Abstract
It is well known that the changes in the breast tissue density are strongly correlated with the risk of breast cancer development and therefore classifying the breast tissue density as fatty, fatty–glandular and dense–glandular has become clinically significant. It is believed that the changes in the tissue density can be captured by computing the texture descriptors. Accordingly, the present work has been carried out with an aim to explore the potential of Laws’ mask texture descriptors for description of variations in breast tissue density using mammographic images. The work has been carried out on the 322 mammograms taken from the MIAS dataset. The dataset consists of 106 fatty, 104 fatty–glandular and 112 dense–glandular images. The ROIs of size 200 × 200 pixels are extracted from the center of the breast tissue, ignoring the pectoral muscle. For the design of a computer aided diagnostic system for three class breast tissue density classification, Laws’ texture descriptors have been computed using Laws’ masks of different resolutions. Five statistical features i.e. mean, skewness, standard deviation, entropy and kurtosis have been computed from all the Laws’ texture energy images generated from each ROI. The feature space dimensionality reduction has been carried out by using principal component analysis. For the classification task kNN, PNN and SVM classifiers have been used. After carrying out exhaustive experimentation, it has been observed that PCA–SVM based CAD system design yields the highest overall classification accuracy of 87.5 %, with individual class accuracy values of 84.9, 84.6 and 92.8 % for fatty, fatty–glandular and dense–glandular image classes respectively. These results indicate the usefulness of the proposed CAD system for breast tissue density classification.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Breast density
- Mammograms
- Laws’ mask texture descriptors
- Principal component analysis
- KNN classifier
- PNN classifier
- SVM classifier
1 Introduction
The most commonly diagnosed disease among women that has become a major health concern for the past few decades is breast cancer [1–3]. For the women in United Kingdom, the lifetime risk of being diagnosed with breast cancer is 1 in 8 [4]. The study in [5] reported 1.67 million new cases of breast cancer worldwide in the year 2012. It has been strongly advocated by many researchers in their study that increased breast density is strongly correlated to the risk of developing breast cancer [5–15]. The association between increased breast density and breast cancer risk can be explained on the basis of effects that are caused by the hormones mitogens and mutagens. The mitogens are known to affect the size of the cell population in the breast and cell proliferation while mutagens increase the likelihood of damage to these cells. Due to increased cell population, there is an increase in reactive oxygen species (ROS) production and lipid peroxidation. The products of lipid peroxidation; malondialdehyde (MDA) and isoprostanes catalyze the proliferation of cells [14].
Breast cancer has a very high mortality rate but the chances of survival are significantly improved if it is detected at an early stage. Different imaging modalities like MRI, computerized tomography, ultrasound, etc. are used in the diagnosis of breast abnormalities but mammography is considered to be the best choice for detection due to its higher sensitivity [14, 16–24]. Mammography is an X–ray imaging technique used to detect any abnormalities in the breast. There are two types of mammography examination:
-
(a)
Screening Mammography: Screening mammography is used to check for breast abnormalities in asymptomatic women. This examination is used to detect breast cancer at an early stage when there are no symptoms present.
-
(b)
Diagnostic Mammography: Diagnostic mammography is performed when either a patient has complaint of some lumps in the breast, pain or any abnormality is detected during the screening process. It helps in determining whether the symptoms indicate the presence of a malignancy and is also used to find the exact location of the abnormalities.
On the basis of density, breast tissue can be classified into the following classes:
-
(a)
Two–class classification: Fatty tissue (F)/Dense tissue (D).
-
(b)
Three–class classification: Fatty tissue (F)/Fatty–glandular tissue (FG)/Dense–glandular tissue (DG).
-
(c)
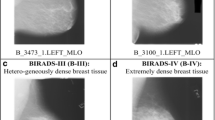
Four–class BI–RADS classification: Almost entirely fatty tissue (B–I)/some fibro–glandular tissue (B–II)/heterogeneously dense breast tissue (B–III)/extremely dense breast tissue (B–IV).
The typical fatty tissue being translucent to X–rays appears dark on a mammogram whereas the dense tissues appear bright on the mammograms. The fatty–glandular breast tissue is an intermediate stage between fatty and dense tissues therefore a typical fatty–glandular breast tissue appears dark with some bright streaks on the mammogram. The mammograms are visually analyzed by the radiologists to identify and differentiate between different density patterns of the breast tissue. The typical breast tissue density patterns are easy to identify and analyze. This analysis is however subjective and depends on the experience of the radiologist. The appearances of atypical cases of the breast tissue density patterns are highly overlapping and differentiating between these atypical cases through visual analysis is considered to be a highly daunting task for the radiologists. The sample images depicting the typical and atypical cases of breast tissue density patterns are shown in Figs. 1 and 2, respectively.
In order to provide the radiologists with a second opinion tool for validating their diagnosis and identify the atypical mammographic images correctly, various computer aided diagnostic (CAD) systems have been developed in the past for breast tissue density classification. A brief description of the related studies is tabulated in Table 1.
From the extensive literature survey presented in Table 1, it can be observed that most of the related studies are based on the pre–processing of mammograms to extract the segmented breast tissue (SBT) after removing the pectoral muscle and the background while very few studies have been carried out that report CAD system designs based on ROIs extracted from the breast. It has also been shown in [47] that the ROIs extracted from the center of the breast result in highest performance as this region of the breast is densest. The ROI extraction method results in the elimination of an extra step of pre–processing to obtain the SBT after removing the background and pectoral muscle.
