Abstract
Somatic embryogenesis (SE), the process through which already differentiated cells reverse their developmental programme and become embryogenic, requires drastic changes in the transcriptome of the explant cells. Among the various factors that underlie this developmental switch, genes encoding transcription factors (TFs), which constitute the sequence-specific DNA-binding proteins, are widely accepted as playing a central function in the gene expression regulation. In recent years, intensive analysis of the global transcriptomes of plant cells that are undergoing embryogenic transition and the use of Arabidopsis (a model in plant genomics) in studies on the genetic control of SE have substantially contributed to the identification of SE regulators. A survey of SE-associated transcriptomes illustrated the combinational effects of stress and hormone signalling that are related to the in vitro environment that is imposed during a culture. Accordingly, among the TFs that are considered to be essential in SE induction, those that are involved in stress and hormone plant responses and especially flower development were found to be most frequent. This chapter provides a comprehensive review of the current knowledge about the TFs that are involved in the induction of SE in plant explants that are cultured in vitro. In addition to a general characterisation of the TF transcriptomes that are associated with SE induction in different plants, the individual TF genes with documented functions in the regulation of SE are presented with a special reference to their possible targets and the TF-controlled molecular mechanisms that underlie SE induction.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Somatic Embryo
- Somatic Embryogenesis
- Embryogenic Culture
- Auxin Biosynthesis
- Somatic Embryogenesis Induction
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Global Characteristics of the SE-Associated TF Transcriptomes
The reprogramming of already differentiated somatic cells towards embryonic development requires a substantial modification of a cell’s transcriptomes. Embryogenic transition induced in somatic cells involves the repression or activation of numerous genes and thus transcription factors (TFs) that have a key function in the regulation of gene expression seem to play a crucial role in this process. The large number (6–10 %) of TF-coding genes that have been found in plant genomes imply the transcriptional regulation to play an even more important role in plant than in animal development (Riechmann et al. 2000).
Most of the available data on SE-involved transcriptomes was provided by global analytical approaches and among these microarray analysis has been intensively applied to investigate the embryogenic cultures of different plants including oil palm (Low et al. 2008), Medicago truncatula (Mantiri et al. 2008a), potato (Sharma et al. 2008), rice (Chakrabarty et al. 2010) and cucumber (Wiśniewska et al. 2012). Besides microarrays, EST sequencing in wheat (Singla et al. 2007) and RNA-seq in Arabidopsis (Wickramasuriya and Dunwell 2015) and cotton (Yang et al. 2012) have been applied in order to reveal SE-related transcriptomes. The microarray-based data showed that 1–12 % of all of the genes that were significantly modulated during embryogenic induction in different plants were found to encode TFs. However, the number of SE-involved TF genes based on microarray data appears to be seriously underestimated. A much more accurate evaluation of the TF genes that are involved in SE might provide approaches that are focused specifically on the analysis of TF transcriptomes. Accordingly, a multi-parallel qRT-PCR analysis of almost 1,900 TF genes of Arabidopsis showed that 1,768 (94 %) TF genes were expressed during SE induction and a large fraction of these (41 %) was found to have undergone a significant modulation of transcription (Gliwicka et al. 2013).
The examination of SE-related transcriptomes indicated that a common set of TF genes that encode the proteins representing MYB, MADS, AP2/ERF, bHLH, C2H2, WRKY, NAC and HB families is engaged in SE induction in different plants. Members of SE-involved TF families belong to various functional groups and the TF genes that are engaged in the transcriptional regulation of hormone and stress responses as well as those that control plant developmental processes, predominantly embryo and flower development, have been found to be the most frequent (Thibaud-Nissen et al. 2003; Che et al. 2006; Hosp et al. 2007; Sharma et al. 2008; Gliwicka et al. 2013; Wickramasuriya and Dunwell 2015). Interestingly, the TFs that control the development of the plant generative organs were found to be especially frequent among SE-modulated transcripts, thus suggesting that there are some similarities in the genetic regulation of generative and embryogenic transitions (Thibaud-Nissen et al. 2003; Sharma et al. 2008; Gliwicka et al. 2013).
The TFs that control zygotic embryogenesis (ZE) constitute the obvious candidates for SE regulators due to the anticipated similarities of an SE to its zygotic counterpart (Dodeman et al. 1997). Expression of at least 500 genes has been reported to control ZE in Arabidopsis and among the genes-encoded TFs, the members of ABI3VP1, AP2/ERF, ARF, C3H and Dof families have been indicated (Tzafrir et al. 2004), but these data also appear to be underestimated. A recent study on the ZE transcriptome of Arabidopsis showed that at least 60 % of Arabidopsis genes are expressed during seed development and up to 5 % of these were identified as TF-encoded (Belmonte et al. 2013). In support of the expected similarities in SE- and ZE-related transcriptomes, an analysis of an embryogenic culture in rice revealed that out of 242 rice homologues of the genes that are essential for ZE in Arabidopsis, 87 % were expressed during SE (Su et al. 2007). Further support for some type of convergence of the genetic determinants that control SE and ZE was provided by the observation that most of the genes that are differentially regulated during SE in various plants were revealed to represent the major TF families that are engaged in ZE (Singla et al. 2007; Imin et al. 2008; Chakrabarty et al. 2010; Wisniewska et al. 2012; Gliwicka et al. 2013; Wickramasuriya and Dunwell 2015).
In contrast to the numerous TF genes that have a differential expression in the embryogenic cultures of different plants and are thus assumed to contribute to SE induction (Singla et al. 2007; Sharma et al. 2008; Mantiri et al. 2008a; Chakrabarty et al. 2010; Wiśniewska et al. 2012), only a small number has been experimentally proven to control SE transition. Conclusively for the mechanism that is involved in SE induction, among the TF genes that have validated functions in SE, those that are related to hormone and stress responses have been indicted to be prevalent (Zavattieri et al. 2010; Fehér 2015).
The overrepresentation of the TFs that is related to hormone responses among the SE-modulated genes that have been reported in different plants reflects the common belief about the essential role of plant growth regulators in the control of the morphogenic pathways, including SE induced in plants in vitro (Jiménez 2005; Fehér et al. 2003). In Arabidopsis, 43 % of SE-modulated TF genes have been annotated to be hormone-related (Gliwicka et al. 2013). Besides the TFs that are involved in the metabolism and signalling of auxin, the genes that are related to cytokinin (CK), abscisic acid (ABA), jasmonic acid (JA), ethylene (ET), gibberellin (GA) and brassinosteroids (BR) have been reported in Arabidopsis and other plants (Singla et al. 2007; Imin et al. 2008; Chakrabarty et al. 2010; Wisniewska et al. 2012; Gliwicka et al. 2013; Wickramasuriya and Dunwell 2015). In support of the essential role of the genetic regulation of hormone metabolism and signalling in embryogenic transition, the mutations affected the level and sensitivity of different hormones (IAA, ABA, GA and ethylene) and the inhibitors of hormone metabolism or signalling have been shown to negatively impact SE induction in Arabidopsis (Gaj et al. 2006; Bai et al. 2013; Nowak et al. 2015).
Relevant to the common use of auxin treatment in SE induction in different plants (Gaj 2004) and the key role of IAA in the control of plant development (reviewed in Vanneste and Friml 2009), auxin signalling and metabolism have been postulated as being crucial for in vitro induced SE (Jiménez 2005; Fehér et al. 2003). In accordance, auxin-related TFs have been found to be the most frequent among the hormone-related genes that are involved in SE induction (Yang et al. 2012; Gliwicka et al. 2013; Wickramasuriya and Dunwell 2015). The TF genes that have a regulatory role in SE include the core regulators of auxin signalling, AUX/IAA and ARF genes (Rensing et al. 2005; Su et al. 2007; Wu et al. 2009; Yang et al. 2012; Gliwicka et al. 2013). A differential expression of IAA16, IAA29, IAA30, IAA31 and ARF1, ARF2, ARF3, ARF5, ARF6, ARF8 and ARF11 was observed during SE induction in Arabidopsis and the mutants in these genes were substantially defective in the embryogenic response (Gliwicka et al. 2013; Wójcikowska and MDG, in preparation for publication). AUX/IAA–ARF-mediated auxin responses were also assumed to operate in the embryogenic cultures of other plants including cotton, rice, Cyclamen persicum and Gossypium hirsutum (Rensing et al. 2005; Su et al. 2007; Wu et al. 2009; Yang et al. 2012). This induction of AUX/IAA and ARF expression that is commonly associated with SE parallels a substantial function of auxin signalling in the control of zygotic embryo development (Sato and Yamamoto 2008; Rademacher et al. 2011, 2012).
In spite of the notable progress that has recently been made in deciphering the auxin-mediated regulation of plant development, our knowledge about the functions of ARFs in different developmental processes is still fragmentary and the results that have been obtained in various experimental approaches are frequently inconclusive (Rademacher et al. 2011). Within ARFs, the ARF5 encoded so-called MONOPTEROS (MP) protein is functionally the best characterised and the role of MP in the mediation of auxin signal has been documented in a number of developmental processes including ZE (reviewed in Möller and Weijers 2009). In addition to auxin signalling, ARF5 was also reported to control the polar auxin transport by targeting the auxin efflux carrier, PIN1 (PIN-FORMED1) (Wenzel et al. 2007). Our recent results provided some evidence about the engagement of ARF5 in SE induction including the auxin-stimulated, strong accumulation of its transcripts in the explant parts that are involved in SE and the significantly impaired embryogenic potential of the afr5 mutant and the overexpressor line (Wójcikowska and MDG, in preparation for publication). However, the mechanism of ARF5 action in embryogenic transition remains to be demonstrated and a prerequisite for an understanding of the biological function of ARFs in somatic cells that are undergoing SE induction is the identification of their target genes, especially those that are directly controlled.
