environment – a Python library for working with atoms. J Phys Condens Matter 29:273002
Holliday GL, Bartlett GJ, Almonacid DE, O’Boyle NM, Murray-Rust P, Thornton JM, Mitchell JBO (2005) MACiE: a database of enzyme reaction mechanisms. Bioinformatics 21:4315
Huskinson B, Marshak MP, Suh C, Er S, Gerhardt MR, Galvin CJ, Chen X, Aspuru-Guzik A, Gordon RG, Aziz MJ (2014) A metal-free organic-inorganic aqueous flow battery. Nature 505:195
Russel D. Johnson II (1999) Computational chemistry comparison and benchmark database. NIST Standard Reference Database Number 101 Release 18, Oct 2016
Jacob CR, Beyhan SM, Bulo RE, Gomes ASP, Götz AW, Kiewisch K, Sikkema J, Visscher L (2011) PyADF - A scripting framework for multiscale quantum chemistry. J Comput Chem 32:2328
Jain A, Ong SP, Hautier G, Chen W, Richards WD, Dacek S, Cholia S, Gunter D, Skinner D, Ceder G, Persson KA (2013) Commentary: the materials project: a materials genome approach to accelerating materials innovation. APL Mater 1:11002
Janz D, van der Westhuizen J, Hernández-Lobato JM (2017) Actively learning what makes a discrete sequence valid. ArXiv:1708.04465
Jaques N, Gu S, Bahdanau D, Hernández-Lobato JM, Turner RE, Eck D (2016) Sequence Tutor: conservative Fine-Tuning of Sequence Generation Models with KL-control Proceedings.Mlr.Press
Jin W, Coley C, Barzilay R, Jaakkola T (2017) Predicting organic reaction outcomes with Weisfeiler-Lehman network ArXiv:1709.04555. https://arxiv.org/abs/1709.04555
Kaiser J (2005) Science resources. Chemists want NIH to curtail database. Science 308:774
Kanal IY, Hutchison GR (2017) Rapid computational optimization of molecular properties using genetic algorithms: searching across millions of compounds for organic photovoltaic materials ArXiv:1707.02949. https://arxiv.org/abs/1707.02949
Karpathy A (2015)
Kearnes S, McCloskey K, Berndl M, Pande V, Riley P (2016) Molecular graph convolutions: moving beyond fingerprints. J Comput Aided Mol Des 30:595
Killoran N, Lee LJ, Delong A, Duvenaud D, Frey BJ (2017) Generating and designing DNA with deep generative models ArXiv:1712.06148
Kirklin S, Saal JE, Meredig B, Thompson A, Doak JW, Aykol M, Rühl S, Wolverton C (2015) The Open Quantum Materials Database (OQMD): assessing the accuracy of DFT formation energies. Npj Comput Mater 1:15010
Klintenberg M, Derenzo SE, Weber MJ (2002) Potential scintillators identified by electronic structure calculations. Nucl Instruments Methods Phys Res Sect A Accel Spectrometers, Detect Assoc Equip 486:298
Kowalski JA, Su L, Milshtein JD, Brushett FR (2016) Recent advances in molecular engineering of redox active organic molecules for nonaqueous flow batteries. Curr Opin Chem Eng 13:45
Kusner MJ, Paige B, Hernández-Lobato JM (2017) Grammar variational autoencoder arXiv:1703.01925. https://arxiv.org/abs/1703.01925
Landis DD, Hummelshoj JS, Nestorov S, Greeley J, Dulak M, Bligaard T, Norskov JK, Jacobsen KW (2012) The Computational materials repository. Comput Sci Eng 14:51
Leung P, Shah AA, Sanz L, Flox C, Morante JR, Xu Q, Mohamed MR, Ponce de León C, Walsh FC (2017) Recent developments in organic redox flow batteries: a critical review. J Power Sources 360:243
Lin L (2015) Materials databases infrastructure constructed by first principles calculations: a review. Mater Perform Charact 4:MPC20150014
Lin K, Gómez-Bombarelli R, Beh ES, Tong L, Chen Q, Valle A, Aspuru-Guzik A, Aziz MJ, Gordon RG (2016) A redox-flow battery with an alloxazine-based organic electrolyte. Nat Energy 1:16102
Linstrom PJ, Mallard WG (2001) The NIST Chemistry WebBook: a chemical data resource on the Internet. J Chem Eng Data 46:1059
Liu Z, Li Y, Han L, Li J, Liu J, Zhao Z, Nie W, Liu Y, Wang R (2015) PDB-wide collection of binding data: current status of the PDBbind database. Bioinformatics 31:405
Lopez SA, Pyzer-Knapp EO, Simm GN, Lutzow T, Li K, Seress LR, Hachmann J, Aspuru-Guzik A (2016) The Harvard organic photovoltaic dataset. Sci Data 3:160086
