Abstract
Plant growth promoting rhizobacteria (PGPR) are extensively studied for their antagonistic effect on soil microbes and their induction of systemic resistance towards root and foliar pathogens. Induced resistance tends to increase basal resistance in host in order to provide broad spectrum protection against a wide array of pathogens in nature. A large number of Pseudomonads and Bacillus spp. have been identified with the potential to induce systemic resistance in various hosts. While certain strains result in induced resistance in a wide host range, some show specificity indicating specific plant–microbe interactions. Induced systemic resistance (ISR) is induced by PGPR through secreted determinants. Determinants such as lipopolysaccharides, siderophores, antibiotic, and enzymes have been reported to effectively elicit ISR in host. ISR signals are transmitted locally and systemically. In ISR, jasmonate (JA) and ethylene (ET) regulate the signal transduction for induction while salicylic acid (SA) remains crucial in the transduction of systemic acquired resistance (SAR). Although ISR and SAR both elicit induced responses in host, their signals, signaling pathways, and genes activated vary. Hence JA/ET-induced defense-related genes will be activated in ISR, and SA-dependent defense genes will be activated in SAR. These genes are effective on different sets of pathogens (necrotrophs and biotrophs). Both mechanisms contribute towards the protected nature of the host. In this chapter, the mechanisms, pathways, signals, importance, similarities, and dissimilarities of both these systems are elaborated.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Induced systemic resistance (ISR)
- Systemic acquired resistance (SAR)
- Plant immunity
- Plant growth promoting rhizobacteria (PGPR)
15.1 Introduction
Plants are dependent on nutrient acquisition from soil. Plant roots secrete a considerable measure of chemicals into the rhizosphere which influences growth, development, and acclimatization to environmental stresses (Vallad and Goodman 2004; Van Loon and Glick 2004). The microbial population within the rhizospheric region will similarly contribute chemical constituents that affect the microbial population as well as the plant. The dynamic nature of the rhizospheric microflora allows for an interplay between pathogenic and beneficial microorganisms. This therefore results in the organisms interacting via synergistic or antagonistic interactions (Beardon et al. 2014) where signals are being exchanged between the microorganisms and the root systems that effectively form an active belowground association (Weller et al. 2002; Van Loon and Bakker 2005). These belowground interactions are functional as long as the microbial–plant systems are kept alive to buffer the activity in the rhizospheric environment. These root microbe interactions can result in variation in effect against soil pathogens, microbial propagation, and colonization of the roots (Somers et al. 2004; De Vleesschauwer et al. 2009).
Beneficial organisms such as PGPR and plant growth promoting fungi (PGPF) control plant diseases through suppression of pathogenic soil organisms and induction of host systemic resistance. The presence of these organisms consistently induces resistance in the host beyond basal levels which acts to protect against a host of non-beneficial organisms in its surrounding. Acinetobacter, Azospirilium, Rhizobium, Pseudomonas, and Bacillus have been reported as efficient inducers of systemic resistance in leguminous and nonleguminous plants. In addition, Trichoderma spp., Penicillium simplicissimum, Piriformospora indica, Phoma sp., non-pathogenic Fusarium oxysporum, and arbuscular mycorrhizal fungi have been listed as PGPF that have successfully suppressed diseases in several plant systems (Bakker et al. 2013).
While a large number of these strains interact and produce beneficial outcomes on varying hosts, certain strains have shown specificity indicating that the plant–microbial interaction are regulated by host variety, soil conditions, and microbial populations. Certain rhizospheric organisms produce determinants such as lipopolysaccharides, siderophores, lytic enzymes, exopolysaccharides, lipopeptides, and others (Nadarajah 2016a). These determinants trigger pathways that result in the activation of defense-related genes and responses downstream. Jasmonic acid (JA) and ethylene (ET) regulate rhizobacterial-induced systemic resistance (ISR), while systemic acquired resistance (SAR) is controlled by SA (Van Loon and Glick 2004; Haas and Défago 2005). Though both mechanisms induce host systemic resistance, they remain distinct.