2 LEAFY COTYLEDON Genes—Master Regulators of the Embryogenic Development in Plants
The LEC group of genes includes the LEC1, LEC2 and FUS3-encoded TFs that have a major role in the control of the morphogenesis and the maturation phases during ZE (Harada 2001). LEC1 encodes the CCAAT box-binding factor HAP3 subunit (Lotan et al. 1998) while LEC2 and FUS3 encode proteins that have a plant-specific B3 domain, which binds a highly conserved RY motif and regulates the expression of ZE-specific genes (Stone et al. 2001; Braybrook et al. 2006). The observation that the overexpression of LEC2 and LEC1 resulted in developmental disorders in plants that included the formation of callus and somatic embryos on seedlings suggested SE-related functions of LECs (Lotan et al. 1998; Stone et al. 2001). In support of the proposed role of the LECs in the embryogenic transition of somatic cells, the lec mutants were found to be strongly defective in SE but not in the shoot organogenesis that was induced in vitro (Gaj et al. 2005). In addition, a key role of LEC TFs in the establishment of a cellular environment that promotes embryo development supported the activity of the LEC genes, which has commonly been observed in the embryogenic cultures of different plants (Zuo et al. 2002b; Harding et al. 2003; Yazawa et al. 2004; Ikeda et al. 2006; Fambrini et al. 2006; Guo et al. 2013; Zhang et al. 2014; Zhu et al. 2014a). The complex interactions between LEC genes and hormone metabolism that were revealed to control the maturation phase in zygotic embryo development (reviewed in Jia et al. 2014) provided a clue as to how LEC genes might support SE induction. The LEC-mediated control of auxin, ABA and GA metabolism observed during ZE seemed to especially be of importance for the promotion of SE (Braybrook and Harada 2008).
2.1 LEC2
Among the LEC genes, the LEC2-mediated mechanism of SE induction has been the most intensively investigated. As a result, the auxin-related functions of LEC2 in the embryogenic transition of somatic cells have been documented. Similar to the regulatory link that was observed between the LEC2 gene and YUC genes involved in auxin biosynthesis in Arabidopsis seedlings (Stone et al. 2008), LEC2 was found to stimulate YUC1, YUC4 and YUC10 transcripts in in vitro cultured explants (Wójcikowska et al. 2013). As a result of the LEC2-mediated activation of the YUC pathway of auxin biosynthesis, a significant increase of IAA content was demonstrated in explant tissue that was undergoing embryogenic transition (Wójcikowska and Gaj 2015). The activation of YUC genes was also reported in the embryogenic callus of Arabidopsis in which somatic embryos were induced in response to the removal of auxin from a medium (Bai et al. 2013). LEC2 was postulated to directly target YUC4 in planta (Stone et al. 2008); however, further analyses are necessary to reveal the mode of regulatory interaction that has been observed between LEC2 and YUC genes upon SE induction in vitro. Collectively, LEC2 contributes to SE induction via an increase in the endogenous auxin levels that in turn results in the activation of the auxin-responsive genes that are operating in the SE-inductive network. Genetic components of this network and their complex interactions remain to be determined. Revealing how similar the molecular pathways that are triggered by endogenous versus exogenous auxin during SE induction will also be challenging.
In addition to the regulation of auxin metabolism, LEC2 may be involved in the control of auxin signalling. To support this, the LEC2-mediated activation of the key components of the auxin-response pathway, members of AUX/IAA family (IAA1, IAA17, IAA30 and IAA31), was reported in Arabidopsis seedlings (Braybrook et al. 2006; Stone et al. 2008). Relevant to these observations and important for the predicted functions of AUX/IAA genes in SE, mutations in two of these genes, iaa30 and iaa31, were observed to seriously impair the embryogenic potential of in vitro cultured explants (Gliwicka et al. 2013). In addition to the postulated regulatory interactions with the auxin metabolism and signalling, the possible involvement of LEC2 in auxin polar transport cannot be ruled out as the upregulation of auxin efflux facilitators, PIN1 and PIN2, was observed in transgenic tobacco plants that overexpressed LEC2 (Guo et al. 2013). PIN proteins are believed to direct plant developmental responses to environmental and endogenous signals through the control of the polar cell-to-cell transport of auxin (Habets and Offringa 2014), and relevantly, a key function of the auxin efflux carriers in ZE was documented (Friml et al. 2003). The findings that the explants of a pin1 mutant were unable to undergo embryogenic induction in vitro (Su et al. 2009) and that the inhibitors of the auxin polar transport severely impaired the embryogenic response of explants in different plants (Venkatesh et al. 2009; Palovaara et al. 2010) provided further support for the involvement of PINs in SE induction.
The complex LEC2-mediated crosstalk between hormones is assumed to be associated with the mechanism of SE induction considering that in tissues that overexpress LEC2, the increase of auxin content has been related to the extensive changes in the accumulation of cytokinins, ABA and SA (Wójcikowska and Gaj 2015). Furthermore, a link between LEC2 and ethylene may be also expected considering the LEC2-stimulated expression of the ACS4 gene that is engaged in the synthesis of an ethylene precursor (Braybrook et al. 2006), and the regulatory relationship that has been indicated between the LEC2 and ERF022 genes involved in the ethylene biosynthesis/signalling (Nowak et al. 2015).
The observation that the overexpression of the YUC genes alone is not sufficient to induce SE provided some additional insight into the hormone-related functions of LEC2 in SE (Zhao et al. 2001). This implies that only SE-competent cells can respond to the auxin signal. Thus, relevant to the LEC2 function in the maturation phase of ZE, the gene was proposed to enable somatic cells to become capable of responding to the SE-inductive signal by lowering the GA content (Braybrook and Harada 2008). In this GA-related regulatory circuit, LEC2 directly activates AGL15, which in turn activates GA2ox6 resulting in a reduced GA level coupled with the enhanced potential for the formation of somatic embryos (Wang et al. 2004). The report on the negative impact of exogenous GA3 on the embryogenic response of Arabidopsis explants supports the inverse relation between the GA level and a tissue’s capacity for SE (Gaj et al. 2006). The LEC2-mediated establishment of a proper balance between GA and ABA levels promotes the accumulation of the storage reserves that was proposed to enhance the embryogenic competence in cells (Braybrook and Harada 2008). The fact that the ectopic expression of AtLEC2 has been reported to induce the maturation processes in transgenic Arabidopsis, tobacco and Theobroma cacao tissue (Stone et al. 2008; Guo et al. 2013; Zhang et al. 2014) and that the high expression of the genes encoding storage proteins, including CRA1 and OLEO4, was found to be associated with the embryogenic potential of Arabidopsis (Stone et al. 2008; Gliwicka et al. 2012) support this speculation.
Considering that LEC2 positively impacts auxin accumulation and in turn, auxin activates its expression (Ledwoń and Gaj 2009; Wójcikowska et al. 2013), the revealing of the genetic components of a regulatory feedback loop that seems to operate between auxin and LEC2 is required for the full understanding of the LEC2-controlled mechanism of embryogenic transition. In line with this notion, an auxin-responsive AuxRE element was identified in LEC2 promoter region, which implies the involvement of ARFs in the regulation of LEC2 expression. Among the potential regulators of LEC2, there are several ARFs (ARF1, ARF2, ARF3, ARF5, ARF6, ARF8, ARF11) that are differentially expressed in SE of Arabidopsis (Wójcikowska and MDG, in preparation for publication).
In the search for the genetic regulators of LEC2 in SE, the proteins that are indicated to directly inhibit the LEC2 expression in planta should be considered including, TT8 (TRANSPARENT TESTA8), ASIL1 (ARABIDOPSIS 6B-INTERACTING PROTEIN1-LIKE1) and PRC (POLYCOMB REPRESSIVE COMPLEXES) (Gao et al. 2009; He et al. 2013; Chen et al. 2014). In addition, miRNA166 was reported to indirectly control LEC2 expression through the regulation of PHABULOSA (PHB) and PHAVOLUTA (PHV) (Tang et al. 2012). Whether similar regulatory interactions exist during SE induction is as yet unknown.
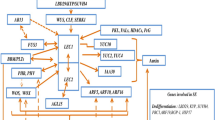
In summary, a possible model of the LEC2-controlled and hormone-related pathways that seem to underlie the embryogenic transition in somatic cells can be proposed (Fig. 5.1).
Model for the LEC2 role in SE induction through gene interactions with hormones. The interactions that need verification are indicated with dashed lines. 1 Ledwoń and Gaj (2009); 2 Wójcikowska et al. (2013); 3 Bai et al. (2013); 4 Curaba et al. (2004); 5 Braybrook et al. (2006); 6 Nowak et al. (2015); 7 Braybrook et al. (2006); 8 Tang et al. (2012). SSP-seed storage protein
2.2 LEC1
Expression of LEC1 has been associated with SE that is induced in vitro and in planta (Lotan et al. 1998; Yazawa et al. 2004; Garcês et al. 2007; Alemanno et al. 2008; Ledwoń and Gaj 2009; Guo et al. 2013; Nic-Can et al. 2013; Zhu et al. 2014a). Similar to LEC2 and meaningful for the possible role of LEC1 in the SE-inductive mechanism, the encoded TF is involved in auxin metabolism/signalling. Among the candidate targets of LEC1, the YUC10 gene, which is involved in auxin biosynthesis and the members of the Aux/IAA family (IAA5, IAA16, IAA19), were postulated (Junker et al. 2012). In addition to auxin, the regulatory relations between LEC1 and the metabolism/signalling of other hormones including ABA, JA and BR were implicated as underlying the function of LEC1 in zygotic and somatic embryogenesis (reviewed in Junker and Bäumlein 2012; Junker et al. 2012).