Lopez SA, Sanchez-Lengeling B, de Goes Soares J, Aspuru-Guzik A (2017) Design Principles and Top Non-Fullerene Acceptor Candidates for Organic Photovoltaics. Joule 1:857
Lowe DM (2012) Extraction of chemical structures and reactions from the literature. PhD Thesis, Cambridge University, PhD.35691, https://doi.org/10.17863/CAM.16293
Lubbers N, Smith JS, Barros K (2017) Hierarchical modeling of molecular energies using a deep neural network. J Chem Phys 148:241715
Martsinovich N, Troisi A (2011) High-throughput computational screening of chromophores for dye-sensitized solar cells. J Phys Chem C 115:11781
Mattingly CJ, Colby GT, Forrest JN, Boyer JL (2003) The Comparative Toxicogenomics Database (CTD). Environ Health Perspect 111:793
Mayeshiba T, Wu H, Angsten T, Kaczmarowski A, Song Z, Jenness G, Xie W, Morgan D (2017) The MAterials Simulation Toolkit (MAST) for atomistic modeling of defects and diffusion. Comput Mater Sci 126:90
Merkys A, Mounet N, Cepellotti A, Marzari N, Gražulis S, Pizzi G (2017) A posteriori metadata from automated provenance tracking: integration of AiiDA and TCOD J Cheminform 9:56. https://doi.org/10.1186/s13321-017-0242-y
Meyer EF (1997) The first years of the Protein Data Bank. Protein Sci 6:1591
Mueller J, Gifford D, Jaakkola T (2017) Sequence to better sequence: continuous revision of combinatorial structures. ICML 70:2536
Nakata M, Shimazaki T (2017) PubChemQC Project: a large-scale first-principles electronic structure database for data-driven chemistry. J Chem Inf Model 57:1300
Nath SR, Kurup SS, Joshi KA (2016) PyGlobal: a toolkit for automated compilation of DFT-based descriptors. J Comput Chem 37:1505
O’boyle NM, Tenderholt AL, Langner KM (2008) cclib: a library for package-independent computational chemistry algorithms. J Comput Chem 29:839
Olivares-Amaya R, Amador-Bedolla C, Hachmann J, Atahan-Evrenk S, Sanchez-Carrera RS, Vogt L, Aspuru-Guzik A (2011) Accelerated computational discovery of high-performance materials for organic photovoltaics by means of cheminformatics. Energy Environ Sci 4:4849
Olivecrona M, Blaschke T, Engkvist O, Chen H (2017) Molecular De Novo Design through Deep Reinforcement Learning J Cheminform 9:48. https://doi.org/10.1186/s13321-017-0235-x
Ong SP, Richards WD, Jain A, Hautier G, Kocher M, Cholia S, Gunter D, Chevrier VL, Persson KA, Ceder G (2013) Python Materials Genomics (pymatgen): a robust, open-source python library for materials analysis Comput. Comput Mater Sci 68:314
Ong SP, Cholia S, Jain A, Brafman M, Gunter D, Ceder G, Persson KA (2015) The Materials Application Programming Interface (API): a simple, flexible and efficient API for materials data based on REpresentational State Transfer (REST) principles. Comput Mater Sci 97:209
Ørnsø KB, Pedersen CS, Garcia-Lastra JM, Thygesen KS (2014) Optimizing porphyrins for dye sensitized solar cells using large-scale ab initio calculations. Phys Chem Chem Phys 16:16246
Ortiz C, Eriksson O, Klintenberg M (2009) Data mining and accelerated electronic structure theory as a tool in the search for new functional materials Comput. Comput Mater Sci 44:1042
Pampel H, Vierkant P, Scholze F, Bertelmann R, Kindling M, Klump J, Goebelbecker H-J, Gundlach J, Schirmbacher P, Dierolf U (2013) Making research data repositories visible: the re3data.org Registry. PLoS One 8:e78080
Papadatos G, Davies M, Dedman N, Chambers J, Gaulton A, Siddle J, Koks R, Irvine SA, Pettersson J, Goncharoff N, Hersey A, Overington JP (2016) SureChEMBL: a large-scale, chemically annotated patent document database. Nucleic Acids Res 44:D1220
Park MH, Lee YS, Lee H, Han Y-K (2011) Low Li+ binding affinity: an important characteristic for additives to form solid electrolyte interphases in Li-ion batteries. J Power Sources 196:5109
Park M-S, Kang Y-S, Im D (2015) A high-speed screening method by combining a high-throughput method and a machine-learning algorithm for developing novel organic electrolytes in rechargeable batteries. ECS Trans 68:75