K.I. Laws developed a method for texture analysis where an image was filtered with various two–dimensional masks to find its texture properties which proved to be useful for texture analysis. In this method five kernels namely Level (L), Edge (E), Spot (S), Wave (W) and Ripple (R) are used to form different masks used for filtering purposes [48]. Laws’ mask analysis is considered to be one of the best methods for texture analysis in image processing applications like breast cancer detection [49, 50], classification of liver diseases [51], bone texture analysis [52] etc.
Thus in the present work, considering the effect of ROI size and location on performance of the algorithms, a CAD system design is proposed for three–class classification of different breast tissue density patterns based on their underlying texture characteristics computed using Laws’ mask texture analysis.
The rest of the paper is organised into 3 sections. Section 2 explains the methodology adopted in the present work for three–class breast tissue density classification, giving a brief description of the dataset and proposed CAD system design. The various experiments carried out for classifying the mammographic images are explained in Sect. 3 and the conclusions drawn from the present work are reported in Sect. 4.
2 Methodology
2.1 Description of the Dataset
For the present work mammographic images have been taken from a publicly available database, mini–MIAS (Mammographic Image Analysis Society). The database contains 322 Medio Lateral Oblique (MLO) view mammographic images. The size of each image is 1024 × 1024 pixels with 256 gray scale tones and a 96 dpi horizontal and vertical resolution. The images in the database are divided into three categories on the basis of density, fatty (106 images), fatty–glandular (104 images) and dense–glandular (112 images). The database includes the name of each image in form of a mnemonic with prefix ‘mdb’ and a three digit number. The database also includes nature of the breast tissue, location of abnormality, the radius of the circle enclosing the abnormality and its severity [53].
2.2 Region of Interest (ROI) Selection
The ROI size is selected carefully, considering the fact that with the selected ROI size, there should be a good population of pixels available for calculating the texture properties. For evaluation, the present work considers an ROI of size 200 × 200 pixels extracted manually from the center of the breast tissue for each mammographic image. The ROI size of 200 × 200 has been taken based on the reference in the previous studies [41, 42]. The process of extraction of ROI from the mammographic image is shown in Fig. 3.
2.3 Proposed CAD System Design
The computer aided diagnostic (CAD) systems are nowadays widely used to identify the hidden abnormalities that might be missed by the radiologists during visual analysis hence improving the overall diagnostic accuracy [54–67]. The experimental workflow to design a CAD system for three–class breast tissue density classification is shown in Fig. 4.
The proposed CAD system consists of three modules: feature extraction module, feature space dimensionality reduction module and classification module. From the extracted ROIs, Laws’ texture descriptors are calculated using Laws’ masks of different lengths to form the feature vectors (FVs). In the feature space dimensionality reduction module, to remove the redundant features from the FVs, the principal component analysis (PCA) algorithm has been applied and reduced feature vectors (RFVs) consisting of principal components (PCs) have been computed. In the classification module, the computed FVs and RFVs have been fed to different classifiers namely k–nearest neighbour (kNN), probabilistic neural network (PNN) and support vector machine (SVM) to classify the mammographic images into one of the three classes: fatty, fatty–glandular and dense–glandular according to the density information.
Feature Extraction Module. In the field of medical imaging, the process of feature extraction is used to convert the texture features of an image into mathematical descriptors to quantify the textural properties exhibited by the image. The textural features from images can be extracted using different methods–statistical methods, signal processing based methods and transform domain methods. These methods have been depicted in Fig. 5.
In the present work, a signal processing based technique called Laws’ mask analysis is used. In this technique the images are filtered with specific masks to extract different textural properties from the images. The masks are formed by combinations of different one–dimensional kernels. Five kernels namely Level (L), Edge (E), Spot (S), Wave (W) and Ripple (R) are used to form different masks used in feature extraction process. Further, the length of these kernels can be 3, 5, 7 and 9 [48, 51, 52, 68, 69]. A description of these one–dimensional kernels is given in Table 2.
The one–dimensional kernels shown in Table 2 are convolved with each other to form the two–dimensional masks used for filtering the images to calculate texture features. The resultant two–dimensional masks for each kernel length are shown in Fig. 6. The process of feature extraction using Laws’ mask analysis is depicted in Fig. 7.
The above steps are demonstrated with an example below:
- Step 1:
-
Consider Laws’ mask of length 3. Convolve the extracted ROIs with each of the above nine 2D filters. Suppose the ROI of size 200 × 200 is convolved with the 2D filter S3S3 to form a texture image (TIS3S3).
- Step 2:
-
The mask L3L3 having zero mean is used to form contrast invariant texture images.
- Step 3:
-
The resultant normalized TIs are passed through a 15 × 15 window to derive 9 texture energy images (TEMs).
- Step 4:
-
Out of 9 TEMs, 6 rotationally invariant texture energy images (TRs) are obtained by averaging.
- Step 5:
-
From the derived TRs five statistical parameters–mean, standard deviation, skewness, kurtosis and entropy [51, 68] are computed, thus a total of 30 Laws’ texture features (6 TRs × 5 statistical parameters) are calculated for each ROI. These statistical parameters are defined as:
Proceeding in the similar manner as above, Laws’ texture features can also be computed for the masks of length 5, 7 and 9 as shown in Table 2.
