Abstract
In an environment consisting of harmful microorganisms, survival of plants mainly depends on efficient microbial recognition and rapid defense mechanisms. After infection with a necrotizing pathogen, many plants develop resistance against attack by phytopathogens. This resistance is regarded as systemic acquired resistance, which is a key portion of plant defense against pathogen infection. Induction of acquired resistance in plants occurs mainly by enhancement of the levels of pathogenesis-related proteins and salicylic acid. Some groups of plant-growth-promoting rhizobacteria are involved in an indirect mechanism, either by their antagonistic effect against phytopathogens or by induced systemic resistance (ISR) mechanisms in plants. ISR has been studied with respect to the underlying signaling pathways and to its application in crop protection. The signaling pathway regulating ISR functions independently of salicylic acid, and is mainly dependent on the plant hormones jasmonic acid and ethylene. Apart from these, NPR1, a defensive regulatory protein, is also involved in both systemic and acquired resistance in plants. In this chapter, the molecular and genetic relationship between basal resistance and induced resistance is highlighted.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Plant diseases have always been major problems. Various abiotic and biotic factors affect plant productivity worldwide. Biotic factors such as viroids and higher organisms are plant pathogen parasites that cause diseases. Plant disease resistance is a prerequisite for modern agriculture; the dynamics of plant–microbe interaction has an immense and positive effect worldwide for the management and control of plant diseases. Since the twentieth century, it has been reported that plants have an innate ability to combat infection, recover from diseases, and evade future infections (Chester 1933). Colonization on host plants and thereby utilization of the reserves of plant stores are the features of phytopathogens. Detection of pathogens and the defensive response of the plant is in the form of secretion of antimicrobial compounds and other stress responses. The abilities of a pathogen to induce a disease in a host plant is usually the exception as plants are capable of recognizing the invading pathogens and establishing a successful defense mechanism. In contrast, some pathogens can successfully produce diseases in plants as they are able to evade suppression of the host defense mechanism (Borrás-Hidalgo 2004). Plant–pathogen interaction results either in a disease condition in the host plant or in a resistance mechanism which prevents the spread and multiplication of the nonhost pathogen. Plants depend on an innate immunity to defend themselves. Innate immunity includes two interrelated branches: pathogen-associated molecular pattern (PAMP)/microbe-associated molecular pattern (MAMP)-triggered immunity and effector-triggered immunity (Euglem and Somssich 2007). PAMPs/MAMPs are identified by host cell-surface-localized pattern recognition receptors and activate plant immunity. PAMP-triggered immunity restricts pathogen proliferation. Since the last decade, plant–pathogen association has been known to lead to the development of multiple mechanisms of surveillance in plants (Zhang and Zhou 2010). By delivering virulence effector proteins into host cells, pathogens adapt themselves into the host plant on inhibiting PAMP-triggered immunity (Abramovitch et al. 2006). To overcome this, plants developed immune receptors known as resistance proteins, which either directly or indirectly detect pathogen-specific effector protein activities inside the plant cell and trigger disease resistance, which results in effector-triggered immunity, which is very specific and mostly leads to hypersensitive responses (Tsuda et al. 2009).
There are mainly two possible kinds of plant resistance mechanisms: active and passive. The active resistance mechanism of the plant depends on the defense mechanism induced only after an attack by a pathogen, whereas the passive resistance mechanism relies only on constitutively expressed defenses. Active defense against an incompatible pathogen is in the form of induced resistance that is categorized by a highly localized defense expression such as the hypersensitive response and phytoalexins (Hammerschmidt and Nicholson 1999). Induced resistance, depending on the mode of expression, can be of two types: local and systemic. Local induced resistance means resistance is induced only in the specific tissue where the attack by the pathogen occurs, whereas systemic induced resistance occurs in a part of a plant that is spatially separated by an induction point (Hammerschmidt 1999). Local systemic resistance involves induction of certain pathogenesis-related (PR) proteins to stop the proliferation of the challenging pathogen; in the case of systemic resistance, the induction of cells away from the induction site occurs, which permits the cells to defend themselves against the challenge by what is called “priming” (Conrath et al. 2002). On the basis of the type of inducing agent and the host signaling pathways, induced resistance is characterized into two forms: systemic acquired resistance (SAR) and induced systemic resistance (ISR) (van Loon et al. 1998). SAR develops subsequent to a localized necrosis, and is dependent on salicylic acid (SA) signaling and on the expression of PR proteins. ISR develops systemically because of plant root colonization by plant-growth-promoting rhizobacteria (PGPR) and plant-growth-promoting fungi. Moreover, besides induced resistance in plants, rhizobacteria are also known to be involved in an indirect mechanism by acting as biocontrol agents (Akhtar and Siddiqui 2010; Glick 2012). Biocontrol activity include nutrient competition, exclusion of niches, and production of antifungal metabolites (lytic enzymes) as chief modes of the mechanism (Lugtenberg and Kamilova 2009) against phytopathogens.
2 Indirect Mechanisms
2.1 Role of PGPR in Suppression of Disease Caused by Phytopathogens
There are several biocontrol methods for the control of soilborne phytopathogens and plant diseases. These methods involve an attempt either to increase soil antagonist activity with pathogens (Gamliel et al. 2000) or to protect plants by the use of bioinoculants (Compant et al. 2005; Akhtar and Siddiqui 2009; Akhtar et al. 2010). Increasing the soil fertility may also reduce the efficacy of bioinoculants, whose niches may be reduced (Hoitink and Boehm 1999). The mechanisms by which PGPR control the damage to plants resulting from pathogen invasion include siderophore secretion, physical displacement, and production of antibiotics, enzymes, and a variety of molecules that inhibit phytopathogen growth (Niranjan Raj et al. 2006). One of the major mechanisms that can control the proliferation of phytopathogens is the production of siderophores with a very much higher affinity for iron than fungal pathogens (Siddiqui et al. 2007; Sayyed and Chincholkar 2009; Sayyed and Patel 2011; Glick 2012; Sayyed et al. 2013; Shaikh et al. 2014; Shaikh and Sayyed 2015). Another effective mechanism is the production of antibiotics, which are deleterious to the metabolism or growth of other pathogens (Doornbos et al. 2012). A large number of antibiotics have been identified, produced by members of Pseudomonads, Bacillus, Stenotrophomonas, and Streptomyces (Compant et al. 2005).