In addition to hormones, the sugar-related functions of LEC1 were reported to promote embryonic cell identity and in support of this, sucrose was demonstrated to modulate the penetrance of embryogenic traits in a turnip mutant that ectopically expressed LEC1 (Casson and Lindsey 2006). Given the fact that sugars are proposed to act as morphogens that provide positional information in plant development, LEC1 may exert its role in control of SE via the regulation of the sugar metabolism (Rolland et al. 2002). Another clue for the identification of the SE-promoting functions of LEC1 was provided by the observation about the upregulation of the genes encoding the cell wall associated enzymes—hydrolase xyloglucan (XTH9) and expansine (EXP1B) in response to LEC1 overexpression (Junker et al. 2012).
LEC1 is a member of the nuclear factor Y (NF-Y) family of TFs, which are highly conserved in all eukaryotic organisms. The NF-Y heterotrimer consists of three subunits NF-YA, NF-YB and NF-YC while LEC1 represents NF-YB9 TF (Mu et al. 2008). Importantly, for the understanding of the LEC1-related regulatory interactions that can operate during SE, the overexpression of LEC1 upregulates NF-YA1, NF-YA5 and NF-YA9 and, in turn, the overexpression of NF-YA1 and NF-YA9 positively regulates the expression of embryo- or seed-specific genes including LEC1 (Mu et al. 2008, 2013). Similar to LEC1, the overexpression of NF-YA1, NF-YA5, NF-YA6 and NF-YA9 is sufficient to induce the formation of somatic embryos from vegetative tissues (Mu et al. 2013). Moreover, the cooperation of NF-YA5 and LEC1 is involved in the regulation of the genes that are responsible for the zygotic embryo development (Zhao et al. 2009) and whether similar interaction occurs during SE induction remains to be determined.
Collectively, some of the evidence presented above indicates the involvement of LEC1 in SE induction; however, the impact of the encoded TF on the embryogenic response appears to be less pronounced in comparison to LEC2.
2.3 FUS3
In contrast to the other two LEC genes, FUS3 is not upregulated in Arabidopsis explants induced towards SE, and overexpression of FUS3 does not lead to the formation of somatic embryos (Ledwoń and Gaj 2011). However, the existence of a pathway that involves an LEC2-induced increase in auxin levels that promotes FUS3 activity was proposed (Braybrook and Harada 2008), thus suggesting that FUS3 might be involved in an LEC2-controlled mechanism of SE induction.
Numerous hormone-related genes are expressed in response to the activation of FUS3 and among them the YUC genes of auxin biosynthesis, AUX/IAAs and ARFs, which encode the key components of auxin signalling, and the genes that are related to the biosynthesis of ABA, CK and BR were reported (Yamamoto et al. 2010; Wang and Perry 2013). Moreover, the relation of FUS3 to GA was documented and the encoded TF, similar to two other LEC TFs, may enhance the competence for SE induction via the repression of AtGA3ox2, thereby resulting in a reduced level of bioactive GA (Curaba et al. 2004).
FUS3 was also reported to regulate vegetative phase transitions by negatively modulating ethylene-regulated genes in Arabidopsis, and among the downregulated genes those involved in ethylene biosynthesis (ACS6) and signalling (ERF1, ERF104, ESE3, EDF4) were reported (Lumba et al. 2012). In support of the ethylene-related function of FUS3, the ethylene level was found to correlate with the expression of GmFUS3 in SE of soybean (Zheng et al. 2013). The role of the ethylene-associated activities of FUS3 in SE induction requires further study.
3 SE-Related Functions of AGL15
AGL15 encodes one of the MADS domain proteins that it is believed to play key roles in the regulation of the developmental processes in eukaryotes (reviewed in Smaczniak et al. 2012). TF with MADS domain selectively binds to a consensus DNA sequence, the CArG (C-A/T rich-G) motif, to either activate or repress the expression of the targeted genes (West et al. 1997). The SE-related function of AGL15 was postulated in Arabidopsis due to the somatic embryo-promoting effect of AGL15 overexpression that was observed in the seedlings and in immature zygotic embryos that were cultured in vitro (Harding et al. 2003; Thakare et al. 2008). The AGL15 protein was found to accumulate during the early stages of ZE in Brassica napus, Zea mays and A. thaliana, in the somatic embryos of Medicago sativa and in a microspore culture of B. napus (Perry et al. 1999). The role of AGL15 in the promotion of embryogenic responses was reported as being related to the GA metabolism and as a result, the GA2ox6 that encodes gibberellin oxidase was identified among the targets of AGL15 (Thakare et al. 2008). It is postulated that AGL15 controls SE via the downregulation of the level of biologically active GA, and the inhibitory effect of GA on cell division may account for the requirement of a low level of this hormone during the early stages of SE (Wang et al. 2004). Besides GA, AGL15 seems to control the metabolism of ethylene. Recently, At5g61590, a member of AP2/ERF family and an orthologue of MtSERF1, which is involved in SE induction in M. truncatula, was identified as being a direct target of AGL15 (Zheng et al. 2013). It was shown that At5g61590 (DEWAX–Decrease Wax Biosynthesis) acts as a repressor of the biosynthesis of cuticular wax (Go et al. 2014). In addition, the ethylene biosynthesis genes, ACC SYNTHASE (ACS) and ACC OXIDASE (ACO), are expressed in response to AGL15 (Zheng et al. 2013). Stress-related functions of AGL15 were postulated in soybean and the enhanced embryogenic response of explants that was observed upon the overexpression of GmAGL15 was suggested to be the result of the activation of the genes that are involved in stress response (Zheng and Perry 2014).
AGL15 is believed to be a component of the SERK1 (SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1) complex (Karlova et al. 2006), which was proposed to mark the cells that are competent in SE (Hecht et al. 2001). SERK1 was assumed to interact with BRI1 (BRASINOSTEROID-INSENSITIVE1), and thus the function of AGL15 in BR signalling might be inferred (Aker and de Vries 2008).
AGL15 was also hypothesised to be involved in chromatin repression via its interaction with SIN3/HDAC (SWI-INDEPENDENT3/HISTONE DEACETYLASE), HDA (Hill et al. 2008) and topless (TPL) and topless-related (TPR) proteins (Causier et al. 2012). AGL15 has also been suggested to contribute to chromatin repression by increasing the efficiency of the formation of a repressive complex or the recruitment of a corepressor (Fernandez et al. 2015). Further analysis is needed to identify the targets of the epigenetic repression that is mediated by AGL15 during embryogenic induction.
Besides protein-encoded targets, the genes encoding microRNA156 (MIR156a and MIR156c), which are involved in the suppression of the SQUAMOSA promoter binding protein-like 3 (SPL3) transcription factor that promotes the floral transition in Arabidopsis, have recently been indicated to be AGL15-controlled (Wu and Poethig 2006; Serivichyaswat et al. 2015). Considering the apparent similarities in the genetic control of floral and embryogenic induction (El Ouakfaoui et al. 2010), a regulatory interaction between AGL15 and the floral suppressor, miR156, may be anticipated during SE induction. In support of this, the significant repression of the miR156 accumulation was observed during SE induction in Arabidopsis (Szyrajew and MDG, data unpublished). However, the existence of a regulatory link between the AGL15 and MIR156 genes in an embryogenic culture remains to be validated, and the identification of the miR156-regulated mRNAs upon SE induction is required in order to reveal whether the mechanism of embryogenic transition is convergent with floral induction.
4 Stress-Related TFs
The embryogenic transition of the somatic cells was postulated to manifest a general response of the cultured tissue to stress conditions imposed in vitro such as wounding or the 2, 4-D treatment of explants (Fehér et al. 2003; Karami and Saidi 2010). In line with this postulate, various stress factors (osmotic, salt, water and heavy metals) were shown to replace or enhance the hormone treatment that was used for SE induction in different plant species (Kamada et al. 1993; Patnaik et al. 2005). Relevant to the assumed role of stress in SE induction, numerous (4–12 %) stress-related mRNAs have been found among those that were differentially expressed during SE induction (Legrand et al. 2007; Chakrabarty et al. 2010; Yang et al. 2012; Jin et al. 2014; Wickramasuriya and Dunwell 2015). In conjunction with the stress-regulated mechanism of SE induction, almost 40 % of the differentially expressed TF genes in an embryogenic culture of Arabidopsis were annotated to stress-related functions (Gliwicka et al. 2013). Some of these genes were subjected to closer inspection including NTL8, DREB2F, ATHB-12, LBD20 and MYB74, and the SE-impaired phenotypes that were observed in the plants that carried mutations in these genes strongly support the notion that SE induction shares a mechanism at the molecular level that is relevant to general stress responses (Gliwicka et al. 2013).
4.1 ERF Genes and the Ethylene Response
In SE induced in Arabidopsis, AP2/ERF genes constituted a substantial part (10 %) of the stress-related TF genes that had a differential expression (Gliwicka et al. 2013). Similar to these data that are based on qRT-PCR analysis, RNA-seq analysis of the SE-related transcriptome of an embryogenic culture in Arabidopsis indicated that almost 40 % of the highly stimulated TF genes represented the genes encoding AP2/ERF TFs (Wickramasuriya and Dunwell 2015). In addition to Arabidopsis, the differential activity of numerous ERF genes was described in embryogenic cultures of wheat (Singla et al. 2007), M. truncatula (Imin et al. 2008), rice (Chakrabarty et al. 2010), cucumber (Wisniewska et al. 2012) and Hevea brasiliensis (Piyatrakul et al. 2012). Given that ERF TFs were assumed to regulate stress responses, and especially ethylene-related pathways (Nakano et al. 2006), the common expression of ERFs during SE induction implies the involvement of ethylene in SE induction.