While an array of microorganisms have been identified as potential biocontrol agents, only a handful have had their mechanisms elucidated (Heil and Bostock 2002; Choudhary et al. 2007). Mutants have proven to be a wonderful tool in studying the role of determinants in the mechanism of disease suppression as seen in the repression of F. oxysporum f. sp. raphani by P. putida WCS358 (Raaijmakers et al. 1995). Similarly cucumber plants challenged with Colletotrichum orbiculare showed inhibition of anthracnose symptoms post inoculation with several strains of PGPR (Wei et al. 1991). These experiments imply that the area of antagonistic influence of PGPR is not confined to the rhizosphere, but develops from below ground into above ground defense elicitations. Hence various studies and experimentations have concluded that the heightened level of resistance in planta was mediated through an immune response that was activated in response to rhizobacteria-ISR. As extensive reviews of these organisms have been presented elsewhere (Van Loon et al. 1998; Pozo and Azcon-Aguilar 2007), this chapter ventures to present the mechanisms, signaling pathways, comparisons between ISR and SAR, in addition to differences between these defense mechanisms that collectively work to defend plants against their hostile environment (Kloepper et al. 2004; Van Loon and Bakker 2006; De Vleesschauwer et al. 2008, 2009; Walters et al. 2013).
15.2 Induced Systemic Resistance (ISR): The Mechanism
ISR and SAR are two major players in induced plant resistance. While both contribute towards resistance, one major difference between these systems lies in the inducers, where contrary to ISR, SAR is induced in response to pathogens which results in subsequent protection from infections against a broad host of attackers (Walters et al. 2013; Pieterse et al. 2014). Further ISR and SAR are not just expressed within the locality of induction but are transmitted to distant tissues through systemic spread of signal molecules (Van Loon et al. 2008). ISR like SAR is regulated by signaling pathways that are interlinked and regulated by signal molecules/hormones (Pieterse et al. 2012). In the following segments, ISR and the contribution of ISR in agricultural practices, specifically in disease suppression, will be discussed.
15.3 Pathogen-Induced SAR
SAR has been studied locally and systemically in various plant systems. The local response includes the production of physical and chemical responses such as structural modification to the cell walls, production of phytoalexins and pathogenesis-related (PR) proteins, and hypersensitive response (HR) (Hunt and Ryals 1996; Lamb and Dixon 1997; Van Loon 1997; Van Loon and Van Strien 1999; Métraux 2002; Durrant and Dong 2004; Conrath et al. 2006). Although HR is produced in both compatible and incompatible gene-for-gene interactions (Hammond-Kosack and Jones 1997; Ellis et al. 2002), at the molecular and cellular level, HR is dispersed through uninfected tissues to trigger systemic resistance in the whole plant (Stone et al. 2000). While changes such as lignification and callous deposition are brought about post infection, the systemic transmission results in PR protein production prior to infection (Sticher et al. 1997; Dong 2004). This rapid response of distant tissue is referred to as conditioning which involves systemic signal molecule(s) such as SA. SA and other related inducers (2,6-dichloroisonicotinic acid [INA] or benzothiadiazole [BTH]) are able to promote superoxide production in the cell resulting in the production of reactive oxygen species (ROS), which ultimately activates downstream host defense enzymes such as phenylalanine ammonia-lyase (PAL) and lipoxygenase (LOX) (Katz et al. 1998; Thulke and Conrath 1998; Kauss et al. 1999; Conrath et al. 2002). Another player in the induction of pathogen-derived resistance, β-aminobutyric acid (BABA), retains effective induction even in plants with impaired SA, JA, and ET pathways (Zimmerli et al. 2000). However BABA is only able to protect mutants insensitive to JA and ET but remains ineffective in rescuing mutants defective in SAR signaling.
15.3.1 SAR Signaling
Endogenous SA has been experimentally proven to induce SAR (Van Loon and Antoniw 1982; Van Loon et al. 2008) resulting in increase of SA post induction in local and distant tissues through phloem transport (Malamy et al. 1990; Métraux et al. 1990; Verberne et al. 2003; Durrant and Dong 2004; Van Loon et al. 2008). The salicylate hydrolase defective mutant, NahG, that reduces SA to catechol leaving it incapable of inducing SAR was used to study the role of SA in SAR. The lack of SAR in these plants may be “rescued” through the application of exogenous INA and BTH (Ryals et al. 1996; Sticher et al. 1997; Conrath et al. 2002). Subsequently in establishing the mobile signal(s) involved in SAR, there are two possibilities: (1) SA is not the mobile signal as the rootstock-scion experiment showed induction of SAR despite no accumulation of SA in the NahG rootstock; and (2) SA as a versatile signal that is transported to distal tissues ensuing SA generation in distant tissues. Further, the presence of SA in the phloem of plants has been linked to the transport of this signal molecule within the plant to distal organs thus lending towards SAR. The overexpression of salicylate hydroxylase focused in phloem tissue of tobacco resulted in diminished SAR thus supporting a role for SA in systemic signaling (Mur et al. 2000). Another compound, methyl salicylate (MeSA) was observed in tobacco to elicit defense response. As such, MeSA was proposed as a component that acts with SA in in planta communication and signaling. It is therefore likely that SA as well as other systemic signals (azelaic acid, diterpenoid dehydroabietinal, glycerol-3-phosphate-dependent factor, pipecolic acid) could be involved in SAR (Shulaev et al. 1995; Seskar et al. 1998; Pieterse et al. 2014). The effective function of SA is dependent on the presence of an ankyrin protein that changes the oligomeric state of NPR1 to monomers (Cao et al. 1997). Pathogenesis-related (PR) proteins are produced from the interaction between NPR1 and transcription factors (Dong 2004). PR proteins are affected by SAR and therefore are suitable markers to study induced resistance (Kessmann et al. 1994) and remain the hallmark of SAR induction.