The brief description of FVs computed using Laws’ mask analysis as used in the present work are described in Table 3.
Feature Space Dimensionality Reduction Module. Some of the computed feature vectors (FVs) may contain redundant or correlated features which must be eliminated. In the present work, the principal component analysis (PCA) algorithm has been used to obtain optimal attributes for the classification task [70–74]. The main steps followed in the PCA algorithm are shown in Fig. 8.
The optimal number of PCs resulting in highest classification accuracy for training dataset is used for obtaining reduced feature vectors (RFVs) as described in Table 4.
Classification Module. Classification is a technique used in machine learning to predict the class of an unknown data instance based on the training data set, containing instances whose class membership is known. In the present work, classifiers like kNN, PNN and SVM are used to classify the instances of the testing dataset. Before feeding the extracted FVs and RFVs to the classifiers, the features are normalised in the range [0, 1] by using min–max procedure to avoid any bias caused by unbalanced feature values.
-
(1)
k–nearest neighbour (kNN) classifier: The kNN classifier is used to estimate the class of an unknown instance from its neighbouring instances. It tries to assemble together the instances of the training feature vector into separate classes based on distance metric. The class of an unknown instance is decided by a majority vote of its neighbouring instances in the training dataset [71, 75–77]. There are many distance metrics that can be used in kNN classification such as Manhattan distance, Minkowski distance, Hamming distance, Mahalanobis distance etc., but Euclidean distance is the most commonly used distance metric. In order to design an efficient kNN classifier the optimal value of k is required. In the present work, the optimal values of (a) the parameter k and (b) the number of PCs to be retained are determined by exhaustive experimentation with k \( { \in }\left\{ {1,2, \ldots ,9,10} \right\} \) and number of PCs \( { \in }\left\{ {1,2, \ldots ,14,15} \right\} \). In case the accuracy values are same for more than one value of the parameter k, smallest value of k is selected for the classification task.
-
(2)
Probabilistic neural network (PNN) classifier: The PNN classifier belongs to a class of supervised (feed–forward) neural network classifiers used for determining the probability of class membership of an instance [78–80]. The PNN architecture has four layers: input layer, pattern layer, summation layer and output layer. The input layer consists of ‘n’ neurons which accept the primitive values. The results obtained in the input unit are transmitted to the hidden units of the pattern layer where the response of each unit is calculated. In the pattern layer, there are ‘p’ neurons, one for each class. The pdf (probability density function) of each class is defined in the pattern layer on the basis of training data and the optimized kernel width parameter S p also called the spread parameter. The summation layer sums the values of each hidden unit to get response in each category. To obtain the class of the unknown instance, decision layer selects the maximum response from all categories. The optimal choice of S p is crucial for the classification task. In order to design an efficient PNN classifier the optimal value of S p is required. In the present work, the optimal values of (a) the spread parameter S p , and (b) the number of PCs to be retained are determined by exhaustive experimentation with \( S_{p} { \in }\left\{ {1,2, \ldots ,9,10} \right\} \) and number of PCs \( { \in }\left\{ {1,2, \ldots ,14,15} \right\} \).
-
(3)
Support vector machine (SVM) classifier: In the present work, SVM classifier has been implemented using one-against-one (OAO) approach for multiclass SVM provided by LibSVM library [81]. The Gaussian radial basis function kernel has been used for non-linear mapping of training data into higher dimensional feature space. In order to design an efficient SVM classifier, the optimal value of C and \( \gamma \) are obtained by grid-search procedure i.e., for each combination of (C, \( \gamma \)) such that, \( {\text{C}}\,{ \in }\,\left\{ {2^{ - 4} ,\,2^{ - 3} , \ldots ,2^{15} } \right\} \) and \( \gamma \,{ \in }\,\left\{ {2^{ - 12} ,\,2^{ - 11} , \ldots ,2^{4} } \right\} \) the 10-foldcross validation accuracy is obtained for training data. The combination of C and \( \gamma \) yielding the maximum training accuracy are selected for generating the training model. Further, the optimal number of PCs to be retained is determined by repeating the experiments with different number of PCs \( { \in }\,\left\{ {1,2, \ldots ,14,\,15} \right\} \) [73, 82–85].
Classification Performance Evaluation. The performance of the CAD system for breast tissue density classification can be measured using overall classification accuracy (OCA) and individual class accuracy (ICA). These values can be calculated using the confusion matrix (CM).
3 Results
The performance of the proposed CAD system to classify the mammographic images based on their density has been evaluated by conducting various experiments. A description of these experiments is given in Table 5.
3.1 kNN Classifier Results: Experiment I
This experiment evaluates the classification performance of different FVs using the kNN classifier. The results are reported in Table 6.
From Table 6, it is observed that for FV1, FV2, FV3 and FV4, the OCA values are 83.2, 83.8, 86.9 and 78.8 %, respectively. For fatty class the ICA values are 83.0, 81.1, 86.7 and 73.5 % for FV1, FV2, FV3 and FV4, respectively. For fatty–glandular class, the ICA values are 82.6, 84.6, 84.6 and 82.6 % for FV1, FV2, FV3 and FV4, respectively. For the dense–glandular class the ICA values are 83.9, 85.7, 89.2 and 80.3 % for FV1, FV2, FV3 and FV4, respectively. For testing dataset with 161 instances, in case of FV1, the total misclassified instances are 27 (27/161), for FV2, the total misclassified instances are 26 (26/161), for FV3, the total misclassified instances are 21 (21/161) and for FV4, the total misclassified instances are 34 (34/161).