Soilborne microorganisms are capable of producing extracellular enzymes such as cellulases chitinases, lipases, β-1-3-glucanases, and proteases, thereby hydrolyzing a wide variety of polymeric compounds, including proteins, chitin, hemicelluloses, and cellulose, which hinders the growth of pathogens (Markovich and Kononova 2003). These enzymes together with antibiotics play an important role in the defense against phytopathogenic fungi as a antagonistic effect (Fogliano et al. 2002). PGPR that produce such enzymes have been shown to have a biocontrol effect against fungi, including Sclerotium rolfsii, Botrytis cinerea, Phytophthora spp., Fusarium oxysporum, Pythium ultimum, and Rhizoctonia solani (Glick 2012).
2.2 Systemic Acquired Resistance
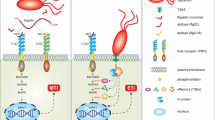
On primary invasion by pathogens in plants, tissue necrosis at the site of infection by pathogens is activated by SAR (Ryals et al. 1996). SAR is known to be associated with a PR gene expression which results in accumulation of PR proteins involved in antimicrobial action and thereby induces resistance. Expression of the PR-1 protein is a molecular marker for SAR induction. The PR-1 proteins are usually appear as a result of SA accumulation (van Loon et al. 2006). It was demonstrated that a transgenic plant expressing the bacterial salicylate hydroxylase gene (nahG) was incapable of accumulating SA. Plants expressing nahG do not show an SAR response as they convert SA to inactive catechol (Lawton et al. 1996). Likewise, the SA-production-deficient mutants sid1 and sid2 do not show SAR after infection with a necrotizing pathogen, which indicates that SA is necessary and sufficient for the induction of SAR (Verhagen et al. 2006). However, SA action requires the protein NPR1, also known as “NIM1,” an ankyrin repeat family protein structurally (Cao et al. 1997). In the presence of SA, oligomers of NPR1 in the cytoplasm are reduced to monomers by redox reactions and interact with specific TGA transcription factors for the expression of gene codings for PRs (Dong 2004). NPR1 is a master regulatory protein identified through genetic screens for SAR-compromised mutants in Arabidopsis thaliana (Dong 2004; Pieterse and van Loon 2004). It significantly increases the binding of TGA2 to SA promoter elements in the Arabidopsis PR-1 gene (Despres et al. 2000). Subramaniam et al. (2001) showed interactions between NPR1 and TGA2 by using a protein fragment complementation assay in vivo, and demonstrated that the SA-induced interaction is strictly localized in the nucleus. The steps involved in SAR signaling are shown in Fig. 1.
SAR signaling induced by Phytopathogens in plants (modified from Pieterse and Van Loon 2004)
2.3 Induced Systemic Resistance
In the last three decades, various reports have confirmed a beneficial effect of root-colonizing bacteria such as PGPR on plant development and disease resistance (Kloepper et al. 1980). A specific recognition factor between the plant and the systemic-resistance-inducing rhizobacteria is needed for the induction of resistance. For instance, Pseudomonas fluorescens and Pseudomonas putida perform differently on different host species, as Arabidopsis responds to P. putida, whereas carnation and radish do not (van Wees et al. 1997). Conversely, radish is responsive to P. fluorescens, whereas Arabidopsis is not (Leeman et al. 1995a). Plant-growth-promoting activities of PGPR can directly affect plant growth but are mostly related to a biocontrol activity of soil microorganisms which can be depend on a few mechanisms, including nutrient competition, siderophore-mediated competition for iron, and antibiotic production (Patel et al. 2012). Van Peer et al. (1991) demonstrated that PGPR also reduce pathogen infections on aboveground parts of plants such as stems and leaves. Some biochemical compounds of PGPR affect the complementary receptors on the plant surface for the successful elicitation of systemic resistance. Root colonization of ISR-triggering bacteria results in a discriminating level of resistance against a wide range of pathogens, and no defense mechanisms are frequently triggered in aboveground plant tissues on the recognition of the resistance-inducing signal. The phenomenon of expressing the basal defense responses faster on pathogen attack on the tissues is known as “priming” (Conrath et al. 2002). Priming shows efficient resistance strategies that assist the plant to efficiently react to any invader by enhancing infection-induced cellular defense responses (Beckers and Conrath 2007). Pieterse et al. (2000) demonstrated that in Arabidopsis, P. fluorescens WCS417r-mediated ISR functions require components of the jasmonic acid (JA) and ethylene response pathways but not SA. Like SAR, P. fluorescens WCS417r-mediated ISR relies on NPR1.
It is well known that only a few PGPR strains trigger ISR by ethylene-, JA-, and NPR1-dependent pathways. Rhizobacteria-mediated systemic resistance is actively efficient against a vast range of fungal phytopathogens in many plant species (van Loon et al. 1998). Certain bacterial-derived compounds have been implicated in elicitation of ISR (van Loon and Bakker 2006). Bacterial cell wall determinants such as flagella and lipopolysaccharides (LPS), secondary metabolites such as siderophores and antibiotics (Bakker et al. 2003; Iavicoli et al. 2003), and a bacterial flagellin receptor have been recognized as bacterial elicitors (Gómez-Gómez and Boller 2000). The prominent homologies with recognition mechanisms for PAMPs in the innate immune response of plants demonstrated that rhizobacteria are recognized as general immunity mechanisms (Nurnberger et al. 2004). The important steps involved in the ISR mechanisms are summarized in Fig. 2.
