Abstract
Azotobacter vinelandii is a soil bacterium that undergoes differentiation to form cysts resistant to desiccation. Alginate, polyhydroxybutyrate (PHB) and alkylresorcinols (AR) are structural components of the cysts. The synthesis of these compounds has been shown to be under control of the Gac/Rsm signal transduction pathway. This pathway includes eight small non-coding RNAs (RsmZ1-7 and RsmY ), and the translational repressor protein RsmA. Binding of RsmA to Rsm-RNAs inhibits its repressor activity. Here, we review and discuss the roles of sRNAs in the formation of cysts and other processes in A. vinelandii. We also report a search for genes encoding small RNAs in the A. vinelandii genome . Two new sRNAs potentially related to the control of cyst components synthesis were identified in this search.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Azotobacter vinelandii is a soil nitrogen fixing soil bacterium related to Pseudomonas species to the extent, that it has been proposed to belong to this genus (Ozen and Ussery 2012; Rediers et al. 2004). Additionally 50 % of A. vinelandii genes encode proteins that show a similarity of 67 % or higher with a Pseudomonas homologues (Gonzalez-Casanova et al. 2014). In spite of close relationship, A. vinelandii shows important phenotypic features that are not observed in most Pseudomonas species, including the ability to fix nitrogen under aerobic conditions, a high respiration rate, and to undergo a differentiation process to form desiccation resistant cysts. A. vinelandii produce the exo-polysaccharide alginate , the intracellular polyester polyhydroxybutyrate (PHB), and the phenolic lipids alkylresorcinols (AR) and alkylpyrones (AP). Alginate, PHB, AR, and AP are structural components of cysts. Mature cysts consist of a cell known as central body, covered by a capsule composed of two layers named the intine and exine. Numerous PHB granules are always observed within the central body, alginate is a major component of the capsule, and AR and AP that are synthesized upon encystment induction replace the phospholipids of the cyst membranes and are components of the exine (Segura et al. 2014).
The expression of genes involved in the synthesis of cyst components mentioned above has been shown to be under the control of small non-coding RNAs (sRNAs) (Hernandez-Eligio et al. 2012; Manzo et al. 2011; Muriel-Millan et al. 2014; Romero et al. 2016). sRNAs are present in diverse organisms (i.e. bacteria, archaea and eukaryotes). In prokaryotes sRNAs have been linked to specialized housekeeping functions, to several stress situations, to pathogenicity and in catabolic carbon repression . The roles played by most sRNAs in the regulation of bacterial gene expression are exerted at a post-transcriptional level, where they activate or repress translation , or by stabilizing or destabilizing mRNAs (Waters and Storz 2009).
Four classical sRNAs, named Ffs, SsrA, RnpB SsrS play essential housekeeping roles and the genes encoding these RNAs have been identified in most bacterial genomes (Sonnleitner and Haas 2011). Their presence in A. vinelandii has not been reported.
Most sRNAs regulate gene expression at the level of translation mainly by two general mechanisms: The most common mechanism involves short, imperfect base- pairing interactions with the 5’ end of mRNAs (Brantl 2002; Storz et al. 2011). The sRNA-mRNA base pairing mechanism requires a chaperon protein called Hfq. One example is the Escherichia coli RyhB RNA and Pseudomonas PrrF1-PrrF2 sRNAs (Masse and Gottesman 2002; Wilderman et al. 2004), These RNAs are involved in iron homeostasis , repressing the expression of some genes encoding nonessential proteins that use iron (Salvail and Masse 2012). RyhB regulates its targets by the formation of sRNA-mRNA complexes stabilized by the chaperone Hfq. A RhyB/PrrF homolog, known as ArrF ( Azotobacter regulatory RNA involving Fe) is present in the A. vinelandii genome, and is involved in iron homeostasis and also in the control of PHB synthesis (Jung and Kwon 2008; Muriel-Millan et al. 2014). Other sRNA that interacts with Hfq is RgsA (Park et al. 2013). Recently a homolog of RgsA has been characterized in A. vinelandii, in this bacterium RgsA is an important factor to resist oxidative stress, and plays a role in biofilm formation (Huerta et al. 2016).
The other regulatory mechanism involves binding of the sRNAs to a small protein that acts as translational repressor. One example involves the regulatory sRNAs named CsrB and CsrC in E. coli or RsmZ , RsmY and RsmX in Pseudomonas species. These sRNAs bind to the CsrA and RsmA proteins respectively. These proteins repress translation by binding their target mRNAs. The proteins encoded in mRNAs that are targets the CsrA/RsmA, are involved in numerous and diverse functions (Lapouge et al. 2008). Seven rsmZ genes and one rsmY as well as the gene encoding the RsmA protein are present in the A. vinelandii genome . Although two RsmY RNAs: named RsmY1 and RsmY2 were first reported (Manzo et al. 2011; Setubal et al. 2009), they were later shown to constitute a single gene (Hernandez-Eligio et al. 2012). In A. vinelandii the Rsm-RNAs are involved in regulating the synthesis of the cyst components alginate , PHB and AR (Hernandez-Eligio et al. 2012; Manzo et al. 2011; Romero et al. 2016).