The observed modulation of ethylene-related ERF genes during SE induction may result in part from the auxin treatment of explants. To support this assumption, auxin was documented as influencing ethylene signalling and metabolism and the complex interactions between auxin and ethylene were recently demonstrated in the regulation of plant development (Stepanova et al. 2007; reviewed in Muday et al. 2012). In accordance with in planta development, the ethylene level was also shown to affect auxin biosynthesis and distribution in an embryogenic culture of Arabidopsis (Bai et al. 2013).
In contrast to dozens of the ethylene-related genes that are differentially expressed during embryogenic transition, the functions of only a few of them have been experimentally proven as being related to SE.
4.1.1 MtSERF1
MtSERF1 gene was identified as promoting SE induction in M. truncatula and its high expression, which was observed in globular somatic embryos, was found to be ethylene-induced (Mantiri et al. 2008a). The orthologues of MtSERF1 were also reported to positively impact the embryogenic responses in cultures of Arabidopsis (Zheng et al. 2013) and H. brasiliensis (Piyatrakul et al. 2012). Importantly for the SERF1-mediated mechanism of SE induction, an Arabidopsis orthologue of MtSERF1 (At5g61590) was recently identified as being a direct target of AGL15 (Zheng et al. 2013), which has an essential role in SE induction (see paragraph 3). In addition, interactions between MtSERF1 and the members of HD-ZIP III family, PHB, PHV and REV (REVOLUTA), which are regulators of early zygotic embryo development in Arabidopsis, were suggested, and the encoded TF was proposed as linking the stress response to development during SE induction (Mantiri et al. 2008b).
4.1.2 ERF022
ERF022 expression was observed to be drastically inhibited during SE induction in Arabidopsis and a significantly impaired embryogenic response of the ERF022 mutant was found to be associated with increased ethylene production (Gliwicka et al. 2013; Nowak et al. 2015). Further analysis indicated the negative impact of ERF022 on the biosynthesis and signalling of ethylene and the candidate target genes including ACS7 involved in ethylene biosynthesis, and ERF1, which is an essential element in the ethylene signal transduction pathway (Nowak et al. 2015). The inhibitory effect of ethylene on SE induction that was observed in Arabidopsis was postulated as resulting from the negative impact of ethylene on biosynthesis and the local distribution of auxin (Bai et al. 2013). In support of this postulate, auxin accumulation following the LEC2-mediated activation of the YUC-dependent pathway of IAA production was found to be required for SE induction in an Arabidopsis culture (Wójcikowska et al. 2013). The interactions between ERF022 and LEC2, which is the key regulator of auxin-dependent embryogenic transition in Arabidopsis, were also demonstrated to be important for the role of ERF022 in the genetic network that underlies SE induction (Nowak et al. 2015).
4.2 TFs that Control LEA Accumulation
LEA proteins are accumulated in the late stages of ZE and the increased expression of LEA and other genes that encode storage proteins was found to be associated with SE induced in numerous plants including soybean (Thibaud-Nissen et al. 2003), maize (Che et al. 2006), rape (Hosp et al. 2007), potato (Sharma et al. 2008) and Arabidopsis (Gliwicka et al. 2012). As a consequence, it was postulated that the increased tolerance to stress that is caused by an accumulation of storage proteins promotes the induction of embryogenic development (Stone et al. 2008).
Consistent with this hypothesis, several TF genes that are presented below appear to promote SE via accumulation of the storage proteins. Among such genes are those encoding the proteins of the MYB family that regulate the transcription of the target genes through a highly conserved DNA-binding domain, which is homologous to animal c-MYB (Dubos et al. 2010). The functions of two of the MYB genes, MYB118 and MYB115, support the proposition that a storage protein-related mechanism might be considered in SE promotion. MYB118 and MYB115 were indicated to have stress-related functions, and their SE-promoting activity in the seedlings and root explants of Arabidopsis was reported (Wang et al. 2008). Relevant to the concept on the positive relation between the storage proteins and the embryogenic potential of tissue, MYB118 and MYB115 were documented to positively control LEA (LATE EMBRYOGENESIS ABUNDANT) genes, including EM1, EM6, EM10, LEA76 and ECP63 in ZE (Zhang et al. 2009). In addition to the stimulation of LEA production, MYB118/MYB115 were recently reported to negatively control bezonyloxy glusosinolate biosynthesis, which is a secondary metabolite produced in response to stress (Zhang et al. 2015). This finding provides further support to the stress-related functions of MYB115/118 in SE induction.
Another TF gene, bHLH109, which is a member of bHLH family, is also assumed to promote SE induction in Arabidopsis via the activation of LEA genes. The strong activation of bHLH109 expression was found to be associated with SE induction, and the overexpression of this gene was indicated as enhancing the embryogenic response of Arabidopsis explants (Gliwicka et al. 2013). Recently, it was postulated that bHLH109 might operate in SE as an activator of the LEA gene ECP63, and the TF genes that were annotated to stress-related functions (At5g61620, bZIP4 and bZIP43) were indicated to be among the potential regulators of bHLH109 (Nowak and Gaj 2016).
Collectively, the identification of the TF genes that control the LEA proteins of stress protective function among the SE regulators provided new evidence that the cell responses to stress that are imposed under in vitro conditions underlie the promotion of SE.
4.3 WIND1
WOUND INDUCED DEDIFFERENTIATION 1 (WIND1/RAP2.4) of the AP2/ERF TF superfamily, which positively regulates cell dedifferentiation in Arabidopsis, was found to be induced by wounding (Iwase et al. 2011). An elevated WIND1 expression was demonstrated to be sufficient to promote unorganised cell proliferation and the redifferentiation of the callus into roots, shoots and embryos on a hormone-free medium. WIND1-overexpressing explant cells were demonstrated to reacquire pluripotency and the modulation of the cytokinin biosynthesis/signalling through ARR-dependent signalling pathway was proposed as being associated with the SE-promoting functions of WIND1 (Iwase et al. 2011). Other molecular elements that link the WIND1-mediated initial wound response to the control of cell dedifferentiation needs to be revealed.
5 PLETHORA Genes—The Integrators of Hormonal Inputs
The AINTEGUMENTA-LIKE (AIL) family of TF genes, which have an AP2/ERF domain, includes the AINTEGUMENTA (ANT), BABY BOOM (BBM/PLT4) and PLETHORA genes. All of these are expressed in young, dividing tissue and play central roles in different developmental processes including embryogenesis (Elliott et al. 1996; Klucher et al. 1996; Boutilier et al. 2002; Aida et al. 2004; Galinha et al. 2007). The importance of the AIL function and its relation to auxin in zygotic embryo development was indicated (Aida et al. 2004; Blilou et al. 2005). Besides auxin, AIL genes were reported to be related to ABA, GA and JA signalling and thus, PLT/BBM genes were postulated to integrate multiple hormonal inputs in the plant development and to act as ‘hubs in a plethora of networks’ (Horstman et al. 2014).
Similar to BBM/PLT4, other PLETHORA TFs (PLT1, PLT2, PLT3, EMK/PLT5 and PLT7) have also been indicted to exert the SE-related functions because the somatic embryo formation in response to their overexpression was observed (Tsuwamoto et al. 2010; Horstman 2015). Although our knowledge about the PLT-mediated induction of SE is rather fragmentary, a recent finding that PLT3, EMK/PLT5 and PLT7 stimulate auxin biosynthesis through the activation of YUC genes (YUC1 and YUC4) to control phyllotaxis (Pinon et al. 2013) implies a possible role of PLTs in the auxin-related mechanism of SE induction. An EMK/PLT5-controlled induction of SE may also be related to GA and an encoded TF was reported to negatively impact GA biosynthesis in the control of the storage protein accumulation in Arabidopsis seeds (Sundaram et al. 2013).
Regulatory interactions between the PLT genes and other TFs that play key roles in SE may be expected. In support for this assumption, the activation of the LEC genes (LEC1, LEC2, FUS3) that have essential functions in SE (Gaj et al. 2005) was found to be associated with the overexpression of PLT2 and SE induction (Horstman 2015). The effect of PLT2 overexpression is dose dependent, and its high expression exclusively leads to the formation of a somatic embryo (Horstman 2015). Along with the central role of PLT2 in the embryonic root development during ZE (Horstman et al. 2014), the gene was recently shown to be involved in the formation of the root stem cell niche in the embryogenic callus (Su et al. 2015).
5.1 BBM
BBM/PLT4, the best characterised PLT gene, which was identified in the microspore-derived embryogenesis in B. napus, was found to produce somatic embryos on a hormone-free medium as a result of its overexpression, and relevant to this observation, it was suggested that the encoded TF may stimulate the production of auxin or increase a cell’s sensitivity to this hormone (Boutilier et al. 2002). The identification of BBM-binding sequences during SE in Arabidopsis revealed the targets that are related to the biosynthesis (YUC3, 8, TAA1), transport (PIN1, 4) and signalling (ARF2, 10, IAA2, 7, 28) of auxin. Among the direct targets of BBM during SE, the LEC genes that have documented SE-promoting functions were also proposed, which implies a linkage between the BBM- and LEC-mediated SE pathways (Horstman 2015).
The phenotypes that are related to BBM overexpression were indicated to be dosage- and context dependent, and accordingly, a model of AIL functions has recently been proposed (Horstman 2015). According to this model, SE induction requires a high level of AIL transcripts and the mode of the embryogenic pathway that is triggered depends on the developmental stage of the seedling. Direct SE is induced when BBM is activated before or during seed germination, whereas post-germination activation of the gene leads to the indirect pathway of SE induction. Analysis of BBM targets revealed the gene’s involvement in the positive control of cell division, cell wall modification and the differentiation of plant organs (Passarinho et al. 2008; Nic-Can et al. 2013).