ISR signaling mediated by Rhizobacteria in plants (adapted from Pieterse et al. 2001)
3 Signaling in Rhizobacteria-Mediated ISR
3.1 SA-Independent Signaling
It has been found that there is no connection between resistance induced by P. fluorescens WCS417r (the Dutch reference strain) and accumulation of messenger RNA of certain PR genes ensures SAR and SA action (van Wees et al. 1997). It is known that ISR is not related to changes in endogenous SA content (Pieterse et al. 2000). ISR mediated by P. fluorescens WCS417 in SA-nonaccumulating Arabidopsis nahG transformants (van Wees et al. 1997) is an independent mechanism and is regulated by signaling pathways different from those for pathogen-induced SAR. The Arabidopsis ethylene mutant etr1 and JA mutant jar1 were studied for their capacity to mount ISR. Both root-colonizing mutant strains of P. fluorescens WCS417r failed to induce systemic resistance against P. syringae pv. tomato (Pieterse et al. 1998), showing ISR signaling is dependent on these phytohormones. Certain well-identified ethylene-signaling mutants do not show enhanced resistance on root treatment with P. fluorescens WCS417r against P. syringae pv. tomato (Knoester et al. 1999), indicating that an ethylene signaling response is necessary for the development of ISR, whereas on leaf infiltration by P. fluorescens WCS417r, systemic resistance was observed, indicating ethylene is required for rhizobacterial induction (Knoester et al. 1999). Moreover, it is known that the activation of ISR in the host by P. fluorescens WCS417r depends on the responsiveness to ethylene and JA but not an increased level of these defense regulators. The sensitivity to ethylene and JA is enhanced as a result of ISR elicitation. This has been supported by two hypotheses. The first is that in plants showing ISR the ability to convert 1-aminocyclopropane-1-carboxylate (ACC) to ethylene is significantly enhanced, providing an ability to produce ethylene on pathogen invasion (Hase et al. 2003). The expression of ethylene and JA responsive genes is greater in induced plants than in noninduced plants after pathogen attack (Pozo et al. 2008). Thus, it seems that induced plants are more sensitive to the recognition of pathogen-induced ethylene and JA, thus resulting in an enhanced and potent response to subsequent pathogen invasion (Conrath et al. 2006). However, elucidation of the ISR signal-transduction pathway resulted in the conclusion that NPR1 acts downstream of the JA- and ethylene-dependent steps (Pieterse et al. 1998).
3.2 SA-Dependent Signaling
It has become clear that only a few rhizobacteria triggering ISR are facilitated by ethylene/JA, although it has been found that rhizobacteria-facilitated systemic resistance is not regulated by SA. The role of SA was first reported in Pseudomonas aeruginosa 7NSK2 and its mutant producing SA. Induction of SA-dependent resistance by Pseudomonas strains is similar to the resistance response of Tobacco mosaic virus, which is not expressed in nahG tobacco (De Meyer et al. 1999), whereas resistance to B. cinerea cannot be triggered in nahG tomato (Audenaert et al. 2002). Systemic resistance induced by some Bacillus strains requires SA but not JA and NPR1, although some strains of Bacillus sp. operate through an ethylene/JA-dependent mechanism and require NPR1 similarly to P. fluorescens WCS417r (Barriuso et al. 2008). ISR in Arabidopsis against Verticillium dahliae in response to root treatment with Paenibacillus alvei K165 requires an SA-dependent mechanism of resistance (Tjamos et al. 2005). Similarly, Domenech et al. (2007) reported an SA- and ethylene-dependent pathway in Bacillus strain N1137 induces systemic resistance to Xanthomonas campestris in Arabidopsis. However, Djavaheri (2007) also reported that ISR against Turnip crinkle virus in Arabidopsis is SA and NPR1 dependent in P. fluorescens.
4 Expression of ISR in Plants
After challenge introduction of a pathogen, expression of ISR is almost similar to that of SAR, in which the severity of disease is decreased with reduced pathogen growth and colonization of induced tissues, confirming that the plant is able to fight the pathogen (van Loon 2000). However, a decrease in disease incidence may protect the plant and increase the yield of the crop. Induced proteins on induction of systemic resistance can be taken as reliable markers for the induced state (van Wees et al. 1999).
There was an increase in the activities of stress-related enzymes such as peroxidase, glucanase, polyphenol oxidase, chitinase, and phenylalanine ammonia-lyase (PAL) as well as total phenolic compounds in PGPR-treated plants (van Loon and van Strien 1999). PAL is an important phenolic biosynthesis enzyme, and oxidative enzymes such as polyphenoloxidase and peroxidase have a vital role in lignification of tissue (Barcelo 1997). The activities of PAL, peroxidase, and phenolic content are responsive to changes in the environment and stresses; these changes often occur as a result of rhizobacterial treatments. Root colonization of cucumber by ISR-mediating Pseudomonas chlororaphis O6 against Corynespora cassiicola causes leaf spot, and effective accumulation of transcripts of six distinct genes on challenge inoculation was found (Kim et al. 2004). Induction was not by P. chlororaphis O6 colonization alone but became evident only after pathogen inoculation.
5 PGPR-Mediated ISR for Disease Suppression Under Field Conditions
Besides laboratory and greenhouse evidence, some experimental evidence indicates that systemic resistance by PGPRs can also be useful for plant protection under field conditions (Nandakumar 1998). Serratia marcescens strain 90-166, P. putida 89B-27, and Flavimonas oryzihabitans strain INR-5 showed ISR against angular leaf spot disease and bacterial wilt in field trials (Kloepper et al. 1993). Several PGPRs on application as bacterization of seed, alone, or as seed treatment plus soil drenching at the time of transplantation have protected cucumber plants against anthracnose, angular leaf spot, and bacterial wilt (Zehnder et al. 2001). In rice, treatment with PGPR strain mixtures of P. fluorescens strains Pf1 and PB2 reduces rice sheath blight disease penetration and increases yield under field trials (Nandakumar 1998). Thus, mixture of strains would be more effective than a single strain against a broad range of pathogens and pests (Raupach and Kloepper 2000; Akhtar and Siddiqui 2010).
6 Rhizobacterial Determinant Help in Indirect Mechanisms
A number of bacterial determinants are involved in the ISR by PGPR as summarized in the following sections.