3.2 PNN Classifier Results: Experiment II
This experiment evaluates the classification performance of different FVs using the PNN classifier. The results are reported in Table 7.
From Table 7, it can be observed that for FV1, FV2, FV3 and FV4, the OCA values are 85.0, 81.9, 83.8 and 78.2 %, respectively. For fatty class, the ICA values are 84.9, 83.0, 84.9 and 75.4 % for FV1, FV2, FV3 and FV4, respectively. For fatty–glandular class, the ICA values are 90.3, 84.6, 88.4 and 88.4 % for FV1, FV2, FV3 and FV4, respectively. For the dense–glandular class the ICA values are 80.3, 78.5, 78.5 and 71.4 % for FV1, FV2, FV3 and FV4, respectively. For testing dataset with 161 instances, in case of FV1, the total misclassified instances are 24 (24/161), for FV2 the total misclassified instances are 29 (29/161), for FV3, the total misclassified instances are 26 (26/161) and for FV4, total misclassified instances are 35 (35/161).
3.3 SVM Classifier Results: Experiment III
This experiment evaluates the classification performance of different FVs using the SVM classifier. The results are reported in Table 8.
From Table 8, it can be observed that for FV1, FV2, FV3 and FV4, the OCA values are 86.3, 86.9, 83.2 and 84.4 %, respectively. For fatty class, the ICA values are 86.7, 88.6, 90.5 and 81.1 % for FV1, FV2, FV3 and FV4, respectively. For fatty–glandular class, the ICA values are 78.8, 78.8, 65.3 and 82.6 % for FV1, FV2, FV3 and FV4, respectively. For the dense–glandular class the ICA values are 92.8, 92.8, 92.8 and 89.2 % for FV1, FV2, FV3 and FV4, respectively. For testing dataset with 161 instances, in case of FV1, the total misclassified instances are 22 (22/161), for FV2 the total misclassified instances are 21 (21/161), for FV3, the total misclassified instances are 27 (27/161) and for FV4, total misclassified instances are 25 (25/161).
3.4 PCA–kNN Classifier Results: Experiment IV
This experiment evaluates the classification performance of different RFVs using the PCA–kNN classifier. The results are reported in Table 9.
From Table 9, it can be observed that for RFV1, RFV2, RFV3 and RFV4, the OCA values are 85.0, 81.9, 86.9 and 79.5 %, respectively. For fatty class, the ICA values are 77.3, 77.3, 83.0 and 69.8 % for RFV1, RFV2, RFV3 and RFV4, respectively. For fatty–glandular class, the ICA values are 90.3, 88.4, 86.5 and 88.4 % for RFV1, RFV2, RFV3 and RFV4, respectively. For the dense–glandular class the ICA values are 87.5, 80.3, 91.0 and 80.3 % for RFV1, RFV2, RFV3 and RFV4, respectively. For testing dataset with 161 instances, in case of RFV1, the total misclassified instances are 24 (24/161), for RFV2 the total misclassified instances are 29 (29/161), for RFV3, the total misclassified instances are 21 (21/161) and for RFV4, total misclassified instances are 33 (33/161).
3.5 PCA–PNN Classifier Results: Experiment V
This experiment evaluates the classification performance of different RFVs using the PCA–PNN classifier. The results are reported in Table 10.
From Table 10, it can be observed that for RFV1, RFV2, RFV3 and RFV4, the OCA values are 83.8, 77.6, 85.0 and 74.5 %, respectively. For fatty class, the ICA values are 84.9, 75.4, 86.7 and 75.4 % for RFV1, RFV2, RFV3 and RFV4, respectively. For fatty–glandular class, the ICA values are 90.3, 88.4, 90.3 and 86.5 % for RFV1, RFV2, RFV3 and RFV4, respectively. For the dense–glandular class the ICA values are 76.7, 69.6, 78.5 and 62.5 % for RFV1, RFV2, RFV3 and RFV4, respectively. For testing dataset with 161 instances, in case of RFV1, the total misclassified instances are 26 (26/161), for RFV2 the total misclassified instances are 36 (36/161), for RFV3, the total misclassified instances are 24 (24/161) and for RFV4, total misclassified instances are 41 (41/161).
3.6 PCA–SVM Classifier Results: Experiment VI
This experiment evaluates the classification performance of different RFVs using the PCA–SVM classifier. The results are reported in Table 11.
From Table 11, it can be observed that for RFV1, RFV2, RFV3 and RFV4, the OCA values are 87.5, 85.7, 86.3 and 85.7 %, respectively. For fatty class the ICA values are 84.9, 81.1, 86.7 and 83.0 % for RFV1, RFV2, RFV3 and RFV4, respectively. For fatty–glandular class, the ICA values are 84.6, 86.5, 78.8 and 82.6 % for RFV1, RFV2, RFV3 and RFV4, respectively. For the dense–glandular class the ICA values are 92.8, 89.2, 92.8 and 91.0 % for RFV1, RFV2, RFV3 and RFV4, respectively. For testing dataset with 161 instances, in case of RFV1, the total misclassified instances are 20 (20/161), for RFV2 the total misclassified instances are 23 (23/161), for RFV3, the total misclassified instances are 22 (22/161) and for RFV4, total misclassified instances are 23 (23/161).