Other example of a regulatory mechanism involving sRNAs and a translation repressor protein has been reported in Pseudomonas species, where the sRNA named CrcZ is involved in carbon catabolic repression . Here the small protein Crc binds the CrcZ RNAs, and also to the 5’ leader of mRNAs having the sequence GGG to repress their translation. Hfq is required for the binding of the small Crc protein to its target mRNAs or to CrcZ sRNA (for a Review see (Sonnleitner and Haas 2011)). Two copies of CrcZ are present in the A. vinelandii genome (Filiatrault et al. 2013).
Here, we present a review and a discussion of the mechanisms by which sRNAs control the synthesis of cyst components and report the results of a search for sRNAs encoded in the A. vinelandii genome.
2 Search of Genes Encoding sRNAs in the A. vinelandii Genome
In order to identify genes for sRNAs in the A. vinelandii DJ genome, we generated a database file of its intergenic regions. The intergenic sequences were analyzed with the INFERNAL algorithm using the Rfam database as comparison pattern (http://rfam.xfam.org/), using the tool “batch sequence search” of this server. The query intergenic sequences were obtained from the A. vinelandii DJ genome sequence (http://img.jgi.doe.gov/). The analysis identified 30 potential sRNAs. This search also revealed the presence of 10 riboswitches or in cis RNA element, 15 self-catalytic ribozymes, a CRISPR element, and 3 RNA thermometers (ROSE) element 3 (data not shown). The 30 sRNAs, their Rfam identifiers, and their position in the chromosome are listed in Table 4.1, and include the four sRNAs ubiquitous in bacteria: SsrS, SsrA, Ffs and RnpB (Sonnleitner and Haas 2011). SsrS or 6S is an RNA that specifically associates with RNA polymerase holoenzyme containing the sigma70 factor. This interaction represses expression from sigma70-dependent promoters during stationary phase (Wassarman 2007); SsrA, also known as 10Sa and tmRNA (transfer-messenger RNA), participates in a mechanism known as trans-translation. The tmRNA forms a ribonucleoprotein complex (tmRNP) together with Small Protein B (SmpB), Elongation Factor Tu (EF-Tu), and ribosomal protein S1. The tmRNA and its associated proteins bind to bacterial ribosomes, which have stalled in the middle of protein biosynthesis; it recycles the stalled ribosome, adds a proteolysis-inducing tag to the unfinished polypeptide, and facilitates the degradation of the aberrant messenger RNA (Keiler 2015); Ffs is a 4.5S RNA which is a component of a signal recognition particle, that promotes the insertion of membrane proteins into the cytoplasmic membranes (Ulbrandt et al. 1997); RnpB is the 10Sb RNA subunit of the catalytic ribonuclease RNase P that is present in almost all living cells. Transfer RNA (tRNAs ) genes are transcribed as precursors with extra residues at their 5’ and 3’ ends that have to be removed to generate functional tRNAs. In bacteria the RNAse P is responsible for removing the extra 5’ residues (for a review see Kirsebom 2007). Due to its essential nature, it is expected that the functions of these RNAs in A. vinelandii is similar to those described above.
As expected, the previously identified and characterized RsmZ1-7, RsmY (Hernandez-Eligio et al. 2012; Manzo et al. 2011), the ArrF (Jung and Kwon 2008), the RgsA (Huerta et al. 2016), and the two CrcZ sRNAs were found in this search and are listed in Table 4.1. Interestingly a new copy of rsmZ, here named rsmZ8 was also identified, raising to nine the number of Rsm-RNAs present in A. vinelandii.
Homologs to the P. aeruginosa P11, P14, P15, P20 (PhrS), P26, P27, P31 sRNAs, that were predicted by the bioinformatic program SRNApredict2, and confirmed by Northern blot analysis in this bacterium (Livny et al. 2006), were identified in the A. vinelandii genome Table 4.1. P20 (PhrS) promotes the expression of pqsR, an important regulator to establish the quorum sensing mediated by quinones in P. aeruginosa (Sonnleitner and Haas 2011). As A. vinelandii does not possess quorum-sensing systems related with quinones, PhrS must have a different function in this bacterium . The functions of P11, P14, P15, P26, P27, P31 are unknown.
An sRNA homologue to Spot 42 sRNA of E. coli (Beisel and Storz 2011) is present in P. aeruginosa where it was named Pseudomon 1. Recently it was renamed as ErsA and was shown to regulate the expression of the alginate , biosynthetic gene algC at a posttranscriptional level (Ferrara et al. 2015).
IsrK is present in Salmonella typhimurium, the gene encoding this sRNA is located within a virulence island and is a host-induced gene (Padalon-Brauch et al. 2008).