6.1 Siderophores
The presence of iron is the limiting condition for both plant and microorganism growth. Disease suppression is performed by rhizobacteria against soilborne pathogens by the release of iron chelators known as “siderophores” in the rhizosphere for competition. Besides competition for ferric iron, siderophore production also triggers ISR and plays a dual role in disease suppression (Hofte and Bakker 2007). Leeman et al. (1996) reported that pseudobactin, a siderophore, produced by P. fluorescens strain WCS374 was responsible for ISR in radish against Fusarium wilt and not the LPS. Application of purified pseudobactin from P. fluorescens strain WCS374 to the roots of radish induces systemic resistance. Pseudobactins from P. putida strain WCS358 were tested for Ralstonia solanacearum suppression in Eucalyptus urophylla, Erwinia carotovora suppression in tobacco, and B. cinerea suppression in tomato. In all three cases, the purified pseudobactin 358 was as effective as the wild type (van Loon et al. 2008). Some bacteria also produce SA-containing siderophores, which means their SA secretion is the precursor for SA-containing siderophores such as pseudomonine and pyochelin produced by P. fluorescens WCS374r and P. aeruginosa 7NSK2 respectively. SA is not excreted by bacteria under iron-limiting conditions, but is channeled into the production of SA-containing siderophores. Audenaert et al. (2002) described that induction of systemic resistance does not depend on SA produced by P. aeruginosa 7NSK2; rather it depends on the synergistic interaction between the siderophore and pyochelin derived from SA (De Vleesschauwer and Hofte 2009). It has been observed that all the siderophores are not involved in induction of systemic resistance as all siderophores possess different chemical structures produced from various bacterial sources (Höfte 1993).
6.2 Lipopolysaccharides
LPS are made up of three different components: lipid A, core oligopolysaccharide, and an O-linked polysaccharide. LPS have an important function in stabilizing the outer membrane structure of gram-negative bacteria and also play another role of interacting with the outer membrane of the eukaryotic hosts. LPS are important to plants in preventing the hypersensitive response induced in plants by a virulent or nonhost bacterial adhesion to the plasma membrane receptors of plants such as tobacco, pepper, turnips, and Arabidopsis (Dow et al. 2000) referred to as a “localized induced response.” Plant pathogenic LPS have also been proven to induce the rapid burst of NO which is responsible for innate immunity in plants. Besides, LPS present in the outer membranes of cells are the major component of ISR in certain PGPRs. LPS from Burkholderia cepacia with which tobacco leaves were pretreated was associated with the accumulation of PR proteins (Coventry and Dubery 2001) and phosphorylation of ERK-like mitogen-activated protein kinase (Piater et al. 2004) against Phytophthora nicotianae, whereas, in tobacco cell suspensions, it has been observed that LPS enhances a rapid flux of Ca2+ ion in aequorin-transformed cells, which is correlated with the production of reactive oxygen and nitrogen species, alkalinization of the extracellular culture medium, and fast phosphorylation of certain proteins (Gerber et al. 2004) due to signaling and regulatory defense mechanism (Gerber et al. 2006) and changes in the expression of several genes (Sanabria and Dubery 2006). LPS from P. fluorescens strains WCS374 and WCS417 induces resistance in radish against F. oxysporum f. sp. raphani (Leeman et al. 1995b). Whereas the mutant of P. fluorescens strain WCS417 lacking the O-antigen side chain of LPS does not induce resistance in radish, the O-antigen side chain triggers a defense in radish plants. LPS from P. putida WCS358, which has the O-antigen side chain, does not show ISR in radish. Van Wees et al. (1997) showed the O-antigen side chain of P. fluorescens WCS417r elicits a defense mechanism in Arabidopsis. This shows that LPS from rhizobacteria differs with different host plants and it is not the only trait defining the ISR.
6.3 Exopolysaccharides
Exopolysaccharides (EPS) are high molecular weight polysaccharides that are secreted by most bacteria. EPS help in colonization of the bacteria within the host tissue as well as on the plant surface (Denny 1995). Jones et al. (2008) suggested that EPS can be used as signaling molecules for the developmental response in plants or to suppress host defense response for, for example, EPS secreted by the alfalfa-symbiotic bacterium Sinorhizobium meliloti (Mendrygal and González 2000). EPS produced by Pantoea agglomerans YAS34 was associated with plant growth promotion of sunflower (Alami et al. 2000). EPS from plant pathogenic Pantoea agglomerans elicited a quick flux of active oxygen species in tobacco, parsley, wheat, and rice cell culture (Conrath et al. 2006). However, elicitation of ISR by EPS from PGPR has not been reported. EPS from Burkholderia gladioli IN-26, a strain of PGPR, can induce systemic resistance to Colletotrichum orbiculare in cucumber when it infiltrates leaves or is applied via seed soaking at a concentration of 200 ppm (Park et al. 2008). However, Ipper et al. (2008) reported that EPS from Serratia strain Gsm01 at a concentration of 200 ppm on tobacco leaves affected with Cucumber mosaic virus results in accumulation of peroxidase, PAL, and phenols, and an increased level of PR-1b protein expression.
6.4 Antibiotics
Antibiotics are low molecular weight compounds produced by a few microorganisms, and are harmful to the growth and metabolism of other microorganisms. A large diversity of antibiotics exists (e.g., 2,4-diacetylophloroglucinol, pyrrolnitrin, pyoluteorin, and phenazines), and their involvement in biocontrol has been well studied (Haas and Défago 2005). 2,4-Diacetylophloroglucinol is an elicitor of systemic resistance against Hyaloperonaspora parasitica induced by P. fluorescens CHA0, whereas its mutant strain does not show any ISR effect (Iavicoli et al. 2003). Moreover, Siddiqui and Shaukat (2003) have demonstrated that P. fluorescens CHA0 producing 2,4-diacetylophloroglucinol induces resistance in tomato against root-knot nematode Meloidogyne javanica, whereas its mutant strain does not have the capacity to induce resistance. Resistance induced by 2,4-diacetylophloroglucinol follows a signaling route that induces ethylene signaling (Iavicoli et al. 2003). Another antibiotic, pyocyanin (an N-containing heterocyclic blue phenanzine pigment), is also considered a bacterial determinant eliciting ISR (Britigan et al. 1997). Pyocyanin production by P. aeruginosa 7NSK2 along with SA-derivative pyochelin triggered ISR in beans and tomato against B. cinerea (Audenaert et al. 2002). De Vleesschauwer et al. (2006) reported the dual role of pyocyanin in P. aeruginosa 7NSK2-triggered ISR, acting as a positive regulator of resistance to Magnaporthe oryzae while also rendering ornamental plants hypersusceptibile to Rhizoctonia solani.