4 Conclusion
In the present work the efficacy of Laws’ texture features derived using Laws’ masks of different resolutions have been tested for three-class breast tissue density classification. From the results obtained, it can be observed that the RFV1 consisting of first 7 PCs computed by applying PCA algorithm to FV1computed using Laws’ mask of length 3 with SVM classifier is significant to discriminate between the breast tissues exhibiting different density patterns achieving the highest overall classification accuracy of 87.5 %. The proposed CAD system design for the present work is shown in Fig. 9.
The proposed CAD system is different from earlier studies as most of the related studies have pre–processed the mammograms for segmenting the breast tissue by removal of pectoral muscle for their analysis while in the proposed CAD system design a fixed size ROI (200 × 200 pixels) is manually extracted from the center of the breast tissue thus eliminating the pre–processing step.
The high density of the breast tissue tends to mask the lesions present in the breast which may be malignant or benign therefore, it is recommended that if the proposed CAD system design classifies a testing instance to be of high density i.e. belonging to either fatty-glandular class or dense-glandular class, then the radiologists should re-examine that particular mammogram for any the presence of any lesion behind the high density tissue.
References
Breast cancer awareness month in October, World Health Organisation (2012).: http://www.who.int/cancer/events/breast_cancer_month/en/
Jain, A., Singh, S., Bhateja, V.: A robust approach for denoising and enhancement of mammographic breast masses. Int. J. Converg. Comput. 1(1), 38–49 (2013)
Bhateja, V., Misra, M., Urooj, S., Lay-Ekuakille, A.: A robust polynomial filtering framework for mammographic image enhancement from biomedical sensors. IEEE Sens. J. 13(11), 4147–4156 (2013)
Cancer stats: key stats, Cancer Research UK.: http://www.cancerresearchuk.org/cancer–info/cancerstats/keyfacts/
Hassanien, A.E., Moftah, H.M., Azar, A.T., Shoman, M.: MRI breast cancer diagnosis hybrid approach using adaptive ant-based segmentation and multilayer perceptron neural networks classifier. Appl. Softw. Comput. 14, 62–71 (2014)
Hassanien, A.E.: Classification and feature selection of breast cancer data based on decision tree algorithm. Stud Inf. Control 12(1), 33–40 (2003)
Wolfe, J.N.: Risk for breast cancer development determined by mammographic parenchymal pattern. Cancer 37(5), 2486–2492 (1976)
Boyd, N.F., Martin, L.J., Yaffe, M.J., Minkin, S.: Mammographic density and breast cancer risk: current understanding and future prospects. Breast Cancer Res. 13(6), 223–235 (2011)
Boyd, N.F., Martin, L.J., Chavez, S., Gunasekara, A., Salleh, A., Melnichouk, O., Yaffe, M., Friedenreich, C., Minkin, S., Bronskill, M.: Breast tissue composition and other risk factors for breast cancer in young women: a cross sectional study. Lancet Oncol. 10(6), 569–580 (2009)
Boyd, N.F., Rommens, J.M., Vogt, K., Lee, V., Hopper, J.L., Yaffe, M.J., Pater–son, A.D.: Mammographic breast density as an intermediate phenotype for breast cancer. Lancet Oncol. 6(10), 798–808 (2005)
Vachon, C.M., Gils, C.H., Sellers, T.A., Ghosh, K., Pruthi, S., Brandt, K.R., Pankratz, V.S.: Mammographic density, breast cancer risk and risk prediction. Breast Cancer Res. 9(6), 217–225 (2007)
Boyd, N.F., Guo, H., Martin, L.J., Sun, L., Stone, J., Fishell, E., Jong, R.A., Hislop, G., Chiarelli, A., Minkin, S., Yaffe, M.J.: Mammographic density and the risk and detection of breast cancer. New Engl. J. Med. 356(3), 227–236 (2007)
Warren, R.: Hormones and mammographic breast density. Maturitas 49(1), 67–78 (2004)
Boyd, N.F., Lockwood, G.A., Byng, J.W., Tritchler, D.L., Yaffe, M.J.: Mammographic densities and breast cancer risk. Cancer Epidemiology Biomarkers Prev. 7(12), 1133–1144 (1998)
Al Mousa, D.S., Brennan, P.C., Ryan, E.A., Lee, W.B., Tan, J., Mello-Thomas, C.: How mammographic breast density affects radiologists visual search patterns. Acad. Radiol. 21(11), 1386–1393 (2014)
Papaevangelou, A., Chatzistergos, S., Nikita, K.S., Zografos, G.: Breast density: computerized analysis on digitized mammograms. Hellenic J. Surg. 83(3), 133–138 (2011)
Colin, C., Prince, V., Valette, P.J.: Can mammographic assessments lead to consider density as a risk factor for breast cancer? Eur. J. Radiol. 82(3), 404–411 (2013)
Heine, J.J., Carton, M.J., Scott, C.G.: An automated approach for estimation of breast density. Cancer Epidemiol. Biomarkers Prev. 17(11), 3090–3097 (2008)
Zhou, C., Chan, H.P., Petrick, N., Helvie, M.A., Goodsitt, M.M., Sahiner, B., Hadjiiski, L.M.: Computerized image analysis: estimation of breast density on mammograms. Med. Phys. 28, 1056–1069 (2001)