15.3.2 SA Mode in SAR
Catalase and ascorbate peroxidase act as SA-binding proteins that result in the formation of phenolic radicals involved in lipid peroxidation. Lipid peroxidation remains a crucial process in ensuring defense gene activation (Farmer and Mueller 2013), hence requiring the proper execution of its production at the right location and time. Other SA-binding proteins (SAPs) that demonstrate a higher affinity for SA and its analogs were identified (Bakker et al. 2014). While the biological significance of these SAPs remains unresolved, they provide an interesting view in comprehending the method of SA activity. SA- and pathogen-inducible protein kinase (SIPK), a MAP kinase member was isolated and studied in tobacco (Zhang and Klessig 1997; Zhang et al. 2002). Various studies have focused on the upstream regulatory sequences (URS) of PR-1 promoter, one of the terminating reactions in SAR. The TGACG sequence in the URS of PR-1 is perceived by a bZIP family TGA transcription factor (Lebel et al. 1998). TGAs were likewise found to interact with the NPR1 protein, providing a connection between NPR1 and SA-induced PR-1 expression (Lebel et al. 1998; Zhang et al. 1999; Després et al. 2000; Zhou et al. 2000). PR expression is suppressed when SNI1 (negative regulator) binds to the DNA or transcription factors (Li et al. 1999). Other research groups have looked into a SA- and pathogen-inducible WRKY DNA-binding elements that recognize specific sequences on the promoter sequence of chitinase gene in tobacco. This study discovered that protein phosphorylation is essential for the function of WRKY DNA-binding components, thus underscoring the function of kinases in SA signaling (Yang et al. 1999).
15.3.3 SAR-Signaling Network
Ding et al. (2015) conducted a genetic screen of SA mutants via biosensor technique (Marek et al. 2010) which identified upstream (EDS1, PAD4, NDR1) and downstream components (e.g., NPR1), transcription factors (CBP60g, SARD1), and metabolic enzymes (EPS1, PBS3) that are crucial for SA signaling (Cao et al. 1997; Ryals et al. 1997; Jirage et al. 1999; Zhang et al. 2010; Wang et al. 2011; Yezhang et al. 2015). These screens identified two leucine rich repeats (LRR—NBS; LRR and TRI:NBS:LRR) as effectors of signal transduction post infection by avirulent pathogens (Glazebrook 1999). These pathways in the end merge at the DND1 protein which controls the development of HR cell death (Clough et al. 2000) (Fig. 15.1). In the event of being induced by a virulent pathogen, PAD4 is activated resulting in phytoalexin production (Jirage et al. 1999). This activation further results in the downstream activation of SID1 and SID2 that controls SA generation (Nawrath and Metraux 1999). Studies on the SID1 gene has shown that it is associated with the SOS response of the cell. The SOS response is elicited upon introduction of stress into the cell system. While EDS5/SID1 expression is independent of SA, EDS5/SID2 genes encode a MATE transporter and an ICS1 enzyme which are crucial in the SA synthesis (Ding et al. 2015) (Fig. 15.1). The sln mutants together with eds5, sid2, and pad4 are involved in SA accumulation (Ryals et al. 1997; Jirage et al. 1999; Nawrath and Métraux, 1999; Nawrath et al. 2002; Ding et al. 2015). NPR1 acts as a feedback inhibitor of SA biosynthesis following accumulation of SA in response to infection and infestation in NPR1 gene mutants. In addition to components upstream of SA biosynthesis (Clarke et al. 2000; Zhang et al. 2010), NPR1-independent defense responses is triggered by EDS5, PAD4, SID2, and SLN genes (Glazebrook 1999, 2001). The sln1 mutant influences the PR protein expression; hence this goes against the proposed involvement of SA-independent pathway in the regulation of PR gene expression.