Finally, the other sRNAs found in A. vinelandii are; a homologue of t44 of unknown function, found in enteric bacteria and in Pseudomonas (Tjaden et al. 2002), and two C4 RNAs. C4 antisense molecules are related with the presence of P1 and P7 temperate phages in bacteria (Citron and Schuster 1990).
Several of the sRNAs listed in the Table 1 such as IsrK, RgsA , Pseudomon-1 (EsrA), CsrZ and PhrS have been shown to interact and mediate its function with the chaperone Hfq (Ferrara et al. 2015; Park et al. 2013; Sonnleitner and Blasi 2014; Sonnleitner and Haas 2011; Zhang et al. 2003). Therefore, in A. vinelandii Hfq the predicted product of gene Avin7540 could mediate the activity of the same RNAs.
3 RsmZ and RsmY SRNAs Involved in Synthesis of Cyst Components and Cyst Resistance to Desiccation
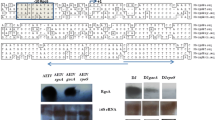
In many γ-proteobacteria the conserved two-component GacS-GacA system (named BarA-UvrY in E. coli) controls the expression of one or more genes encoding small RNAs. These RNAs bind small proteins named RsmA or CsrA that act as translational repressors. Thus the GacS-GacA system controls the post-transcriptional regulatory system that involves the translation repressor protein RsmA (CsrA) and the sRNAs RsmZ, RsmY (CsrB) (Lapouge et al. 2008). As in other γ-proteobacteria in A. vinelandii GacA the protein acting as a transcriptional activator is required for the transcription of the genes encoding the eight Rsm-sRNAs (RsmZ1, RsmZ2, RsmZ3, RsmZ4, RsmZ5, RsmZ6, RsmZ7 and RsmY (Hernandez-Eligio et al. 2012; Manzo et al. 2011). In agreement with this requirement, the Rsm-sRNAs genes possess a GacA binding box within their regulatory region (Manzo et al. 2011). In the search carried out in this study, we found an additional rsmZ gene (rsmZ8). Although the nucleotide sequence of RsmZ8 is the least conserved among the RsmZs, its secondary structure is well conserved (data not shown). An inspection of the regulatory region of rsmZ8 revealed the presence of GacA binding sites (Table 4.2).
3.1 Rsm-RNAs and Alginate Production
In A. vinelandii as in Pseudomonas species the genes encoding the enzymes that participate in alginate biosynthesis (with the exception of algC which encodes for the second enzyme of the pathway) constitute a cluster headed by algD , which encodes GDP-mannose dehydrogenase , the key enzyme of the pathway (Campos et al. 1996). Alginate is essential for cyst formation as mutants unable to produce this polysaccharide are impaired in cyst development (Campos et al. 1996; Mejia-Ruiz et al. 1997). Early, we reported that inactivation of the gacS or gacA gene impaired alginate synthesis (Castañeda et al. 2000, 2001).
The RsmZ1 and RsmZ2 RNAs were shown to interact with the RsmA protein by RNA gel mobility shift experiments, and RsmA was shown to bind to a 200 nt transcript, corresponding to the 5’ leader of the algD mRNA that includes a putative RsmA binding site (Manzo et al. 2011). Thus, It was concluded that in the gacA mutant the absence of Rsm RNAs allows , the free RsmA protein to bind the algD mRNA inhibiting its translation . Indeed, transcription of a single Rsm RNA (RsmZ1) from a GacA-independent promoter, restored alginate synthesis to the gacA mutant (Manzo et al. 2011). These studies clearly indicated the important role of the Rsm-RNAs acting as RsmA anti-repressors to allow translation of algD and the synthesis of alginate an essential component of the cyst capsule .
3.2 Rsm-RNAs and Alkylresorcinols Production
In A. vinelandii alkylresorcinols replace the phospholipids in the cysts membranes and are components of the exine, the outer layer of the cyst envelope. Mutants impaired in AR synthesis produce cysts with a defective exine (Segura et al. 2009). The biosynthesis of alkylresorcinols is also under the control of the Gac-Rsm system. This control is exerted at the level of arpR expression. ArpR is the transcriptional activator of the alkylresorcinol biosynthetic operon arsABCD (Romero et al. 2013). Inactivation of gacA impaired the synthesis of AR, and mutations in rsmZ1 diminished the synthesis of these phenolic lipids . Additionally the RsmA protein was shown to bind to a transcript corresponding to the 5’ leader of the arpR mRNA (Romero et al. 2016). Taken together these data indicated the essentiality of Rsm RNAs to release the repression of arpR mRNA translation exerted by RsmA.