The HD-ZIP IV/HOMEODOMAIN GLABROUS (HDG) TFs, which are expressed in the L1 layer of meristems and specify an epidermis identity, were reported within the potential targets of BBM (Takada et al. 2013). BBM and HDGs are co-expressed during early ZE and their transcripts were found to promote cell divisions and differentiation, respectively (Horstman et al. 2015). The antagonistic functions of these genes are also observed in SE where the downregulation of BBM and the overexpression of HDGs result in a reduced embryogenic response of cultured explants. The overexpression of HDG1 leads to the development of highly differentiated cells along the margin of the cotyledons and leaves due to the downregulation of cell proliferation genes including the D-type cyclin CYCD3;1. In contrast, the cotyledons of 35S::BBM transgenic seedlings consist of small, undifferentiated cells that are able to produce somatic embryos. BBM and HDG1 have common target genes that might be antagonistically regulated or co-regulated, i.e. PLT5 is activated by BBM in contrast to HGD1 (Horstman et al. 2015).
6 WOX Genes
The WOX (WUSCHEL-RELATED HOMEOBOX) genes form a plant-specific subclade of the eukaryotic homeobox TF superfamily whose members display the specialised functions that are related to either the promotion of cell division and/or the prevention of the premature cell differentiation. Accordingly, WOXs repress or activate their targets depending on the cell type and developmental stage (reviewed in van der Graaff et al. 2009). Fifteen members of WOX (WUS and WOX1-14) family were identified in the Arabidopsis genome, but only a subset of these has yet been characterised in detail. The activity of WOX genes that is specific to the tissue and the developmental process was reported. Consequently, in order to maintain stem cells in Arabidopsis, WOX5 has to be expressed in RAM (Sarkar et al. 2007) and WOX4 in the cambial meristem (Hirakawa et al. 2010). WOX2, WOX8 and WOX9 transcripts accumulate in the early stages of ZE in Arabidopsis and P. abies to control the polarity of cell divisions (Ueda et al. 2011; Zhu et al. 2014b). WOX9 regulates cell divisions in SAM and acts upstream of WUS (Wu et al. 2005).
Consistent with the WOX activity in ZE (Hacker et al. 2004), the expression of the WOX family members was also indicated to be associated with somatic embryo development. In an embryogenic culture of P. abies, WOX2, WOX8 and WOX9 were transcribed in the early stage of the somatic embryo and later in the development; the expression of PaWOX2 was visible in the basal part of the developing embryo while the PaWOX8/9 transcripts marked the future RAM and the sites of the initiation of the cotyledon (Palovaara et al. 2010). In accordance with this finding, a reduced expression of WOX8 and WOX9 was found to result in the aberrant development of somatic embryos because of the deregulation of the cell divisions that were related to the downregulation of the PaE2F and PaCYCBL genes that control the cell cycle progress (Zhu et al. 2014b).
6.1 WUS and WOX5 in Control of the Apical–Basal Axis of the Embryo
The WUS gene that encodes the WUSCHEL protein was identified as a positive regulator of the stem cells in the SAM formation through the control of the meristematic cell number (Mayer et al. 1998). Parallel to the activation of floral patterning, the encoded TF was also indicated to repress the stem cell regulation and this bifunctional mode of activity placed the WUS TF among the developmental regulators with unique functions (Ikeda et al. 2009). The role of WUS in the promotion of the vegetative-to-embryogenic transition was uncovered in a culture of an Arabidopsis mutant that produced somatic embryos on root explants that were cultured on a hormone-free medium (Zuo et al. 2002a). Moreover, the WUS overexpression was indicated to compensate the requirement of auxin treatment in SE induction in Capsicum chinense and Coffea canephora (Solís-Ramos et al. 2009; Arroyo-Herrera et al. 2008) and to enhance the embryogenic potential in an embryogenic culture of G. hirsutum (Zheng et al. 2014).
WUS together with WOX5 were found to play a key role in the origin of the apical–basal pattern of the shoot–root axis in the zygotic embryos of Arabidopsis and the establishment of SAM and RAM, respectively (Jürgens 2001; Friml et al. 2003). Both genes were also recently demonstrated to specify the establishment of apical–basal polarity during formation of somatic embryos in Arabidopsis; however, some remarkable differences were noticed in comparison to ZE. In contrast to the distinct spatiotemporal separation of the WUS and WOX5 expression that underlies the formation of the opposite embryo poles in early ZE, WUS and WOX5 were simultaneously activated in nearly overlapped callus cells in the embryogenic culture of Arabidopsis, thus implying that the stem cell niches of the SAM and the RAM are developmentally related during SE initiation (Su et al. 2015).
Expression of WUS in SAM regeneration in vitro is positively affected by auxin or cytokinin depending on the mode of the morphogenic pathway that is induced. Auxin was found to stimulate WUS activity during SE induction (Su et al. 2009) and cytokinin was reported to enhance the gene expression in the regenerating shoots of root-derived cultures (Gordon et al. 2009). Some evidence implies that the mechanism of the WUS-mediated hormonal regulation of SE initiation differs from shoot and root regeneration that is induced separately; however, the genetic interactions that determine this difference need further investigations (Su et al. 2015).
In controlling SAM, WUS directly represses the transcription of the ARABIDOPSIS RESPONSE REGULATOR genes (ARR5, ARR6, ARR7 and ARR15), which act in the negative feedback loop of cytokinin signalling (Leibfried et al. 2005). The differential expression of ARRs that is observed during SE of Arabidopsis (Gliwicka et al. 2013) and M. truncatula (Imin et al. 2008) provides the possibility for the role of cytokinin signalling in this process; however, the components of the WUS-controlled initiation of embryonic SAM during SE remain unknown. Cytokinin signals and WUS were postulated to reinforce each other through multiple feedback loops (Gordon et al. 2009) and a high specificity of these interactions might be expected. The regulatory relation between auxin and WUS might also be assumed and in accordance, auxin treatment was found to be essential for the correct regulation of WUS expression during somatic embryo induction in Arabidopsis (Su et al. 2009). In addition, WUS appears not to interact with the auxin metabolism during SE because the content of IAA was not modulated in response to WUS overexpression in the embryogenic callus of cotton (Bouchabké-Coussa et al. 2013). The understanding of the interactions between the endogenous hormones and WUS expression might contribute to the application of this TF in the genetic improvement of plants with a poor capacity for in vitro regeneration, as was demonstrated in C. chinense (Solís-Ramos et al. 2009).
7 Conclusions
The central role of the transcriptional regulation in the control of the embryogenic transition of somatic cells has been recently documented. However, in contrast to the spectacular progress on the identification of TFs that are decisive for the reprogramming of differentiated cells into totipotent stem cells that has been made in animals, much less is known about these master regulators in plant cells. In the last 10 years intensive analysis of the global transcriptomes of plant cells that are undergoing embryogenic transition and the use of Arabidopsis (a model in plant genomics) in studies on the genetic control of SE have substantially contributed to the identification of the TF regulators of SE. As a result, dozens of TF genes that are differentially expressed in embryogenic cultures have been identified that provide a base in searches for other genetic elements of their decisive roles in SE induction. So far, only a small subset of the potential SE regulators has been verified experimentally. The emerging picture of a TF-controlled process of SE induction shows a complex network of genetic interactions in which the transcriptional regulation of hormone and stress responses appears to play a fundamental role (Fig. 5.2). It is also apparent that the majority of the already identified SE-involved TF genes are also critical for the development of zygotic embryos. Thus, the similarities in the regulatory mechanisms that underlie SE and ZE that were expected from early 1990s have recently become evident at the molecular level.
Interaction between SE-related TFs. Arrows indicate the activation of the genes expression. The blunt end shows inhibition of gene expression. The dotted line indicates the suggested interactions. Full-line frame indicates stress-related genes. Dotted frame indicates hormone-related genes. 1 Wu et al. (2005); 2 Mantiri et al. (2008b); 3 Smith and Long (2010); 4 Nowak et al. (2015); 5 Tang et al. (2012); 6 Horstman (2015); 7 Zhang et al. (2009); 8 To et al. (2006); 9 Stone et al. (2008); 10 Thakare et al. (2008); 11 Gazzarrini et al. (2004); 12 Yamamoto et al. (2010); 13 Kagaya et al. (2005); 14 Mu et al. (2013); 15 Nowak and Gaj (2016); 16 Iwase et al. (2011); 17 Leibfried et al. (2005); 18 Palovaara et al. (2010); 19 Su et al. (2015); 20 Curaba et al. (2004); 21 Sundaram et al. (2013); 22 Wang et al. (2004); 23 Zheng et al. (2013); 24 Pinon et al. (2013); 25 Lumba et al. (2012); 26 Wang and Perry (2013); 27 Junker et al. (2012); 28 Wójcikowska et al. (2013); 29 Guo et al. (2013); 30 Braybrook et al. (2006). SSP—seed storage protein
In addition to ZE-associated regulators, TFs that have less obvious functions in the control of SE induction have been recognised. Among them, TF genes that enhance cell tolerance to stress imposed under in vitro conditions including the activators of storage material accumulation such as LEA proteins were found to be essential for embryogenic transition.
In spite of the apparent progress that has been made, it seems that most of the TF genes that have a decisive role in the reprogramming of somatic cells into embryonic ones still remain uncovered. Special efforts should be focused on the identification of the targets of the SE-involved TF genes that operate at the very early stage of embryogenic transition, which is the most intriguing moment in the reprogramming of somatic cells. In order to recognise the early targets of SE-related TFs, new efficient approaches can be applied including the protein-binding microarray coupled with the analysis of co-regulated genes that was recently recommended for exploring the regulatory networks in plants (Franco-Zorrilla et al. 2014). However, given the fact that many variables determine the cellular and developmental context of TF–DNA interactions (Slattery et al. 2013, 2014), the candidate TF targets that are identified using in vitro approaches should be verified as also acting in vivo during SE induction. To meet this requirement, the versatile molecular tools that are available for functional genomics in Arabidopsis might be helpful. In addition to the dissection of SE-specific TFs and their targets, the recognition of TF regulators, especially the chromatin remodelling factors and miRNAs, is a prerequisite for the full understanding of how already differentiated cells become competent to respond to the embryogenic signal that triggers the developmental switch.