Jagannath, H.S., Virmani, J., Kumar, V.: Morphological enhancement of microcalcifications in digital mammograms. J. Inst. Eng. Ser. B (India), 93(3), 163–172 (2012)
Huo, Z., Giger, M.L., Vyborny, C.J.: Computerized analysis of multiple-mammographic views: potential usefulness of special view mammograms in computer-aided diagnosis. IEEE Trans. Med. Imaging 20(12), 1285–1292 (2001)
Yaghjyan, L., Pinney, S.M., Mahoney, M.C., Morton, A.R., Buckholz, J.: Mammographic breast density assessment: a methods study. Atlas J. Med. Biological Sci. 1(1), 8–14 (2011)
Bhateja, V., Urooj, S., Misra, M.: technical advancements to mobile mammography using non-linear polynomial filters and IEEE 21451–1 NCAP information model. IEEE Sens. J. 15(5), 2559–2566 (2015)
Virmani, J., Kumar, V.: Quantitative evaluation of image enhancement techniques. In: Proceedings of International Conference on Biomedical Engineering and Assistive Technology (BEATS), pp. 1–8. IEEE Press, New York (2010)
Miller, P., Astley, A.: Classification of breast tissue by texture analysis. Image Vis. Comput. 10(5), 277–282 (1992)
Karssemeijer, N.: Automated classification of parenchymal patterns in mammograms. Phys. Med. Biol. 43(2), 365–378 (1998)
Blot, L., Zwiggelaar, R.: Background texture extraction for the classification of mammographic parenchymal patterns. In: Proceedings of Conference on Medical Image Understanding and Analysis, pp. 145–148 (2001)
Bovis, K., Singh, S.: Classification of mammographic breast density using a combined classifier paradigm. In: 4th International Workshop on Digital Mammography, 1–4 (2002)
Wang, X.H., Good, W.F., Chapman, B.E., Chang, Y.H., Poller, W.R., Chang, T.S., Hardesty, L.A.: Automated assessment of the composition of breast tissue revealed on tissue–thickness–corrected mammography. Am. J. Roentgenol. 180(1), 257–262 (2003)
Petroudi, S., Kadir T., Brady, M.: Automatic classification of mammographic parenchymal patterns: a statistical approach. In: Proceedings of 25th Annual International Conference of IEEE on Engineering in Medicine and Biology Society, pp. 798–801. IEEE Press, New York (2003)
Oliver, A., Freixenet, J., Bosch, A., Raba, D., Zwiggelaar, R.: Automatic classification of breast tissue. In: Maeques, J.S., et al. (eds.) Pattern Recognition and Image Analysis. LNCS, vol. 3523, pp. 431–438. Springer, Heidelberg (2005)
Bosch, A., Munoz, X., Oliver, A., Marti, J.: Modelling and classifying breast tissue density in mammograms. in: computer vision and pattern recognition. In: IEEE Computer Society Conference, vol. 2, pp. 1552–1558. IEEE Press, New York (2006)
Muhimmah, I., Zwiggelaar, R.: Mammographic density classification using multiresolution histogram information. In: Proceedings of 5th International IEEE Special Topic Conference on Information Technology in Biomedicine (ITAB), pp. 1–6. IEEE Press, New York (2006)
Castella, C., Kinkel, K., Eckstein, M.P., Sottas, P.E., Verdun, F.R., Bochud, F.: Semiautomatic mammographic parenchymal patterns classification using multiple statistical features. Acad. Radiol. 14(12), 1486–1499 (2007)
Oliver, A., Freixenet, J., Marti, R., Pont, J., Perez, E., Denton, E.R.E., Zwiggelaar, R.: A Novel breast tissue density classification methodology. IEEE Trans. Inf. Technol. Biomed. 12, 55–65 (2008)
Subashini, T.S., Ramalingam, V., Palanivel, S.: Automated assessment of breast tissue density in digital mammograms. Comput. Vis. Image Underst. 114(1), 33–43 (2010)
Tzikopoulos, S.D., Mavroforakis, M.E., Georgiou, H.V., Dimitropoulos, N., Theodoridis, S.: A fully automated scheme for mammographic segmentation and classification based on breast density and asymmetry. Comput. Methods Programs Biomed. 102(1), 47–63 (2011)
Li, J.B.: Mammographic image based breast tissue classification with kernel self-optimized fisher discriminant for breast cancer diagnosis. J. Med. Syst. 36(4), 2235–2244 (2012)
Mustra, M., Grgic, M., Delac, K.: Breast density classification using multiple feature selection. Auotomatika 53(4), 362–372 (2012)
Silva, W.R., Menotti, D.: Classification of mammograms by the breast composition. In: Proceedings of the 2012 International Conference on Image Processing, Computer Vision, and Pattern Recognition, pp. 1–6 (2012)
Sharma, V., Singh, S.: CFS–SMO based classification of breast density using multiple texture models. Med. Biol. Eng. Comput. 52(6), 521–529 (2014)
Sharma, V., Singh, S.: Automated classification of fatty and dense mammograms. J. Med. Imaging Health Inform. 5(3, 7), 520–526 (2015)
Kriti., Virmani, J., Dey, N., Kumar, V.: PCA–PNN and PCA–SVM based cad systems for breast density classification. In: Hassanien, A.E., et al. (eds.) Applications of Intelligent Optimization in Biology and Medicine. vol. 96, pp. 159–180. Springer (2015)