3.3 Rsm-RNAs and PHB Production
Numerous PHB granules are present in mature cysts, and mutants unable to synthesize PHB produce cysts that lack PHB granules but resist desiccation (Segura et al. 2003) indicating that PHB is not essential for cyst formation or germination . However, this result does not rule out the possibility that under natural conditions PHB plays an important role in the formation and/or survival of cysts. Encystment in laboratory conditions is induced in Burk’s medium containing 0.2 % of γ-hydroxybutyrate as sole carbon source, thus it is likely that in soil, this compound is provided by the degradation of PHB. Similar to alginate and AR genes, the expression of the PHB biosynthetic and regulatory genes is controlled by the Gac-Rsm pathway, as inactivation of gacA impaired PHB synthesis. The PHB biosynthetic operon phbBAC is activated by the transcriptional activator PhbR. RsmA was shown to interact with RNAs corresponding to the phbB and phbR mRNA leaders. Additionally the stability of phbB and phbR transcripts is increased in a rsmA mutant strain (Hernandez-Eligio et al. 2012). Therefore as in the case of alginate , and AR the Rsm RNAs are also required to antagonize the repressor activity of RsmA on translation of phbR and phbB mRNAs.
3.4 Expression of Hsp20 an Essential Protein for Cyst Resistance to Desiccation Is Regulated at a Posttranscriptional Level
The small heat shock protein Hsp20 is one of the most abundant proteins in A. vinelandii cysts and is present in low levels in vegetative cells (Cocotl-Yañez et al. 2014). However, transcription of hsp20 is higher in vegetative cells than in cysts, but translation is much higher under encystment conditions than in vegetative cells, indicating a post-transcriptional regulation mechanism. Indeed the hsp20 Shine-Dalgarno possesses a potential RsmA binding site that matched the RsmA binding sites present in the algD, phbR, and arpR transcripts (Cocotl-Yañez et al. 2014), suggesting that expression of Hsp20 is under Rsm-RNAs control. Mutations of hsp20 did not affect the synthesis of cyst components nor the morphology of the cyst, however wild type-like cysts formed by the hsp20 mutant were unable to resist desiccation (Cocotl-Yañez et al. 2014).
By the mean of allowing the expression of genes involved in the synthesis of cyst component alginate , AR and PHB as well as the hsp20 gene essential for cyst desiccation resistance, the Rsm-RNAs play an important role in the differentiation process to produce cysts. A model for the control of cyst formation by the Rsm-RNAs is shown in Fig. 4.1.
Model for the control of cyst formation by the Rsm-RNAs. a The binding of RsmA to the Rsm-RNAs inhibits its translation repression function, allowing expression of the AlgD , ArpR PhbR and Hsp20 proteins, the synthesis of alginate , alkylresorcinol and PHB proteins, and the formation of cysts. b Binding of RsmA to the 5’ UTRs of the algD, arpR, phbR and hsp20 mRNAs inhibits their translation
3.5 Role of Rsm-RNAs Reiterated Genes
The presence of multiple copies of Rsm-RNAs in A. vinelandii raises the question about their biological role. One possibility is to provide with a range of levels of these regulatory RNAs (doses). This range could be achieved by differential expression under different conditions. This in turn will result in a wide range of levels of the free RsmA repressor. Indeed although rsmZ1-7 and rsmY genes and probably rsmZ8, are all activated by GacA , their expression levels varies among them (Hernandez-Eligio et al. 2012). This could be related to the position of the GacA binding sequences, respect to the transcription start site, as well as the % of AT (Table 4.2). In fact, regulators with affinity to AT rich sequences like IHF and H-NS have been related with differential expression of Rsm-RNAs genes in Pseudomonas species (Brencic et al. 2009; Humair et al. 2010).
The effect of the RNA structure-affinity on the binding to the protein RsmA can also provide the cells with an even higher range of free RsmA levels. In a model of RsmA (CsrA)-RNAs interaction, the greater number of stem-loop structures, the more RsmA molecules sequestered is expected (Babitzke and Romeo 2007; Heroven et al. 2012). Indeed in the A. vinelandii Rsm-RNAs the numbers of stem-loops structures vary from four to nine (Fig. 4.2, Table 4.2). Differences in their ΔG, and therefore their stability are also predicted (Table 4.2). Other factor affecting the level of Rsm RNAs is the turn over, as their enzymatic degradation is related to their structure (Seyll and Van Melderen 2013).
In summary, we propose that the redundancy of Rsm-RNAs allows the cell to have a wide range of RsmA levels in response to different environmental or metabolic signals including those that induce differentiation. In fact, redundancy in A. vinelandii is higher than in any Pseudomonas species, where the number of Rsm-RNAs varies from two to five (Moll et al. 2010). Thus, the presence of nine Rsm-RNAs in A. vinelandii could be related to their involvement in cyst formation, a feature not observed in its close relatives, the Pseudomonas.