6.5 Flagella
The protein flagellin is the building block of the flagellum, a motility organ. It is recognized by most plants, an indication that detection of flagellin is evolutionarily ancient (Boller and Felix 2009), and is required for root colonization by rhizobacteria (De Weger et al. 1987). Conserved peptides of flagellin are observed in Toll-like receptor-like kinase FLS2 in Arabidopsis (Gómez-Gómez et al. 2001) and in tomato (Robatzek et al. 2007). Flagella from P. putida WCS358 have been widely studied in Arabidopsis, bean, and tomato plants (Meziane et al. 2005) but it was shown that its mutant strain lacking flagella also induced resistance; hence, it was concluded that flagella do not play a role in induction of systemic resistance by P. putida WCS358. Therefore, there must be other bacterial determinants involved in induction of systemic resistance by P. putida WCS358. Plant cells have some receptors which recognize the stretching of 15–22 amino acids of flg22 in a conserved domain, which is a potent elicitor in cell culture of certain plant species such as Arabidopsis, tobacco, potato, and tomato. In tomato, flg22 receptor is active at a concentration of 1 pM and has half-maximal resistance at a concentration of 30 pM (Felix et al. 1999). Chinchilla et al. (2006) reported that flagellin is identified through its interaction with FLS2 in Arabidopsis. Thereafter, it was identified in Nicotiana benthamiana, tomato, Brassica sp., and rice (Takai et al. 2008). FLS2 present on the plasma membrane was internalized on flg22 stimulation (Robatzek et al. 2006).
6.6 Volatile Metabolites
Chemically diverse, volatile metabolites are produced by plants and microorganisms, such as terpenes, indoles, fatty acid derivatives, and molecules from other chemical families (Pare and Tumlinson 1999). Ryu et al. (2004) reported that volatile organic compounds (VOCs) released from PGPR trigger ISR in Arabidopsis using an in vitro Petri plate method against the necrotrophic pathogen Pectobacterium carotovorum subsp. carotovorum using the PGPR strains GB03 and IN937a. The VOCs which were involved were analyzed by gas chromatography–mass spectrometry and were found to be 2,3-butanediol and its precursor 3-hydroxy-2-butanone (Farag et al. 2013). However, it is yet to be investigated whether recognition of VOCs is by aboveground or belowground parts and how plants recognize VOC signals (Pare et al. 2005). Heil and Ton (2008) postulated that in the presence VOCs, changes in transmembrane channels lead to enhanced gene activity.
6.7 Other Compounds
Biosurfactants, most specifically cyclic lipopeptides, act as ISR signaling molecules in plants. Cyclic lipopeptides such as members of the fengycin, iturin, and surfactin families from Bacillus sp. are known to induce resistance mechanisms in plants (Ongena and Jacques 2008). Pure fengycin and surfactins provided ISR-mediated protection in beans against B. cinerea similarly to that induced by B. subtilis S499 (Ongena et al. 2007). Massitolide A cyclic lipopeptide, a member of the viscoin group from P. fluorescens strain SS101, shows direct antagonisms and not ISR, but massitolide A when applied alone reduces lesion areas in tomato but not disease incidence, whereas its mutant strain does not show any such effect (Tran et al. 2007).
Certain compounds, such as N-acyl homoserine lactones (AHL), a class of bacterial quorum-sensing signals from Pseudomonas, assist bacterial cells to regulate gene expression in shoots and roots, and modulate defense and cell growth responses, depending on the population density (Jha and Saraf 2012). P. putida strain IsoF producing four different 3-oxo-AHL molecules with acyl side chains showed marked reduction in tomato damage when plants were challenged with Alternaria, whereas the mutant strain was 50 % as effective as the wild-type strain. A microarray analysis of defense gene expression of tomato leaves on application of N-hexanoyl and N-butanoyl homoserine lactones to the roots showed enhanced production of PR proteins and acidic chitinase (Schuhegger et al. 2006). Pure benzylamine at a concentration of 1 μM induced systemic resistance in plants such as beans and cucumber, indicating that the amino group is involved in the resistance (De Vleesschauwer et al. 2008). N-Dimethyl-N-tetradecyl-N-benzylammonium released by P. putida BTP1 seems to be the bacterial determinant of systemic resistance in cucumber (Ongena et al. 2008).
7 Conclusions and Future Prospects
Induction of resistance in plants has opened a new horizon in disease maintenance and plant protection. It is a promising tool for ecofriendly disease control and sustainable agricultural practices. PGPR help the plant by plant growth promotion mechanisms, biocontrol, and inducing systemic resistance in host plants. SA-dependent and SA-independent pathways are both involved in systemic signaling for defense responses. The variety of rhizobacterial determinants shows their vital role in ISR and their regulation in the rhizosphere against multiple pathogens attacking crops. The various bacterial determinants and their regulation in the rhizosphere to explore the fundamentals of plant–microbe interactions will a hot topic of future research because it may offer an opportunity to use the above-mentioned attributes of PGPR in crop management strategies.