Virmani, J., Kriti.: Breast tissue density classification using wavelet–based texture descriptors. In: Proceedings of the Second International Conference on Computer and Communication Technologies (IC3T–2015), vol. 3, pp. 539–546 (2015)
Kriti., Virmani, J.: Breast density classification using laws’ mask texture features. Int. J. Biomed. Eng. Technol. 19(3), 279–302 (2015)
Kumar, I., Bhadauria, H.S., Virmani, J.: Wavelet packet texture descriptors based four-class BI-RADS breast tissue density classification. Procedia Comput. Sci. 70, 76–84 (2015)
Li, H., Giger, M.L., Huo, Z., Olopade, O.I., Lan, L., Weber, B.L., Bonta, I.: Computerized analysis of mammographic parenchymal patterns for assessing breast cancer risk: effect of ROI size and location. Med. Phys. 31(3), 549–555 (2004)
Laws, K.I.: Rapid texture identification. Proc. SPIE Image Process. Missile Guidance 238, 376–380 (1980)
Mougiakakou, S.G., Golimati, S., Gousias, I., Nicolaides, A.N., Nikita, K.S.: Computer-Aided diagnosis of carotid atherosclerosis based on ultrasound image statistics, laws’ texture and neural networks. Ultrasound Med. Biol. 33, 26–36 (2007)
Polakowski, W.E., Cournoyer, D.A., Rogers, S.K., DeSimio, M.P., Ruck, D.W., Hoffmeister, J.W., Raines, R.A.: Computer-Aided breast cancer detection and diagnosis of masses using difference of gaussians and derivative-based feature saliency. IEEE Trans. Med. Imaging 16(6), 811–819 (1997)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: Prediction of cirrhosis from liver ultrasound b–mode images based on laws’ mask analysis. In: Proceedings of the IEEE International Conference on Image Information Processing, (ICIIP–2011), pp. 1–5. IEEE Press, New York (2011)
Rachidi, M., Marchadier, A., Gadois, C., Lespessailles, E., Chappard, C., Benhamou, C.L.: laws’ masks descriptors applied to bone texture analysis: an innovative and discriminant tool in osteoporosis. Skeletal Radiol. 37(6), 541–548 (2008)
Suckling, J., Parker, J., Dance, D.R., Astley, S., Hutt, I., Boggis, C.R.M., Ricketts, I., Stamatakis, E., Cerneaz, N., Kok, S.L., Taylor, P., Betal, D., Savage, J.: The mammographic image analysis society digital mammogram database. In: Gale, A.G., et al. (eds.) Digital Mammography. LNCS, vol. 1069, pp. 375–378. Springer, Heidelberg (1994)
Doi, K.: Computer-Aided diagnosis in medical imaging: historical review, current status, and future potential. Comput. Med. Imaging Graph. 31(4–5), 198–211 (2007)
Doi, K., MacMahon, H., Katsuragawa, S., Nishikawa, R.M., Jiang, Y.: Computer-Aided diagnosis in radiology: potential and pitfalls. Eur. J. Radiol. 31(2), 97–109 (1997)
Tang, J., Rangayyan, R.M., Xu, J., El Naqa, I., Yang, Y.: Computer-Aided detection and diagnosis of breast cancer with mammography: recent advances. IEEE Trans. Inf Technol. Biomed. 13(2), 236–251 (2009)
Tagliafico, A., Tagliafico, G., Tosto, S., Chiesa, F., Martinoli, C., Derechi, L.E., Calabrese, M.: Mammographic density estimation: comparison among bi–rads categories, semi-automated software and a fully automated one. Breast 18(1), 35–40 (2009)
Giger, M.L., Doi, K., MacMahon, H., Nishikawa, R.M., Hoffmann, K.R., Vyborny, C.J., Schmidt, R.A., Jia, H., Abe, K., Chen, X., Kano, A., Katsuragawa, S., Yin, F.F., Alperin, N., Metz, C.E., Behlen, F.M., Sluis, D.: An intelligent workstation for computer-aided diagnosis. Radiographics 13(3), 647–656 (1993)
Li, H., Giger, M.L., Olopade, O.I., Margolis, A., Lan, L., Bonta, I.: Computerized texture analysis of mammographic parenchymal patterns of digitized mammograms. Int. Congr. Ser. 1268, 878–881 (2004)
Tourassi, G.D.: Journey toward computer aided diagnosis: role of image texture analysis. Radiology 213(2), 317–320 (1999)
Kumar, I., Virmani, J., Bhadauria, H.S.: A review of breast density classification methods. In: Proceedings of 2nd IEEE International Conference on Computing for Sustainable Global Development, (IndiaCom–2015), pp. 1960–1967. IEEE Press, New York (2015)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: Neural network ensemble based CAD system for focal liver lesions from B-Mode ultrasound. J. Digit. Imaging 27(4), 520–537 (2014)
Zhang, G., Wang, W., Moon, J., Pack, J.K., Jean, S.: A review of breast tissue classification in mammograms. In: Proceedings of ACM Symposium on Research in Applied Computation, pp. 232–237 (2011)