4 ArrF Controls Iron Homeostasis and PHB Synthesis in A. vinelandii
Iron acquisition is controlled by the Fur (Ferric uptake repressor) protein, which represses the transcription of many genes, including those associated with iron uptake, like siderophores and receptors (Escolar et al. 1999). Under high concentrations of iron, Fur forms a complex with Fe2+, and binds to “iron boxes” present in its target genes, repressing their transcription . When iron is limited, Fur is unable to bind iron boxes. Fur also represses the transcription of RyhB in E. coli, and of ArrF in A. vinelandii (Jung and Kwon 2008; Masse and Gottesman 2002). RhyB represses the expression of genes encoding non-essential proteins that use iron (Salvail and Masse 2012). RhyB sRNA forms sRNA-mRNA complexes stabilized by Hfq. RhyB also exerts a positive regulation on the expression of shiA. In this case the rhyB-mRNA pairing disrupt an inhibitory structure allowing translation (Prevost et al. 2007).
Iron is essential for different biological processes in A. vinelandii including nitrogen fixation, respiration, and nitrogenase protection (Page and Huyer 1984; Page and von Tigerstrom 1982). ArrF was shown to negatively regulate the expression of sodB gene encoding Fe-containing superoxide dismutase and FeSII protein (Jung and Kwon 2008), which is known to mediate the conformational protection of nitrogenase enzyme against oxygen inactivation. Iron also affects PHB accumulation in this bacterium (Pyla et al. 2009). Under Iron limitation transcription of arrF increased significantly. The expression of PhbR, the transcriptional activator of PHB synthesis also increased post- transcriptionally in iron limitation, suggesting that iron limitation increases translation of phbR through ArrF (Muriel-Millan et al. 2014). The study of the role of ArrF in the control of PHB was carried out in vegetative cells. Thus its role in encystment remains to be investigated.
5 Roles of EsrA sRNA in Pseudomonas
In P. aeruginosa and A. vinelandii the alginate biosynthetic , gene algC is transcribed from a promoter dependent on AlgU , a homologue of the RpoE alternative sigma factor (Gaona et al. 2004; Zielinski et al. 1991). Therefore, inactivation of algU in A. vinelandii abrogates alginate synthesis and encystment (Moreno et al. 1998). In P. aeruginosa the EsrA RNA regulates expression of AlgC at a posttranscriptional level and, esrA as algC is transcribed from a AlgU promoter (Ferrara et al. 2015). As shown in Table 1 an EsrA homolog sharing 78 % of identity with Pseudomonas esrA is present in A. vinelandii. Interestingly conserved −10 and −35 boxes that correspond to an AlgU promoter are present in the regulatory region of the A. vinelandii esrA. These observations suggest that in A. vinelandii ErsA sRNA may be involved in the control of alginate synthesis.
6 CrcZ and Its Role in Carbon Catabolic Repression in Pseudomonas Species
Carbon catabolic repression is a mechanism that allows bacteria to catabolize preferred carbon sources for most efficient growth. In enteric bacteria this feature is controlled by the phosphoenolpyruvate (PEP): carbohydrate phosphotransferase system (carbohydrate PTS) by the mean of controlling cAMP-dependent transcriptional activation (for a review see Lengeler and Jahreis 2009).
In Pseudomonas and in A. vinelandii PTS plays no role in catabolic repression (Filiatrault et al. 2013; Moreno et al. 2015). Instead, in Pseudomonas species aeruginosa, putida, and syringae the control of carbon catabolic repression involves the two-component system CbrBA, the sRNAs CrcX , CrcY or CrcZ , and the Crc protein. The expression of the genes encoding Crc sRNAs is dependent on the alternative sigma factor RpoN and the CbrB transcriptional activator. Crc is a small protein that recognizes and bind mRNAs encoding proteins involved in catabolic pathways of less preferred carbon sources. This binding results in repression of translation. In the presence of Hfq, CrcZ sRNA binds the translation repressor Crc. When Pseudomonas grows on a less preferred carbon source, CbrB activates transcription of CrcZ that sequesters the Crc protein allowing translation of mRNAs that are Crc targets. On the other side, in the presence of a preferred source, CbrAB activity is inhibited, resulting in low levels of CrcZ sRNA and repression of mRNA by Crc.
The crbAB , crc genes, and two copies encoding sRNA CrcZ are present in the A. vinelandii genome (Filiatrault et al. 2013; Setubal et al. 2009). The high conservation of the catabolic repression CbrAB/Crc system genes between Pseudomonas and A. vinelandii suggest the same function in this bacterium . If this was the case, The CbrAB/Crc system is likely to have an effect on the synthesis of alginate and PHB as the precursors for their synthesis (fructose 6P and acetyl-CoA respectively ) are originated from the catabolism of the carbon source provided.
7 Outlook
Several important aspects of the encystment process that undergoes A. vinelandii, such as the control of gene expression that leads to the synthesis of important cyst components have been shown to be under Rsm-RNAs control. However, more details about their individual expression and their interaction with RsmA remain to be investigated. We do not rule out that in addition to the Rsm-RNAs other RNAs like EsrA a regulator of the alginate biosynthetic , gene algC in P. aeruginosa also participate in the control of alginate synthesis, an essential component of mature cysts.