References
Abramovitch RB, Anderson JC, Martin GB (2006) Bacterial elicitation and evasion of plant immunity. Nat Rev Mol Cell Biol 7:601–611
Akhtar MS, Siddiqui ZA (2009) Use of plant growth promoting rhizobacteria for the biocontrol of root-rot disease complex of chickpea. Aust Plant Pathol 38:44–50
Akhtar MS, Siddiqui ZA (2010) Role of Plant growth promoting rhizobacteria in biocontrol of plant diseases and sustainable agriculture. In: Maheshwari DK (ed) Plant growth and health promoting bacteria, vol 18, Microbiology monographs. Springer, Berlin, pp 157–196
Akhtar MS, Shakeel U, Siddiqui ZA (2010) Biocontrol of Fusarium wilt by Bacillus pumilus, Pseudomonas alcaligenes and Rhizobium sp. on lentil. Turk J Biol 34:1–7
Alami Y, Achouak W, Marol C, Heulin T (2000) Rhizosphere soil aggregation and plant growth-promotion of sunflowers by an exopolysaccharide-producing Rhizobium sp. strain isolated from sunflower roots. Appl Environ Microbiol 66:3393–3398
Audenaert K, Pattery T, Cornelis P, Hofte M (2002) Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7NSK2: role of salicylic acid, pyochelin, and pyocyanin. Mol Plant Microbe Interact 15:1147–1156
Bakker PAHM, Ran LX, Pieterse CMJ, van Loon LC (2003) Understanding the involvement of rhizobacteria-mediated induction of systemic resistance in biocontrol of plant diseases. Can J Plant Pathol 25:5–9
Barcelo AR (1997) Lignification in plant cell walls. Int Rev Cytol 176:87–132
Barriuso J, Ramos Solano B, Manero Gutierrez FJ (2008) Protection against pathogen and salt stress by four plant growth-promoting rhizobacteria isolated from Pinus spp. on Arabidopsis thaliana. Phytopathology 98:666–672
Beckers GJM, Conrath U (2007) Priming for stress resistance: from the lab to the field. Curr Opin Plant Biol 10:425–431
Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406
Borrás-Hidalgo O (2004) Basic insight in plant-pathogen interaction. Biotecnol Apl 21:1–4
Britigan BE, Rasmussen GT, Cox CD (1997) Augmentation of oxidant injury to human pulmonary epithelial cells by the Pseudomonas aeruginosa siderophore pyochelin. Infect Immun 65:1071–1076
Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88:57–63
Chester KS (1933) The problem of acquired physiological immunity in plants. Q Rev Biol 8:275–324
Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18:1–12
Compant S, Duffy B, Nowak J, Clément C (2005) Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action and future prospects. Appl Environ Microbiol 71:4951–4959
Conrath U, Pieterse CMJ, Mauch-Mani B (2002) Priming in plant–pathogen interactions. Trends Plant Sci 7:210–216
Conrath U, Beckers GJM, Flors V, Garcia-Agustin P, Jakab G, Mauch F, Newman MA, Pieterse CMJ, Poinssot B, Pozo MJ, Pugin A, Schaffrath U, Ton J, Wendehenne D, Zimmerli L, Mauch-Mani B (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19:1062–1071
Coventry HS, Dubery IA (2001) Lipopolysaccharides from Burkholderia cepacia contributes to an enhanced defense capacity and the induction of pathogenesis-related proteins in Niocotiana tobacum. Physiol Mol Plant Pathol 58:149–158
De Meyer G, Audenaert K, Hofte M (1999) Pseudomonas aeruginosa 7NSK2-induced systemic resistance in tobacco depends on in planta salicylic acid accumulation but is not associated with PR1a expression. Eur J Plant Pathol 105:513–517
De Vleesschauwer D, Hofte M (2009) Rhizobacteria-induced systemic resistance. Adv Bot Res 51:223–281
De Vleesschauwer D, Cornelis P, Höfte M (2006) Redox-active pyocyanin secreted by Pseudomonas aeruginosa 7NSK2 triggers systemic resistance to Magnaporthe grisea but enhances Rhizoctonia solani susceptibility in rice. Mol Plant Microbe Interact 19:1406–1419
De Vleesschauwer D, Djavaheri M, Bakker PAHM, Hofte M (2008) Pseudomonas fluorescens WCS374r-induced systemic resistance in rice against Magnaporthe oryzae is based on pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response. Plant Physiol 148:1996–2012
De Weger LA, van der Vlugt CIM, Wijfjes AHM, Bakker PAHM, Schippers B, Lugtenberg BJJ (1987) Flagella of a plant growth stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J Bacteriol 169:2769–2773
Denny TP (1995) Involvement of bacterial polysaccharides in plant pathogenesis. Annu Rev Phytopathol 33:173–197
Despres C, DeLong C, Glaze S, Liu E, Fobert PR (2000) The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12:279–290
Djavaheri M (2007) Iron-regulated metabolites of plant growth-promoting Pseudomonas fluorescens WCS374: their role in induced systemic resistance. PhD thesis, Utrecht University
Domenech J, Ramos SB, Probanza A, Lucas GJA, Gutierrez MFJ (2007) Elicitation of systemic resistance and growth promotion of Arabidopsis thaliana by PGPRs from Nicotiana glauca: a study of the putative induction pathway. Plant Soil 290:43–50
Dong X (2004) NPR1, all things considered. Curr Opin Plant Biol 7:547–552
Doornbos RF, van Loon LC, Bakker PAHM (2012) Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. Agron Sustain Dev 32:227–243
Dow M, Newman MA, Von Roepenack E (2000) The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu Rev Phytopathol 38:241–261
Euglem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371
Farag MA, Zhang H, Ryu CM (2013) Dynamic chemical communication between plants and bacteria through airborne signals: induced resistance by bacterial volatiles. J Chem Ecol 39:1007–1018
Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18:265–276
Fogliano V, Ballio A, Gallo M, Woo SL, Scala F, Lorito M (2002) Pseudomonas lipopeptides and fungal cell wall degrading enzymes act synergistically in biocontrol. Mol Plant Microbe Interact 15:323–333
Gamliel A, Austerweil M, Kritzman G (2000) Non-chemical approach to soil borne pest management–organic amendments. Crop Prot 19:847–853
Gerber IB, Zeidler D, Durner J, Dubery IA (2004) Early perception responses of Nicotiana tabacum cells in response to lipopolysaccharides from Burkholderia cepacia. Planta 218:647–657
Gerber IB, Laukens K, Witters E, Dubery IA (2006) Lipopolysaccharide-responsive phosphoproteins in Nicotiana tabacum cells. Plant Physiol Biochem 44:369–379
Glick BR (2012) Plant growth promoting bacteria: mechanisms and applications. Scientifica 2013:15
Gómez-Gómez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5:1003–1012