Besides its cognitive value, the efforts that are aimed at the revealing the TF-controlled regulatory network that governs embryonic transition in plants may enable further progress in the genetic improvement of plants (Zuo et al. 2002b). Such perspectives for the use of the TFs that control SE induction in increasing the regeneration potential of some crop species have already been demonstrated for BBM, WUS and LEC2 (Arroyo-Herrera et al. 2008; Deng et al. 2009; Solís-Ramos et al. 2009; Belide et al. 2013; Zheng et al. 2014).
References
Aida M, Beis D, Heidstra R et al (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119:109–120. doi:10.1016/j.cell.2004.09.018
Aker J, de Vries SC (2008) Plasma membrane receptor complexes. Plant Physiol 147:1560–1564. doi:10.1104/pp.108.120501
Alemanno L, Devic M, Niemenak N et al (2008) Characterization of leafy cotyledon1-like during embryogenesis in Theobroma cacao L. Planta 227:853–866. doi:10.1007/s00425-007-0662-4
Arroyo-Herrera A, Gonzalez AK, Moo RC et al (2008) Expression of WUSCHEL in Coffea canephora causes ectopic morphogenesis and increases somatic embryogenesis. Plant Cell Tiss Org 94:171–180. doi:10.1007/s11240-008-9401-1
Bai B, Su YH, Yuan J et al (2013) Induction of somatic embryos in Arabidopsis requires local YUCCA expression mediated by the down-regulation of ethylene biosynthesis. Mol Plant 6(4):1247–1260. doi:10.1093/mp/sss154
Belide S, Zhou XR, Kennedy Y et al (2013) Rapid expression and validation of seed-specific constructs in transgenic LEC2 induced somatic embryos of Brassica napus. Plant Cell Tiss Org 113:543–553. doi:10.1007/s11240-013-0295-1
Belmonte MF, Kirkbride RC, Stone SL et al (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110(5):E435–E444. doi:10.1073/pnas.1222061110
Blilou I, Xu J, Wildwater M et al (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433:39–44. doi:10.1038/nature03184
Bouchabké-Coussa O, Obellianne M, Linderme D et al (2013) Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep 32:675–686. doi:10.1007/s00299-013-1402-9
Boutilier K, Offringa R, Sharma VK et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14:1737–1749. doi:10.1105/tpc.001941
Braybrook SA, Stone SL, Park S et al (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci (USA) 103:3468–3473. doi:10.1073/pnas.0511331103
Braybrook SA, Harada JJ (2008) LECs go crazy in embryo development. Trends Plant Sci 13:624–630. doi:10.1016/j.tplants.2008.09.008
Casson SA, Lindsey K (2006) The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol 42(2):526–541. doi:10.1104/pp.106.080895
Chakrabarty D, Trivedi KP, Shri M et al (2010) Differential transcriptional expression following thidiazuron-induced callus differentiation developmental shifts in rice. Plant Biol 12:46–59. doi:10.1111/j.1438-8677.2009.00213.x
Causier B, Ashworth M, Guo W et al (2012) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 158:423–438. doi:10.1104/pp.111.186999
Che P, Love TM, Frame BR et al (2006) Gene expression patterns during somatic embryo development and germination in maize Hi II callus cultures. Plant Mol Biol 62:1–14. doi:10.1007/s11103-006-9013-2
Chen M, Xuan L, Wang Z et al (2014) TRANSPARENT TESTA8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis. Plant Physiol 165:905–916. doi:10.1104/pp.114.235507
Curaba J, Moritz T, Blervaque R et al (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136:3660–3669. doi:10.1104/pp.104.047266
Deng W, Luo K, Li Z et al (2009) A novel method for induction of plant regeneration via somatic embryogenesis. Plant Sci 177:43–48. doi:10.1016/j.plantsci.2009.03.009
Dodeman VL, Ducreux G, Kreis M (1997) Zygotic versus somatic embryogenesis. J Exp Bot 48:1493–1509. doi:10.1093/jxb/48.8.1493
Dubos C, Stracke R, Grotewold E et al (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581. doi:10.1016/j.tplants.2010.06.005
Elliott RC, Betzner AS, Huttner E et al (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8:155–168. doi:10.1105/tpc.8.2.155
El Ouakfaoui S, Schnell J, Abdeen A et al (2010) Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol Biol 74:313–326. doi:10.1007/s11103-010-9674-8
Fambrini M, Durante C, Cionini G et al (2006) Characterization of LEAFY COTYLEDON1-LIKE gene in Helianthus annuus and its relationship with zygotic and somatic embryogenesis. Dev Genes Evol 216:253–264. doi:10.1007/s00427-005-0050-7
Fehér A, Pasternak TP, Dudits D (2003) Transition of somatic plant cells to an embryogenic state. Plant Cell Tiss Org 74:201–228. doi:10.1023/A:1024033216561
Fehér A (2015) Somatic embryogenesis—stress-induced remodeling of plant cell fate. Biochim Biophys Acta 1849:385–402. doi:10.1016/j.bbagrm.2014.07.005
Fernandez DE, Wang CT, Zheng Y et al (2015) The MADS-domain factors AGL15 and AGL18, along with SVP and AGL24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol 165(4):1591–1603. doi:10.1104/pp.114.242990
Franco-Zorrilla JM, López-Vidriero I, Carrasco JL et al (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci (USA) 111(6):2367–2372. doi:10.1073/pnas.1316278111
Friml J, Vieten A, Sauer M et al (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426:147–153. doi:10.1038/nature02085
Gaj MD (2004) Factors influencing somatic embryogenesis induction and plant regeneration with particular reference to Arabidopsis thaliana (L.) Heynh. Plant Growth Regul 43:27–47. doi:10.1023/B:GROW.0000038275.29262.fb
Gaj MD, Zhang S, Harada JJ et al (2005) LEAFY COTYLEDON genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222:977–988. doi:10.1007/s00425-005-0041-y
Gaj MD, Trojanowska A, Ujczak A et al (2006) Hormone-response mutants of Arabidopsis thaliana (L.) Heynh. impaired in somatic embryogenesis. Plant Growth Regul 49:189–197. doi:10.1007/s10725-006-9104-8
Galinha C, Hofhuis H, Luijten M et al (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449:1053–1057. doi:10.1038/nature06206
Gao MJ, Lydiate DJ, Li X et al (2009) Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21:54–71. doi:10.1105/tpc.108.061309
Garcês HM, Champagne CE, Townsley BT et al (2007) Evolution of asexual reproduction in leaves of the genus Kalanchoë. Proc Natl Acad Sci (USA) 104:15578–15583. doi:10.1073/pnas.0704105104
Gazzarrini S, Tsuchiya Y, Lumba S et al (2004) The transcription factor FUSCA3 controls development timing in Arabidopsis through the hormones gibberellin and abscisic acid. Develop Cell 7:373–385. doi:10.1016/j.devcel.2004.06.017
Gliwicka M, Nowak K, Cieśla E et al (2012) Expression of seed storage product genes (CRA1 and OLEO4) in embryogenic cultures of somatic tissues of Arabidopsis. Plant Cell Tiss Org 109:235–245. doi:10.1007/s11240-011-0089-2
Gliwicka M, Nowak K, Balazadeh S et al (2013) Extensive modulation of the transcription factor transcriptome during somatic embryogenesis in Arabidopsis thaliana. PLoS ONE 8(7):e69261. doi:10.1371/journal.pone.0069261
Go YS, Kim H, Kim HJ et al (2014) Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 26:1666–1680. doi:10.1105/tpc.114.123307
Gordon SP, Chickarmane VS, Ohno C et al (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci (USA) 106:16529–16534. doi:10.1073/pnas.0908122106
Guo F, Liu C, Xia H et al (2013) Induced expression of AtLEC1 and AtLEC2 differentially promotes somatic embryogenesis in transgenic tobacco plants. PLoS ONE 8:e71714. doi:10.1371/journal.pone.0071714
Habets MEJ, Offringa R (2014) PIN-driven polar auxin transport in plant developmental plasticity: a key target for environmental and endogenous signals. New Phytol 203:362–377. doi:10.1111/nph.12831
Hacker A, Groß-Hardt R, Geiges B et al (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131:657–668. doi:10.1242/dev.00963
Harada JJ (2001) Role of Arabidopsis LEAFY COTYLEDON genes in seed development. J Plant Physiol 158:405–409. doi:10.1078/0176-1617-00351
Harding EW, Tang W, Nichols KW et al (2003) Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol 133:653–663. doi:10.1104/pp.103.023499
He C, Huang H, Xu L (2013) Mechanisms guiding Polycomb activities during gene silencing in Arabidopsis thaliana. Front Plant Sci 4:454. doi:10.3389/fpls.2013.00454
Hecht V, Vielle-Calzada JP, Hartog MV et al (2001) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127:803–816. doi:10.1104/pp.010324
Hill K, Wang H, Perry SE (2008) A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deactylase complex components. Plant J 53:172–185. doi:10.1111/j.1365-313X.2007.03336.x
Hirakawa Y, Kondo Y, Fukuda H (2010) TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22:2618–2629. doi:10.1105/tpc.110.076083
Horstman A, Willemsen V, Boutilier K et al (2014) AINTEGUMENTA-LIKE proteins: hubs in a plethora of networks. Trends Plant Sci 19:146–157. doi:10.1016/j.tplants.2013.10.010
Horstman A (2015) BABBY BOOM-induced somatic embryogenesis in Arabidopsis. Dissertation, Wegeningen Univeristy
Horstman A, Fukuoka H, Muino JM et al (2015) AIL and HDG proteins act antagonistically to control cell proliferation. Development 142:454–464. doi:10.1242/dev.117168