Another important subject that remains to be investigated is the behavior of A. vinelandii cysts or vegetative cells in natural soil conditions. It is well known that bacteria are killed by viruses (bacteriophages) , and are predated by other bacteria and eukaryotic organisms as protozoan, amoebas and fungi . Bacteria has developed anti-depredator strategies, one of them is the synthesis of exo-polysaccharides, as mutant strains impaired in exo-polysaccharide production are more susceptible to bacteriovores (Matz et al. 2004). Whether alginate producing , cells of A. vinelandii are more resistant to predators than alginate minus mutants remains to be investigated. However, It has been reported that A. vinelandii cysts (that possess an alginate capsule), are more resistant to predation by Agromyces ramosus than vegetative cells (Casida 1983). Whether cysts are also more resistant to eukaryotic predators like protozoan remains to be tested. Thus, in A. vinelandii sRNAs are essential for the production of exo-polysaccharide and for cyst formation, two features that may play essential roles in the survival of this bacterium in soil.
References
Babitzke P, Romeo T (2007) CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol 10(2):156–163. doi:10.1016/j.mib.2007.03.007
Beisel CL, Storz G (2011) The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell 41(3):286–297. doi:10.1016/j.molcel.2010.12.027
Brantl S (2002) Antisense-RNA regulation and RNA interference. Biochim Biophys Acta 1575(1–3):15–25
Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S (2009) The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73(3):434–445. doi:10.1111/j.1365-2958.2009.06782.x
Campos M, Martinez-Salazar JM, Lloret L, Moreno S, Nunez C, Espin G, Soberon-Chavez G (1996) Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J Bacteriol 178(7):1793–1799
Casida LE (1983) Interaction of Agromyces ramosus with Other Bacteria in Soil. Appl Environ Microbiol 46(4):881–888
Castañeda M, Guzman J, Moreno S, Espin G (2000) The GacS sensor kinase regulates alginate and poly-beta-hydroxybutyrate production in Azotobacter vinelandii. J Bacteriol 182(9):2624–2628
Castañeda M, Sanchez J, Moreno S, Nunez C, Espin G (2001) The global regulators GacA and sigma(S) form part of a cascade that controls alginate production in Azotobacter vinelandii. J Bacteriol 183(23):6787–6793. doi:10.1128/JB.183.23.6787-6793.2001
Citron M, Schuster H (1990) The c4 repressors of bacteriophages P1 and P7 are antisense RNAs. Cell 62(3):591–598
Cocotl-Yañez M, Moreno S, Encarnacion S, Lopez-Pliego L, Castañeda M, Espin G (2014) A small heat-shock protein (Hsp20) regulated by RpoS is essential for cyst desiccation resistance in Azotobacter vinelandii. Microbiology 160(Pt 3):479–487. doi:10.1099/mic.0.073353-0
Escolar L, Perez-Martin J, de Lorenzo V (1999) Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181(20):6223–6229
Ferrara S, Carloni S, Fulco R, Falcone M, Macchi R, Bertoni G (2015) Post-transcriptional regulation of the virulence-associated enzyme AlgC by the sigma(22)-dependent small RNA ErsA of Pseudomonas aeruginosa. Environ Microbiol 17(1):199–214. doi:10.1111/1462-2920.12590
Filiatrault MJ, Stodghill PV, Wilson J, Butcher BG, Chen H, Myers CR, Cartinhour SW (2013) CrcZ and CrcX regulate carbon source utilization in Pseudomonas syringae pathovar tomato strain DC3000. RNA Biol 10(2):245–255. doi:10.4161/rna.23019
Gaona G, Nunez C, Goldberg JB, Linford AS, Najera R, Castañeda M, Guzman J, Espin G, Soberon-Chavez G (2004) Characterization of the Azotobacter vinelandii algC gene involved in alginate and lipopolysaccharide production. FEMS Microbiol Lett 238(1):199–206. doi:10.1016/j.femsle.2004.07.044
Gonzalez-Casanova A, Aguirre-von-Wobeser E, Espin G, Servin-Gonzalez L, Kurt N, Spano D, Blath J, Soberon-Chavez G (2014) Strong seed-bank effects in bacterial evolution. J Theor Biol 356:62–70. doi:10.1016/j.jtbi.2014.04.009
Hernandez-Eligio A, Moreno S, Castellanos M, Castañeda M, Nunez C, Muriel-Millan LF, Espin G (2012) RsmA post-transcriptionally controls PhbR expression and polyhydroxybutyrate biosynthesis in Azotobacter vinelandii. Microbiology 158(Pt 8):1953–1963. doi:10.1099/mic.0.059329-0
Heroven AK, Bohme K, Dersch P (2012) Regulation of virulence gene expression by regulatory RNA elements in Yersinia pseudotuberculosis. Adv Exp Med Biol 954:315–323. doi:10.1007/978-1-4614-3561-7_39