Gómez-Gómez L, Bauer Z, Boller T (2001) Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13:1155–1163
Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3:307–319
Hammerschmidt R (1999) Induced disease resistance: how do induced plants stop pathogens. Physiol Mol Plant Pathol 55:77–84
Hammerschmidt R, Nicholson RL (1999) A survey of plant defense responses to pathogens. In: Agrawal A, Tuzun S (eds) Induced plant defenses against pathogens and herbivores. APS Press, St Paul, pp 55–71
Hase S, Van Pelt JA, Van Loon LC, Pieterse CMJ (2003) Colonization of Arabidopsis roots by Pseudomonas fluorescens primes the plant to produce higher levels of ethylene upon pathogen infection. Physiol Mol Plant Pathol 62:219–226
Heil M, Ton J (2008) Long-distance signalling in plant defence. Trends Plant Sci 13:264–272
Höfte M (1993) Classes of microbial siderophores. In: Barton LL, Hemming BC (eds) Iron chelation in plants and soil microorganisms. Academic, San Diego, pp 3–26
Hofte M, Bakker PAHM (2007) Competition for iron and induced systemic resistance by siderophores of plant growth promoting rhizobacteria. In: Varma A, Chincholkar SB (eds) Microbial siderophores. Springer, Berlin, pp 121–133
Hoitink HAJ, Boehm MJ (1999) Biocontrol within the context of soil microbial communities: a substrate-dependent phenomenon. Annu Rev Phytopathol 37:427–446
Iavicoli A, Boutet E, Buchala A, Metraux JP (2003) Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Mol Plant Microbe Interact 16:851–858
Ipper NS, Cho S, Lee SH, Cho JM, Hur JH, Lim CK (2008) Antiviral activity of the exopolysaccharide produced by Serratia sp. strain Gsm01 against cucumber mosaic virus. J Microbiol Biotechnol 18:67–73
Jha CK, Saraf M (2012) Hormonal signaling by PGPR improves plant health under stress conditions. In: Maheshwari DK (ed) Bacteria in agrobiology: stress management. Springer, Berlin, pp 119–140
Jones KM, Sharopova N, Lohar DP, Zhang JQ, Vanden Bosch KA, Walker GC (2008) Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc Natl Acad Sci U S A 105:704–709
Kim MS, Kim YC, Cho BH (2004) Gene expression analysis in cucumber leaves primed by root colonization with Pseudomonas chlororaphis O6 upon challenge-inoculation with Corynespora cassiicola. Plant Biol 6:105–108
Kloepper JW, Leong J, Teintze M, Schroth MN (1980) Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286:885–886
Kloepper JW, Tuzun S, Liu L, Wei G (1993) Plant growth-promoting rhizobacteria as inducers of systemic disease resistance. In: Lumsden RD, Waughn JL (eds) Pest management: biologically based technologies. American Chemical Society Books, Washington, pp 156–165
Knoester M, Pieterse CMJ, Bol JF, van Loon LC (1999) Systemic resistance in Arabidopsis induced by rhizobacteria requires ethylene-dependent signaling at the site of application. Mol Plant Microbe Interact 12:720–727
Lawton KA, Friedrich L, Hunt M, Weymann K, Delanay T, Staub T, Ryals J (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of systemic acquired resistance signal transduction pathway. Plant J 10:71–82
Leeman M, Van Pelt JA, Den Ouden FM, Heinsbroek M, Bakker PAHM, Schippers B (1995a) Induction of systemic resistance against Fusarium wilt of radish by lipopolysaccharides of Pseudomonas fluorescens. Phytopathology 85:1021–1027
Leeman M, Van Pelt JA, Den Ouden FM, Heinsbroek M, Bakker PAHM, Schippers B (1995b) Induction of systemic resistance by Pseudomonas fluorescens in radish cultivars differing insusceptibility to Fusarium wilt, using a novel bioassay. Eur J Plant Pathol 101:655–664
Leeman M, den Ouden FM, van Pelt JA, Dirkx FPM, Steijl H, Bakker PHAM, Schippers B (1996) Iron availability affects induction of systemic resistance to Fusarium wilt of radish by Pseudomonas fluorescens. Phytopathology 86:149–155
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Markovich NA, Kononova GL (2003) Lytic enzymes of Trichoderma and their role in plant defense from fungal diseases. Appl Biochem Microbiol 39:341–351
Mendrygal KE, González JE (2000) Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J Bacteriol 82:599–606
Meziane H, Van der Sluis I, van Loon LC, Höfte M, Bakker PAHM (2005) Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol Plant Pathol 6:177–185
Nandakumar R (1998) Induction of systemic resistance in rice with fluorescent pseudomonads for the management of sheath blight disease. MSc thesis, Tamil Nadu Agriculture University, Coimbatore
Niranjan Raj S, Shetty HS, Reddy MS (2006) Plant growth promoting rhizobacteria potential green alternative for plant productivity. In: Siddiqui ZA (ed) PGPR: biocontrol and biofertilization. Springer, Dordrecht, pp 197–216
Nurnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198:249–266
Ongena M, Jacques P (2008) Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol 16:115–125
Ongena M, Jourdan E, Adam A, Paquot M, Brans A, Joris B, Arpigny JL, Thonart P (2007) Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ Microbiol 9:1084–1090
Ongena M, Jourdan E, Adam A, Schäfer M, Budzikiewicz H, Thonart P (2008) Amino acids, iron, and growth rate as key factors influencing production of the Pseudomonas putida BTP1 benzylamine derivative involved in systemic resistance induction in different plants. Microb Ecol 55:280–292
Pare PW, Tumlinson JH (1999) Plant volatiles as a defense against insect herbivores. Plant Physiol 121:325–331
Pare PW, Farag MA, Zhang H, Ryu CM, Kloepper JW (2005) Elicitors and priming agents initiate plant defense responses. Photosynth Res 85:149–159
Park MR, Kim YC, Park JY, Han SH, Kim KY, Lee SW, Kim LS (2008) Identification of an ISR related metabolite produced by Pseudomonas chlororaphid O6 against the wild fire pathogen Pseudomonas syringae pv. tabaci in tobacco. J Microbiol Biotechnol 18:1659–1662
Patel D, Jha CK, Tank N, Saraf M (2012) Growth enhancement of chickpea in saline soils using plant growth-promoting rhizobacteria. J Plant Growth Regul 31:53–62
Piater LA, Nurnberger T, Dubery IA (2004) Identification of a lipopolysaccharides responsive erk-like MAP kinase in tobacco leaf tissue. Mol Plant Pathol 5:331–341
Pieterse CMJ, van Loon LC (2004) NPR1: the spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol 7:456–464