Hosp J, Maraschin SDF, Touraev A et al (2007) Functional genomics of microspore embryogenesis. Euphytica 158:275–285. doi:10.1007/s10681-006-9238-9
Ikeda M, Umehara M, Kamada H (2006) Embryogenesis-related genes; its expression and roles during somatic and zygotic embryogenesis in carrot and Arabidopsis. Plant Biotechnol 23:153–161. doi:http://doi.org/10.5511/plantbiotechnology.23.153
Ikeda M, Mitsuda N, Ohme-Takagi M (2009) Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21:3493–3505. doi:10.1105/tpc.109.069997
Imin N, Goffard N, Nizamidin M et al (2008) Genome-wide transcriptional analysis of super-embryogenic Medicago truncatula explant cultures. BMC Plant Biol 8:110–124. doi:10.1186/1471-2229-8-110
Iwase A, Mitsuda N, Koyama T et al (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21:508–514. doi:10.1016/j.cub.2011.02.020
Jia H, Suzuki M, McCarty DR (2014) Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. WIREs Dev Biol 3:135–145. doi:10.1002/wdev.126
Jiménez VM (2005) Involvement of plant hormones and plant growth regulators on in vitro somatic embryogenesis. Plant Growth Regul 47:91–110. doi:10.1007/s10725-005-3478-x
Jin F, Hu L, Yuan D et al (2014) Comparative transcriptome analysis between somatic embryos (SEs) and zygotic embryos in cotton: evidence for stress response functions in SE development. Plant Biotechnol J 12:161–173. doi:10.1111/pbi.12123
Junker A, Monke G, Rutten T et al (2012) Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J 71:427–442. doi:10.1111/j.1365-313X.2012.04999.x
Junker A, Bäumlein H (2012) Multifunctionality of the LEC1 transcription factor during plant development. Plant Signal Behav 7:1718–1720. doi:10.4161/psb.22365
Jürgens G (2001) Apical-basal pattern formation in Arabidopsis embryogenesis. EMBO J 20:3609–3616. doi:10.1093/emboj/20.14.3609
Kagaya Y, Toyoshima R, Okuda R et al (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46:399–406. doi:10.1093/pcp/pci048
Kamada H, Ishikawa K, Saga H et al (1993) Induction of somatic embryogenesis in carrot by osmotic stress. Plant Tiss Cult Lett 10:38–44. doi:http://doi.org/10.5511/plantbiotechnology1984.10.38
Karami O, Saidi A (2010) The molecular basis for stress-induced acquisition of somatic embryogenesis. Mol Biol Rep 37:2493–2507. doi:10.1007/s11033-009-9764-3
Karlova R, Boeren S, Russinova E et al (2006) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18:626–638. doi:10.1105/tpc.105.039412
Klucher KM, Chow H, Reiser L et al (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8:137–153. doi:10.1105/tpc.8.2.137
Ledwoń A, Gaj MD (2009) LEAFY COTYLEDON2 gene expression and auxin treatment in relation to embryogenic capacity of Arabidopsis somatic cells. Plant Cell Rep 28:1677–1688. doi:10.1007/s00299-009-0767-2
Ledwoń A, Gaj MD (2011) LEAFY COTYLEDON1, FUSCA3 expression and auxin treatment in relation to somatic embryogenesis induction in Arabidopsis. Plant Growth Regul 65:157–167. doi:10.1007/s10725-011-9585-y
Legrand S, Hendricks T, Gilbert JL et al (2007) Characterization of expressed sequence tags obtained by SSH during somatic embryogenesis in Cichorium intybus L. BMC Plant Biol 7:27. doi:10.1186/1471-2229-7-27
Leibfried A, To JP, Busch W et al (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438:1172–1175. doi:10.1038/nature04270
Lotan T, Ohto M, Yee KM et al (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93:1195–1205. doi:10.1016/S0092-8674(00)81463-4
Low ET, Alias H, Boon SH et al (2008) Oil palm (Elaeis guineensis Jacq.) tissue culture ESTs: Identifying genes associated with callogenesis and embryogenesis. BMC Plant Biol 8:62. doi:10.1186/1471-2229-8-62
Lumba S, Tsuchiya Y, Delmas F et al (2012) The embryonic leaf identity gene FUSCA3 regulates vegetative phase transitions by negatively modulating ethylene-regulated gene expression in Arabidopsis. BMC Biol 10:8. doi:10.1186/1741-7007-10-8
Mantiri FR, Kurdyukov S, Lohar DP et al (2008a) The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiol 146:1622–1636. doi:10.1104/pp.107.110379
Mantiri FR, Kurdyukov S, Chen SK et al (2008b) The transcription factor MtSERF1 may function as a nexus between stress and development in somatic embryogenesis in Medicago truncatula. Plant Signal Behav 3:498–500. doi:10.4161/psb.3.7.6049
Mayer KFX, Schoof H, Haecker A et al (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95:805–815. doi:10.1016/S0092-8674(00)81703-1
Möller B, Weijers D (2009) Auxin control of embryo patterning. Cold Spring Harb Perspect Biol 1:a001545. doi:10.1101/cshperspect.a001545
Mu J, Tan H, Zheng Q et al (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148:1042–1054. doi:10.1104/pp.108.126342
Mu J, Tan H, Hong S et al (2013) Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant 6:188–201. doi:10.1093/mp/sss061
Muday GK, Rahman A, Binder BM (2012) Auxin and ethylene: collaborators or competitors? Trends Plant Sci 17:181–195. doi:10.1016/j.tplants.2012.02.001
Nakano T, Suzuki K, Fujimura T et al (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432. doi:10.1104/pp.105.073783
Nic-Can GI, Lopez-Torres A, Barredo-Pool F et al (2013) New insights into somatic embryogenesis: LEAFY COTYLEDON1, BABY BOOM1 and WUSCHEL-RELATED HOMEOBOX4 are epigenetically regulated in Coffea canephora. PLoS ONE 8:e72160. doi:10.1371/journal.pone.0072160
Nowak K, Wójcikowska B, Gaj MD (2015) ERF022 impacts the induction of somatic embryogenesis in Arabidopsis through the ethylene-related pathway. Planta 241:967–985. doi:10.1007/s00425-014-2225-9
Nowak K, Gaj MD (2016) Stress-related function of bHLH109 in somatic embryo induction in Arabidopsis. J Plant Physiol 193:119–126. doi:10.1016/j.jplph.2016.02.012
Palovaara J, Hallberg H, Stasolla C et al (2010) Comparative expression pattern analysis of WUSCHEL related homeobox 2 (WOX2) and WOX8⁄9 in developing seeds and somatic embryos of the gymnosperm Picea abies. New Phytol 188:122–135. doi:10.1111/j.1469-8137.2010.03336.x
Passarinho P, Ketelaar T, Xing M et al (2008) BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol Biol 68:225–237. doi:10.1007/s11103-008-9364-y
Patnaik D, Mahalakshmi A, Khurana P (2005) Effect of water stress and heavy metals on induction of somatic embryogenesis in wheat leaf base cultures. Ind J Exp Biol 43:740–745
Perry SE, Lehti MD, Fernandez DE (1999) The MADS-domain protein AGAMOUS-like 15 accumulates in embryonic tissues with diverse origins. Plant Physiol 120:121–129. doi:10.1104/pp.120.1.121
Pinon V, Prasada K, Grigga SP et al (2013) Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc Natl Acad Sci (USA) 110:1107–1112. doi:10.1073/pnas.1213497110
Piyatrakul P, Putranto RA, Martin F et al (2012) Some ethylene biosynthesis and AP2/ERF genes reveal a specific pattern of expression during somatic embryogenesis in Hevea brasiliensis. BMC Plant Biol 12:244–256. doi:10.1186/1471-2229-12-244
Rademacher EH, Möller B, Lokerse AS et al (2011) A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J 68:597–606. doi:10.1111/j.1365-313X.2011.04710.x
Rademacher EH, Lokerse AS, Schlereth A et al (2012) Different auxin response machineries control distinct cell fates in the early plant embryo. Develop Cell 22:211–222. doi:10.1016/j.devcel.2011.10.026
Riechmann JL, Heard J, Martin G et al (2000) Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290:2105–2110. doi:10.1126/science.290.5499.2105
Rensing SA, Lang D, Schumann E et al (2005) EST sequencing from embryogenic Cyclamen persicum cell cultures identifies a high proportion of transcripts homologous to plant genes involved in somatic embryogenesis. J Plant Growth Regul 24:102–115. doi:10.1007/s00344-005-0033-y
Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell 14:S185-S205. doi:10.1105/tpc.010455
Sarkar AK, Luijten M, Miyashima S et al (2007) Conserved factors regulate signaling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446:811–814. doi:10.1038/nature05703
Sato A, Yamamoto KT (2008) Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol Plant 133:397–405. doi:10.1111/j.1399-3054.2008.01055.x
Serivichyaswat P, Ryu HS, Kim W et al (2015) Expression of the floral repressor miRNA156 is positively regulated by the AGAMOUS-like proteins AGL15 and AGL18. Mol Cells 38:259–266. doi:10.14348/molcells.2015.2311
Sharma SK, Millam S, Hedley PE et al (2008) Molecular regulation of somatic embryogenesis in potato: an auxin led perspective. Plant Mol Biol 68:185–201. doi:10.1007/s11103-008-9360-2
Singla B, Akhilesh K, Jitendra PT et al (2007) Analysis of expression profile of selected genes expressed during auxin-induced somatic embryogenesis in leaf base system of wheat (Triticum aestivum) and their possible interactions. Plant Mol Biol 65:677–692. doi:10.1007/s11103-007-9234-z
Slattery M, Voutev R, Ma L et al (2013) Divergent transcriptional regulatory logic at the intersection of tissue growth and developmental patterning. PLoS Genet 9:e1003753. doi:10.1371/journal.pgen.1003753