Huerta JM, Aguilar I, Lopez-Pliego L, Fuentes-Ramirez LE, Castañeda M (2016) The Role of the ncRNA RgsA in the Oxidative Stress Response and Biofilm Formation in Azotobacter vinelandii. Curr Microbiol. doi:10.1007/s00284-016-1003-2
Humair B, Wackwitz B, Haas D (2010) GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl Environ Microbiol 76(5):1497–1506. doi:10.1128/AEM.02014-09
Jung YS, Kwon YM (2008) Small RNA ArrF regulates the expression of sodB and feSII genes in Azotobacter vinelandii. Curr Microbiol 57(6):593–597. doi:10.1007/s00284-008-9248-z
Keiler KC (2015) Mechanisms of ribosome rescue in bacteria. Nat Rev Microbiol 13(5):285–297. doi:10.1038/nrmicro3438
Kirsebom LA (2007) RNase P RNA mediated cleavage: substrate recognition and catalysis. Biochimie 89(10):1183–1194. doi:10.1016/j.biochi.2007.05.009
Lapouge K, Schubert M, Allain FH, Haas D (2008) Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67(2):241–253. doi:10.1111/j.1365-2958.2007.06042.x
Lengeler JW, Jahreis K (2009) Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib Microbiol 16:65–87. doi:10.1159/000219373
Livny J, Brencic A, Lory S, Waldor MK (2006) Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res 34(12):3484–3493. doi:10.1093/nar/gkl453
Manzo J, Cocotl-Yañez M, Tzontecomani T, Martinez VM, Bustillos R, Velasquez C, Goiz Y, Solis Y, Lopez L, Fuentes LE, Nunez C, Segura D, Espin G, Castañeda M (2011) Post-transcriptional regulation of the alginate biosynthetic gene algD by the Gac/Rsm system in Azotobacter vinelandii. J Mol Microbiol Biotechnol 21(3–4):147–159. doi:10.1159/000334244
Masse E, Gottesman S (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA 99(7):4620–4625. doi:10.1073/pnas.032066599
Matz C, Bergfeld T, Rice SA, Kjelleberg S (2004) Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ Microbiol 6(3):218–226
Mejia-Ruiz H, Moreno S, Guzman J, Najera R, Leon R, Soberon-Chavez G, Espin G (1997) Isolation and characterization of an Azotobacter vinelandii algK mutant. FEMS Microbiol Lett 156(1):101–106
Moll S, Schneider DJ, Stodghill P, Myers CR, Cartinhour SW, Filiatrault MJ (2010) Construction of an rsmX co-variance model and identification of five rsmX non-coding RNAs in Pseudomonas syringae pv. tomato DC3000. RNA Biol 7(5):508–516
Moreno R, Hernandez-Arranz S, La Rosa R, Yuste L, Madhushani A, Shingler V, Rojo F (2015) The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ Microbiol 17(1):105–118. doi:10.1111/1462-2920.12499
Moreno S, Najera R, Guzman J, Soberon-Chavez G, Espin G (1998) Role of alteRNAtive sigma factor algU in encystment of Azotobacter vinelandii. J Bacteriol 180(10):2766–2769
Muriel-Millan LF, Castellanos M, Hernandez-Eligio JA, Moreno S, Espin G (2014) Posttranscriptional regulation of PhbR, the transcriptional activator of polyhydroxybutyrate synthesis, by iron and the sRNA ArrF in Azotobacter vinelandii. Appl Microbiol Biotechnol 98(5):2173–2182. doi:10.1007/s00253-013-5407-7
Ozen AI, Ussery DW (2012) Defining the Pseudomonas genus: where do we draw the line with Azotobacter? Microb Ecol 63(2):239–248. doi:10.1007/s00248-011-9914-8
Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S (2008) Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res 36(6):1913–1927. doi:10.1093/nar/gkn050
Page WJ, Huyer M (1984) Derepression of the Azotobacter vinelandii siderophore system, using iron-containing minerals to limit iron repletion. J Bacteriol 158(2):496–502
Page WJ, von Tigerstrom M (1982) Iron- and molybdenum-repressible outer membrane proteins in competent Azotobacter vinelandii. J Bacteriol 151(1):237–242
Park SH, Butcher BG, Anderson Z, Pellegrini N, Bao Z, D’Amico K, Filiatrault MJ (2013) Analysis of the small RNA P16/RgsA in the plant pathogen Pseudomonas syringae pv. tomato strain DC3000. Microbiology 159 (Pt 2):296–306. doi:10.1099/mic.0.063826-0
Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Masse E (2007) The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol 64(5):1260–1273. doi:10.1111/j.1365-2958.2007.05733.x
Pyla R, Kim TJ, Silva JL, Jung YS (2009) Overproduction of poly-beta-hydroxybutyrate in the Azotobacter vinelandii mutant that does not express small RNA ArrF. Appl Microbiol Biotechnol 84(4):717–724. doi:10.1007/s00253-009-2002-z