Pieterse CMJ, van Wees SCM, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10:1571–1580
Pieterse CMJ, Van Pelt JA, Ton J, Parchmann S, Mueller MJ, Buchala AJ, Metraux JP, van Loon LC (2000) Rhizobacteria mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol Mol Plant Pathol 57:123–134
Pozo MJ, Van der Ent S, Van Loon LC, Pieterse CMJ (2008) Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol 180:511–523
Raupach GS, Kloepper JW (2000) Biocontrol of cucumber diseases in the field by plant growth-promoting rhizobacteria with and without methyl bromide fumigation. Plant Dis 84:1073–1075
Robatzek S, Chinchilla D, Boller T (2006) Ligand induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20:537–542
Robatzek S, Bittel P, Chinchilla D, Kochner P, Felix G, Shiu SH, Boller T (2007) Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol Biol 64:539–547
Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8:1808–1819
Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW (2004) Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134:1017–1026
Sanabria NM, Dubery IA (2006) Differential display profiling of the Nicotiana response to LPS reveals elements of plant basal resistance. Biochem Biophys Res Commun 344:1001–1007
Sayyed RZ, Chincholkar SB (2009) Siderophore producing A. feacalis more biocontrol potential vis-à-vis chemical fungicide. Curr Microbiol 58:47–51
Sayyed RZ, Patel PR (2011) Biocontrol potential of siderophore producing heavy metal resistant Alcaligenes sp. and Pseudomonas sp. vis-à-vis organophosphorus fungicide. Indian J Microbiol 51:266–272
Sayyed RZ, Chincholkar SB, Reddy MS, Gangurde NS, Patel PR (2013) Siderophore producing PGPR for crop nutrition and phytopathogen suppression. In: Maheshwari DK (ed) Bacteria in agrobiology: disease management. Springer, Berlin, pp 449–471
Schuhegger R, Ihring A, Gantner S, Bahnweg G, Knappe C, Vogg G, Hutzler P, Schmid M, Van Breusegem F, Eberl L, Hartmann A, Langebartels C (2006) Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ 29:909–918
Shaikh SS, Sayyed RZ (2015) Role of plant growth promoting rhizobacteria and their formulation in biocontrol of plant diseases. In: Arora NK (ed) Plant microbes symbiosis: applied facets. Springer, New Delhi, pp 337–351
Shaikh SS, Patel PR, Patel SS, Nikam SD, Rane TU, Sayyed RZ (2014) Production of biocontrol traits by banana field fluorescent pseudomonads and their comparison with chemical fungicides. Indian J Exp Biol 52:917–920
Siddiqui IA, Shaukat SS (2003) Plant species, host age and host genotype effects on Meloidogyne incognita biocontrol by Pseudomonas fluorescens strain CHA0 and its genetically-modified derivatives. J Phytopathol 151:231–238
Siddiqui ZA, Baghel G, Akhtar MS (2007) Biocontrol of Meloidogyne javanica by Rhizobium and plant growth-promoting rhizobacteria on lentil. World J Microbiol Biotechnol 23:435–441
Subramaniam R, Desveaux D, Spickler C, Michnick SW, Brisson N (2001) Direct visualization of protein interactions in plant cells. Nat Biotechnol 19:769–772
Takai R, Isogai A, Takayama S et al (2008) Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol Plant Microbe Interact 21:1635–1642
Tjamos SE, Flemetakis E, Paplomatas EJ, Katinakis P (2005) Induction of resistance to Verticillium dahliae in Arabidopsis thaliana by the biocontrol agent K-165 and pathogenesis related proteins gene expression. Mol Plant-Microbe Interact 18:555–561
Tran H, Ficke A, Assimwe T, Hofte M, Raaijmakers JM (2007) Role of cyclic Massetolide A inbiological control of Phytophthora infestans in colonization of tomato plants by Pseudomonas fluorescens. New Phytol 175:731–742
Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F (2009) Network properties of robust immunity in plants. PLoS Genet 5:e1000772
van Loon LC (2000) Systemic induced resistance. In: Slusarenko AJ, Fraser RSS, Van Loon LC (eds) Mechanisms of resistance to plant diseases. Kluwer, Dordrecht, pp 521–574
van Loon LC, Bakker PAHM (2006) Induced systemic resistance as a mechanism of disease suppression by rhizobacteria. In: Siddiqui ZA (ed) PGPR: biocontrol and biofertilization. Springer, Dordrecht, pp 39–66
van Loon LC, van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55:85–97
van Loon LC, Bakker PAHM, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483
van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
van Loon LC, Bakker PAHM, Van der Heijat WHW, Wendehenne D, Pugin A (2008) Early responses of tobacco suspension cells to rhizobacterial elicitors of Induced Systemic resistance. Mol Plant-Microbe Interact 21:1609–1621
van Peer R, Niemann GJ, Schippers B (1991) Induced resistance and phytoalexin accumulation in biological control of Fusarium wilt of carnation by Pseudomonas sp. strain WCS417r. Phytopathology 81:728–734
van Wees SCM, Pieterse CMJ, Trijssenaar A, Van’t Westende Y, Hartog F, van Loon LC (1997) Differential induction of systemic resistance in Arabidopsis by biocontrol bacteria. Mol Plant-Microbe Interact 10:716–724
van Wees SCM, Luijendijk M, Smoorenburg I, van Loon LC, Pieterse CMJ (1999) Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol Biol 41:537–549
Verhagen BWM, van Loon LC, Pieterse CMJ (2006) Induced disease resistance signaling in plants. In: Floriculture, ornamental and plant biotechnology, vol 3. Global Science Books, Ikenobe, pp 334–343
Zehnder GW, Murphy JF, Sikora EJ, Kloepper JW (2001) Application of rhizobacteria for induced resistance. Eur J Plant Pathol 107:39–50
Zhang J, Zhou JM (2010) Plant immunity triggered by microbial molecular signatures. Mol Plant Adv 3:783–793
Acknowledgments
We thank the Department of Microbiology, Gujarat University, for encouraging us and helping us with the required facilities and British Petroleum International for financial support.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Patel, S., Sayyed, R.Z., Saraf, M. (2016). Bacterial Determinants and Plant Defense Induction: Their Role as Biocontrol Agents in Sustainable Agriculture. In: Hakeem, K., Akhtar, M. (eds) Plant, Soil and Microbes. Springer, Cham. https://doi.org/10.1007/978-3-319-29573-2_9
Download citation
DOI: https://doi.org/10.1007/978-3-319-29573-2_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-29572-5
Online ISBN: 978-3-319-29573-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)