Park MS, Park I, Kang Y-S, Im D, Doo S-G, Sik Park M, Park I, Kang Y-S, Im D, Doo S-G (2016) A search map for organic additives and solvents applicable in high-voltage rechargeable batteries. Phys Chem Chem Phys 18:26807
Pelzer KM, Cheng L, Curtiss LA (2017) Effects of functional groups in redox-active organic molecules: a high-throughput screening approach. J Phys Chem C 121:237
Pence HE, Williams A (2010) ChemSpider: an online chemical information resource. J Chem Educ 87:1123
Pierce TH, Hohne BA (eds) (1986) Artificial intelligence applications in chemistry (American Chemical Society). Washington, DC
Pineda Flores SD, Martin-Noble GC, Phillips RL, Schrier J (2015) Bio-inspired electroactive organic molecules for aqueous redox flow batteries. 1 Thiophenoquinones. J Phys Chem C 119:21800
Pizzi G, Cepellotti A, Sabatini R, Marzari N, Kozinsky B (2016) AiiDA: automated interactive infrastructure and database for computational science. Comput Mater Sci 111:218
Polishchuk PG, Madzhidov TI, Varnek A (2013) Estimation of the size of drug-like chemical space based on GDB-17 data. J Comput Aided Mol Des 27:675
Pyzer-Knapp EO, Suh C, Gómez-Bombarelli R, Aguilera-Iparraguirre J, Aspuru-Guzik AA, Gomez-Bombarelli R, Aguilera-Iparraguirre J, Aspuru-Guzik AA, Clarke DR (2015) What is high-throughput virtual screening? a perspective from organic materials discovery. Annu Rev Mater Res 45:195
Pyzer-Knapp EO, Simm GN, Aspuru Guzik A (2016) A Bayesian approach to calibrating high-throughput virtual screening results and application to organic photovoltaic materials. Mater Horizons 3:226
Qu X, Jain A, Rajput NN, Cheng L, Zhang Y, Ong SP, Brafman M, Maginn E, Curtiss LA, Persson KA (2015) The Electrolyte Genome project: a big data approach in battery materials discovery. Comput Mater Sci 103:56
Qu X, Zhang Y, Rajput NN, Jain A, Maginn E, Persson KA (2017) Computational design of new magnesium electrolytes with improved properties. J Phys Chem C 121:16126
Ramakrishnan R, Dral PO, Rupp M, von Lilienfeld OA (2014) Quantum chemistry structures and properties of 134 kilo molecules. Sci Data 1:140022
Reymond J-L (2015) The chemical space project. Acc Chem Res 48:722
Ruddigkeit L, van Deursen R, Blum LC, Reymond J-L (2012) Enumeration of 166 billion organic small molecules in the chemical universe database GDB-17. J Chem Inf Model 52:2864
Rupakheti C, Virshup A, Yang W, Beratan DN (2015) Strategy to discover diverse optimal molecules in the small molecule universe. J Chem Inf Model 55:529
Rupakheti C, Al-Saadon R, Zhang Y, Virshup AM, Zhang P, Yang W, Beratan DN (2016) Diverse optimal molecular libraries for organic light-emitting diodes. J Chem Theory Comput 12:1942
Rupp M, Tkatchenko A, Müller K-R, von Lilienfeld OA (2012) Fast and accurate modeling of molecular atomization energies with machine learning. Phys Rev Lett 108:58301
Sanchez-Lengeling B, Outeiral C, Guimaraes GL, Aspuru-Guzik A (2017) Optimizing distributions over molecular space. An objective-Reinforced generative adversarial network for inverse-design chemistry (ORGANIC) chemrxiv:5309668 https://chemrxiv.org/articles/ORGANIC_1_pdf/5309668
Schuchardt KL, Didier BT, Elsethagen T, Sun L, Gurumoorthi V, Chase J, Li J, Windus TL (2007) Basis set exchange: a community database for computational sciences. J Chem Inf Model 47:1045
Schütt KT, Arbabzadah F, Chmiela S, Müller KR, Tkatchenko A (2017) Quantum-chemical insights from deep tensor neural networks. Nat Commun 8:13890
Schütter C, Husch T, Viswanathan V, Passerini S, Balducci A, Korth M (2016) Rational design of new electrolyte materials for electrochemical double layer capacitors. J Power Sources 326:541
Schwaller P, Gaudin T, Lanyi D, Bekas C, Laino T (2018) Found in translation: predicting outcomes of complex organic chemistry reactions using neural sequence-to-sequence models. Chem Sci 9:6091–6098
Segler MHS, Waller MP (2017a) Modelling chemical reasoning to predict and invent reactions. Chem A Eur J 23:6118
Segler MHS, Waller MP (2017b) Neural-symbolic machine learning for retrosynthesis and reaction prediction. Chem A Eur J 23:5966