Chan, H.P., Doi, K., Vybrony, C.J., Schmidt, R.A., Metz, C., Lam, K.L., Ogura, T., Wu, Y., MacMahon, H.: Improvement in radiologists’ detection of clustered micro-calcifications on mammograms: the potential of computer-aided diagnosis. Instigat. Radiol. 25(10), 1102–1110 (1990)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: Prediction of liver cirrhosis based on multiresolution texture descriptors from B-Mode ultrasound. Int. J. Converg. Comput. 1(1), 19–37 (2013)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: A rapid approach for prediction of liver cirrhosis based on first order statistics. In: Proceedings of the IEEE International Conference on Multimedia, Signal Processing and Communication Technologies, pp. 212–215. IEEE Press, New York (2011)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: Prediction of cirrhosis based on singular value decomposition of gray level co–occurrence matrix and a neural network classifier. In: Proceedings of Development in e–systems Engineering (DESE–2011), pp. 146–151 (2011)
Vince, D.G., Dixon, K.J., Cothren, R.M., Cornhill, J.F.: Comparison of texture analysis methods for the characterization of coronary plaques in intravascular ultrasound images. Comput. Med. Imaging Graph. 24(4), 221–229 (2000)
Seng, G.H., Chai, Y., Swee, T.T.: Research on laws’ mask texture analysis system reliability. Reasearch J. Appl. Sci. Eng. Technol. 7(19), 4002–4007 (2014)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: Characterization of primary and secondary malignant liver lesions from B-Mode ultrasound. J. Digit. Imaging 26(6), 1058–1070 (2013)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: A comparative study of computer–aided classification systems for focal hepatic lesions from B–mode ultrasound. J. Med. Eng. Technol. 37(44), 292–306 (2013)
Romano, R., Acernese, F., Canonico, R., Giordano, G., Barone, F.: A principal components algorithm for spectra normalisation. Int. J. Biomed. Eng. Technol. 13(4), 357–369 (2013)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: PCA–SVM based CAD system for focal liver lesions using B–mode ultrasound images. Def. Sci. J. 63(5), 478–486 (2013)
Kumar, I., Bhadauria, H.S., Virmani, J., Rawat, J.: Reduction of speckle noise from medical images using principal component analysis image fusion. In: Proceedings of 9th International Conference on Industrial and Information Systems, pp. 1–6. IEEE Press, New York (2014)
Yazdani, A., Ebrahimi, T., Hoffmann, U.: Classification of EEG signals using dempster shafer theory and a k–nearest neighbor classifier. In: Proceedings of 4th International IEEE EMBS Conference on Neural Engineering, pp. 327–330 (2009)
Amendolia, S.R., Cossu, G., Ganadu, M.L., Galois, B., Masala, G.L., Mura, G.M.: A comparative study of k–Nearest neighbor, support vector machine and multi-layer perceptron for thalassemia screening. Chemom. Intell. Lab. Syst. 69(1–2), 13–20 (2003)
Wu, Y., Ianakiev, K., Govindaraju, V.: Improved kNN classification. Pattern Recogn. 35(10), 2311–2318 (2002)
Specht, D.F.: Probabilistic neural networks. Neural Netw. 1, 109–118 (1990)
Specht, D.F., Romsdahl, H.: Experience with adaptive probabilistic neural network and adaptive general regression neural network. In: Proceedings of the IEEE International Conference on Neural Networks, pp. 1203–1208. IEEE Press, New York (1994)
Georgiou, V.L., Pavlidis, N.G., Parsopoulos, K.E., Vrahatis, M.N.: Optimizing the performance of probabilistic neural networks in a bioinformatics task. In: Proceedings of the EUNITE 2004 Conference, pp. 34–40 (2004)
Chang, C.C., Lin, C.J.: LIBSVM, a library of support vector machines. ACM Trans. Intell. Syst. Technol. 2(3), 27–65 (2011)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: SVM–based characterization of liver cirrhosis by singular value decomposition of GLCM matrix. Inter. J. Artif. Intell. Soft Comput. 3(3), 276–296 (2013)
Hassanien, A.E., Bendary, N.E., Kudelka, M., Snasel, V.: Breast cancer detection and classification using support vector machines and pulse coupled neural network. In: Proceedings of 3rd International Conference on Intelligent Human Computer Interaction (IHCI 2011), pp. 269–279 (2011)
Virmani, J., Kumar, V., Kalra, N., Khandelwal, N.: SVM–Based characterization of liver ultrasound images using wavelet packet texture descriptors. J. Digit. Imaging 26(3), 530–543 (2013)
Azar, A.T., El–Said, S.A.: Performance analysis of support vector machine classifiers in breast cancer mammography recognition. Neural Comput. Appl. 24, 1163–1177 (2014)
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Kriti, Virmani, J. (2016). Comparison of CAD Systems for Three Class Breast Tissue Density Classification Using Mammographic Images. In: Dey, N., Bhateja, V., Hassanien, A. (eds) Medical Imaging in Clinical Applications. Studies in Computational Intelligence, vol 651. Springer, Cham. https://doi.org/10.1007/978-3-319-33793-7_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-33793-7_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-33791-3
Online ISBN: 978-3-319-33793-7
eBook Packages: EngineeringEngineering (R0)