Rediers H, Vanderleyden J, De Mot R (2004) Azotobacter vinelandii: a Pseudomonas in disguise? Microbiology 150(Pt 5):1117–1119. doi:10.1099/mic.0.27096-0
Romero Y, Guzman J, Moreno S, Cocotl M, Espin G, Castañeda M, Vences M, Segura D (2016) The GacS/A-RsmA signal transduction pathway controls the synthesis of alkylresorcinol lipids that replace membrane phospholipids during encystment of Azotobacter vinelandii SW136. PLoS ONE 11, e0153266. doi:10.1371/journal.pone.0153266
Romero Y, Moreno S, Guzman J, Espin G, Segura D (2013) Sigma factor RpoS controls alkylresorcinol synthesis through ArpR, a LysR-type regulatory protein, during encystment of Azotobacter vinelandii. J Bacteriol 195(8):1834–1844. doi:10.1128/JB.01946-12
Salvail H, Masse E (2012) Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip Rev RNA 3(1):26–36. doi:10.1002/wrna.102
Segura D, Cruz T, Espin G (2003) Encystment and alkylresorcinol production by Azotobacter vinelandii strains impaired in poly-beta-hydroxybutyrate synthesis. Arch Microbiol 179(6):437–443. doi:10.1007/s00203-003-0553-4
Segura D, Nunez C, Espín G (2014) Azotobacter Cysts. In: Ltd JWS (ed) Encyclopedia of life sciences. Chichester, West Sussex, UK. doi:10.1002/9780470015902.a0000295.pub2
Segura D, Vite O, Romero Y, Moreno S, Castañeda M, Espin G (2009) Isolation and characterization of Azotobacter vinelandii mutants impaired in alkylresorcinol synthesis: alkylresorcinols are not essential for cyst desiccation resistance. J Bacteriol 191(9):3142–3148. doi:10.1128/JB.01575-08
Setubal JC, dos Santos P, Goldman BS, Ertesvag H, Espin G, Rubio LM, Valla S, Almeida NF, Balasubramanian D, Cromes L, Curatti L, Du Z, Godsy E, Goodner B, Hellner-Burris K, HeRNAndez JA, Houmiel K, Imperial J, Kennedy C, Larson TJ, Latreille P, Ligon LS, Lu J, Maerk M, Miller NM, Norton S, O’Carroll IP, Paulsen I, Raulfs EC, Roemer R, Rosser J, Segura D, Slater S, Stricklin SL, Studholme DJ, Sun J, Viana CJ, Wallin E, Wang B, Wheeler C, Zhu H, Dean DR, Dixon R, Wood D (2009) Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J Bacteriol 191(14):4534–4545. doi:10.1128/JB.00504-09
Siu FY, Spanggord RJ, Doudna JA (2007) SRP RNA provides the physiologically essential GTPase activation function in cotranslational protein targeting. RNA 13(2):240-250. doi:10.1261/rna.13540
Seyll E, Van Melderen L (2013) The ribonucleoprotein Csr network. Int J Mol Sci 14(11):22117–22131. doi:10.3390/ijms141122117
Sonnleitner E, Blasi U (2014) Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet 10(6):e1004440. doi:10.1371/journal.pgen.1004440
Sonnleitner E, Haas D (2011) Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl Microbiol Biotechnol 91(1):63–79. doi:10.1007/s00253-011-3332-1
Storz G, Vogel J, Wassarman KM (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43(6):880–891. doi:10.1016/j.molcel.2011.08.022
Tjaden B, Saxena RM, Stolyar S, Haynor DR, Kolker E, Rosenow C (2002) Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res 30(17):3732–3738
Ulbrandt ND, Newitt JA, Bernstein HD (1997) The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88(2):187–196
Wassarman KM (2007) 6S RNA: a regulator of transcription. Mol Microbiol 65(6):1425–1431. doi:10.1111/j.1365-2958.2007.05894.x
Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136(4):615–628. doi:10.1016/j.cell.2009.01.043
Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML (2004) Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci USA 101(26):9792–9797. doi:10.1073/pnas.0403423101
Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S (2003) Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50(4):1111–1124
Zielinski NA, Chakrabarty AM, Berry A (1991) Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J Biol Chem 266(15):9754–9763
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Castañeda, M., López-Pliego, L., Espín, G. (2016). Azotobacter vinelandii Small RNAs: Their Roles in the Formation of Cysts and Other Processes. In: Leitão, A., Enguita, F. (eds) Non-coding RNAs and Inter-kingdom Communication. Springer, Cham. https://doi.org/10.1007/978-3-319-39496-1_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-39496-1_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-39494-7
Online ISBN: 978-3-319-39496-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)