Segler MHS, Preuss M, Waller MP (2017) Learning to plan chemical syntheses ArXiv:1708.04202
Shin Y, Liu J, Quigley JJ, Luo H, Lin X (2014) Combinatorial design of copolymer donor materials for bulk heterojunction solar cells. ACS Nano 8:6089
Shu Y, Levine BG (2015) Simulated evolution of fluorophores for light emitting diodes. J Chem Phys 142:104104
Sinai S, Kelsic E, Church GM, Nowak MA (2017) Variational auto-encoding of protein sequences. arXiv:1712.03346. https://arxiv.org/abs/1712.03346
Smith JS, Isayev O, Roitberg AE (2017) ANI-1, A data set of 20 million calculated off-equilibrium conformations for organic molecules. Sci Data 4:170193
Snyder JC, Rupp M, Hansen K, Müller K-R, Burke K (2012) Finding density functionals with machine learning. Phys Rev Lett 108:253002
Teunissen JL, De Proft F, De Vleeschouwer F (2017) Tuning the HOMO-LUMO energy gap of small diamondoids using inverse molecular design. J Chem Theory Comput 13:1351
Thygesen KS, Jacobsen KW (2016) Making the most of materials computations. Science 354:180
van Deursen R, Reymond J-L (2007) Chemical space travel. Chem Med Chem 2:636
Vanderveen JR, Patiny L, Chalifoux CB, Jessop MJ, Jessop PG, Vanderveen JR, Patiny L, Chalifoux CB, Jessop MJ, Jessop PG (2015) A virtual screening approach to identifying the greenest compound for a task: application to switchable-hydrophilicity solvents. Green Chem 17:5182
Virshup AM, Contreras-García J, Wipf P, Yang W, Beratan DN (2013) Stochastic voyages into uncharted chemical space produce a representative library of all possible drug-Like compounds. J Am Chem Soc 135:7296
Waller MP, Dresselhaus T, Yang J (2013) JACOB: an enterprise framework for computational chemistry. J Comput Chem 34:1420
Wang R, Fang X, Lu Y, Wang S (2004) The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J Med Chem 47:2977
Ward AL, Doris SE, Li L, Hughes MA, Qu X, Persson KA, Helms BA (2017) Materials genomics screens for adaptive ion transport behavior by redox-Switchable microporous polymer membranes in lithium–Sulfur batteries. ACS Cent Sci 3:399
Wei JN, Duvenaud D, Aspuru-Guzik A (2016) Neural networks for the prediction of organic chemistry reactions. ACS Cent Sci 2:725
Wei X, Pan W, Duan W, Hollas A, Yang Z, Li B, Nie Z, Liu J, Reed D, Wang W, Sprenkle V (2017) Materials and systems for organic redox flow batteries: status and challenges. ACS Energy Lett 2:2187
Weininger D (1988) SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Model 28:31
Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J (2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 34:D668
Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46:D1074
Yuan G, Gygi F (2010) ESTEST: a framework for the validation and verification of electronic structure codes. Comput Sci Discov 3:15004
Xu Z, Wang S, Zhu F, Huang J (2017) Seq2seq Fingerprint: an unsupervised deep molecular embedding for drug discovery. ACM-BCB ‘17. In: Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, pp 285–294. https://doi.org/10.1145/3107411.3107424
Acknowledgments
AAG acknowledges support from The Department of Energy, Office of Basic Energy Sciences under award de-sc0015959. He also thanks Dr. Anders Frøseth for his generous support of this work. RGB acknowledges the Toyota Career Development Chair for financial support.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Section Editor information
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this entry
Cite this entry
Gómez-Bombarelli, R., Aspuru-Guzik, A. (2020). Machine Learning and Big-Data in Computational Chemistry. In: Andreoni, W., Yip, S. (eds) Handbook of Materials Modeling. Springer, Cham. https://doi.org/10.1007/978-3-319-44677-6_59
Download citation
DOI: https://doi.org/10.1007/978-3-319-44677-6_59
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-44676-9
Online ISBN: 978-3-319-44677-6
eBook Packages: Physics and AstronomyReference Module Physical and Materials ScienceReference Module Chemistry, Materials and Physics