Slattery M, Zhou T, Yang L et al (2014) Absence of a simple code: how transcription factors read the genome. Trends Biochem Sci 39:381–399. doi:10.1016/j.tibs.2014.07.002
Smaczniak C, Immink RGH, Angenent GC et al (2012) Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139:3081–3098. doi:10.1242/dev.074674
Smith ZR, Long JA (2010) Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464:423–426. doi:10.1038/nature08843
Solís-Ramos LY, González-Estrada T, Nahuath-Dzib S et al (2009) Overexpression of WUSCHEL in Capsicum chinense causes ectopic morphogenesis. Plant Cell Tiss Org 96:279–287. doi:10.1007/s11240-008-9485-7
Stepanova AN, Yun J, Likhacheva AV et al (2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19:2169–2185. doi:10.1105/tpc.107.052068
Stone SL, Kwong LW, Yee KM et al (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci (USA) 98:11806–11811. doi:10.1073/pnas.201413498
Stone SL, Braybrook SA, Paula SL et al (2008) Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc Natl Acad Sci (USA) 105:3151–3156. doi:10.1073/pnas.0712364105
Su N, He K, Jiao Y et al (2007) Distinct reorganization of the genome transcription associates with organogenesis of somatic embryo, shoots, and roots in rice. Plant Mol Biol 63:337–349. doi:10.1007/s11103-006-9092-0
Su YH, Zhao XY, Liu YB et al (2009) Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J 59:448–460. doi:10.1111/j.1365-313X.2009.03880.x
Su YH, Liu YB, Bai B et al (2015) Establishment of embryonic shoot–root axis is involved in auxin and cytokinin response during Arabidopsis somatic embryogenesis. Front Plant Sci Plant Evol Develop 5:792. doi:10.3389/fpls.2014.00792
Sundaram S, Kertbundit S, Shakirov EV et al (2013) Gene networks and chromatin and transcriptional regulation of the phaseolin promoter in Arabidopsis. Plant Cell 25:2601–2617. doi:10.1105/tpc.113.112714
Takada S, Takada N, Yoshida A (2013) ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development 140:1919–1923. doi:10.1242/dev.094417
Tang X, Bian S, Tang M et al (2012) MicroRNA–mediated repression of the seed maturation program during vegetative development in Arabidopsis. PLoS Genet 8:e1003091. doi:10.1371/journal.pgen.1003091
Thakare D, Tang W, Hill K et al (2008) The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiol 146:1663–1672. doi:10.1104/pp.108.115832
Thibaud-Nissen F, Shealy RT, Khanna A et al (2003) Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol 132:118–136. doi:10.1104/pp.103.019968
To A, Valon C, Savino G et al (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18:1642–1651. doi:10.1105/tpc.105.039925
Tsuwamoto R, Pokoi S, Takahata Y (2010) Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase. Plant Mol Biol 73:481–492. doi:10.1007/s11103-010-9634-3
Tzafrir I, Pena-Muralla R, Dickerman A et al (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135:1206–1220. doi:10.1104/pp.104.045179
Ueda M, Zhang Z, Laux T (2011) Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Develop Cell 20:264–270. doi:10.1016/j.devcel.2011.01.009
van der Graaff E, Laux T, Rensing SA (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10:248–259. doi:10.1186/gb-2009-10-12-248
Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136:1005–1016. doi:10.1016/j.cell.2009.03.001
Venkatesh K, Rani AR, Baburao N et al (2009) Effect of auxins and auxin polar transport inhibitor (TIBA) on somatic embryogenesis in groundnut (Arachis hypogaea L.). Afr J Plant Sci 3:288–293
Wang H, Caruso LV, Downie AB et al (2004) The embryo MADS domain protein AGAMOUS-Like15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 16:1206–1219. doi:10.1105/tpc.021261
Wang XC, Niu QW, Teng C et al (2008) Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res 19:224–235. doi:10.1038/cr.2008.276
Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161:1251–1264. doi:10.1104/pp.112.212282
Wenzel CL, Schuetz M, Yu Q et al (2007) Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J 49:387–398. doi:10.1111/j.1365-313X.2006.02977.x
West AG, Shore P, Sharrocks AD (1997) DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol Cell Biol 17:2876–2887. doi:10.1128/MCB.17.5.2876
Wickramasuriya AM, Dunwell JM (2015) Global scale transcriptome analysis of Arabidopsis embryogenesis in vitro. BMC Genom 16:301. doi:10.1186/s12864-015-1504-6
Wiśniewska A, Grabowska A, Pietraszewska-Bogiel A et al (2012) Identification of genes up-regulated during somatic embryogenesis of cucumber. Plant Physiol Biochem 50:54–64. doi:10.1016/j.plaphy.2011.09.017
Wójcikowska B, Jaskóła K, Gąsiorek P et al (2013) LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissue of Arabidopsis via YUCCA-mediated auxin biosynthesis. Planta 238:425–440. doi:10.1007/s00425-013-1892-2
Wójcikowska B, Gaj MD (2015) LEAFY COTYLEDON2-mediated control of the endogenous hormone content: implications for the induction of somatic embryogenesis in Arabidopsis. Plant Cell Tiss Org 121:255–258. doi:10.1007/s11240-014-0689-8
Wu XL, Dabi T, Weigel D (2005) Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr Biol 15:436–440. doi:10.1016/j.cub.2004.12.079
Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133:3539–3547. doi:10.1242/dev.02521
Wu X, Li F, Zhang C et al (2009) Differential gene expression of cotton cultivar CCRI24 during somatic embryogenesis. J Plant Physiol 166:1275–1283. doi:10.1016/j.jplph.2009.01.012
Yamamoto A, Kagaya Y, Usui H et al (2010) Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol 51:2031–2046. doi:10.1093/pcp/pcq162
Yang X, Zhang X, Yuan D et al (2012) Transcript profiling reveals complex auxin signaling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol 12:110. doi:10.1186/1471-2229-12-110
Yazawa K, Takahata K, Kamada H (2004) Isolation of the gene encoding Carrot leafy cotyledon1 and expression analysis during somatic and zygotic embryogenesis. Plant Physiol Biochem 42:215–223. doi:10.1016/j.plaphy.2003.12.003
Zavattieri MA, Frederico AM, Lima M et al (2010) Induction of somatic embryogenesis as an example of stress-related plant reactions. Elect J Biotechnol 13:1–9. doi:10.2225/vol13-issue1-fulltext-4
Zhang Y, Cao G, Qu JL et al (2009) Involvement of an R2R3-MYB transcription factor gene AtMYB118 in embryogenesis in Arabidopsis. Plant Cell Rep 28:337–346. doi:10.1007/s00299-008-0644-4
Zhang Y, Clemens A, Maximova SN et al (2014) The Theobroma cacao B3 domain transcription factor TcLEC2 plays a dual role in control of embryo development and maturation. BMC Plant Biol 14:106. doi:10.1186/1471-2229-14-106
Zhang Y, Li B, Huai D et al (2015) The conserved transcription factors, MYB115 and MYB118, control expression of the newly evolved benzoyloxy glucosinolate pathway in Arabidopsis thaliana. Front Plant Sci 6:1–19. doi:10.3389/fpls.2015.00343
Zhao Y, Christensen SK, Fankhauser C et al (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291:306–309. doi:10.1126/science.291.5502.306
Zhao B, Ge L, Liang R et al (2009) Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol 10:29–41. doi:10.1186/1471-2199-10-29
Zheng Q, Zheng Y, Perry SE (2013) AGAMOUS-Like15 promotes somatic embryogenesis in Arabidopsis and soybean in part by the control of ethylene biosynthesis and response. Plant Physiol 161:2113–2127. doi:10.1104/pp.113.216275
Zheng W, Zhang X, Yang Z et al (2014) AtWuschel promotes formation of the embryogenic callus in Gossypium hirsutum. PLoS ONE 9:e87502. doi:10.1371/journal.pone.0087502
Zheng Q, Perry SE (2014) Alterations in the transcriptome of soybean in response to enhanced somatic embryogenesis promoted by orthologs of AGAMOUS-Like15 and AGAMOUS-Like18. Plant Physiol 164:1365–1377. doi:10.1104/pp.113.234062
Zhu S, Wang J, Ye J et al (2014a) Isolation and characterization of LEAFY COTYLEDON 1-LIKE gene related to embryogenic competence in Citrus sinensis. Plant Cell Tiss Org 119:1–13. doi:10.1007/s11240-014-0509-1
Zhu T, Moschou PN, Alvarez JM et al (2014b) WUSCHEL-RELATED HOMEOBOX 8/9 is important for proper embryo patterning in the gymnosperm Norway spruce. J Exp Bot 65(22):6543–6552. doi:10.1093/jxb/eru371
Zuo J, Niu QW, Frugis G et al (2002a) The WUSCHEL gene promotes vegetative- to -embryonic transition in Arabidopsis. Plant J 30:349–359. doi:10.1046/j.1365-313X.2002.01289.x
Zuo J, Niu QW, Ikeda Y et al (2002b) Marker-free transformation: increasing transformation frequency by the use of regeneration-promoting genes. Curr Opin Biotech 13:173–180. doi:10.1016/S0958-1669(02)00301-4
Acknowledgements
This work was supported by a research grant of the National Centre for Science in Poland (Grant N 2013/09/B//NZ2/03233)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Nowak, K., Gaj, M.D. (2016). Transcription Factors in the Regulation of Somatic Embryogenesis. In: Loyola-Vargas, V., Ochoa-Alejo, N. (eds) Somatic Embryogenesis: Fundamental Aspects and Applications. Springer, Cham. https://doi.org/10.1007/978-3-319-33705-0_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-33705-0_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-33704-3
Online ISBN: 978-3-319-33705-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)