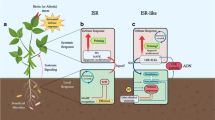
Schematic representation of the components involved in the activation of SAR and ISR. These protein perceive and transmit the signal in SAR and ISR. Solid lines and arrows are confirmed connections. Dashes imply further study required. Solid arrows indicate the transmission from local site of elicitation to distant tissue transmission. Abbreviations: NB-LRR Nucleotide-Binding-Leucine-Rich Repeat, PCD Programmed Cell Death, PRR Pattern-Recognition Receptor, PTI PAMP-Triggered Immunity, SA Salicylic Acid, TF Transcription Factor, PAD Phytoalexin Deficient, EDS Enhanced Disease Susceptibility, SID SA Induction-Deficient, DND Defense, No Death, NDR Non-Race Specific Disease Resistance, DIR Defective In Induced Resistance, FMO Flavin-Dependent Monooxygenase, NPR Non-Expressor PR, TGA TGACG-binding protein (TF), MYB Myeloblastosis (TF), and MYC TF regulator
15.4 Rhizobacteria-ISR
The microorganisms within the rhizosphere of the soil have an important role in the general well-being and health of plants. The bacteria and fungi within the rhizosphere can either be in a beneficial relationship or negatively affect the plant or microbial population (Nadarajah 2016a, b). Among the functions attributed to these organisms are the ability to participate in the nutrient cycles, nutrient acquisition, and management of biotic (Bakker et al. 2007; Van der Heijden et al. 2008; Khan et al. 2009; Kraiser et al. 2011; Berendsen et al. 2012) and abiotic stresses (Yang et al. 2004). These functions are associated with a staggeringly dynamic and complex microbiome within the rhizosphere (Berendsen et al. 2012; Hartmann et al. 2009; Raaijmakers et al. 2009). Through the utilization of molecular techniques, it is expected that the repertoire of microbiome identified that are in interaction with plants and associated with ISR against biotic and abiotic stresses will increase. Studies will not be limited to only identification of new rhizospheric microorganisms but to the mechanisms, key players, and pathways involved in these processes.
15.4.1 Beneficial Microbes and the SAR Pathway
While SAR is a complex mechanism of systemic resistance in plants, ISR prompts a more complex defense in response to non-pathogenic rhizobacteria. Due to the systemic response elicited by ISR, it was at one point assumed to be SAR. This misconception was debunked by Hoffland et al. (1995) who established that ISR against F. oxysporum was induced by P. fluorescens WCS417r in radish without the trademark PR production as seen in SAR. Similar findings were described by Pieterse et al. (1996) in Arabidopsis. This was further corroborated when NahG resulted in an ISR response post treatment with WCS417r-ISR, indicating the involvement of an SA-independent pathway and separating this process from SAR (Pieterse et al. 1996). The same is seen in response to P. putida WCS358r (Raaijmakers et al. 1995; Van Wees et al. 1999) and Serratia marcescens 90-166 where the loss of SA production induced resistance against Colletotrichum orbiculare and P. syringae pv. tomato (Press et al. 1997). However when strain 7NKS2 was used in treatment of NahG mutants in Arabidopsis and tomato, ISR was abolished against TMV and Botrytis cinerea (De Meyer et al. 1999a; Audenaert et al. 2002). Ryu et al. (2003) also encountered similar findings in Arabidopsis, with B. pumilus SE34 against P. syringae pv maculicola. Further, Maurhofer et al. (1998) observed SA-dependent SAR elicitation post treatment with P. fluorescens P3 overexpressing SA biosynthesis gene cluster of P. aeruginosa PAO1. Additionally P. fluorescens SS101, Paenibacillus alvei K165 (Tjamos et al. 2005; Van de Mortel et al. 2012), and Trichoderma PGPF (Mathys et al. 2012; Martínez-Medina et al. 2013) were also reported to induce SA-dependent SAR. The requirement for SA in 7NKS2 was substantiated through utilization of bacterial mutants defective in SA production (De Meyer and Höfte 1997; De Meyer et al. 1999b; Audenaert et al. 2002). However following this finding, further experiments were conducted by Van Loon and Bakker (2005) who inferred that SA-independent pathways is the main regulatory pathway in rhizobacteria-mediated ISR (De Meyer et al. 1999a). Although some PGPR produce SA but it is still not the main signal involved in elicitation of ISR (Ran et al. 2005; Djavaheri et al. 2012). The SA produced however binds with siderophores in iron-limiting condition and thus is not directly involved in SAR. Meanwhile, in cases where SA is produced by PGPR, ROS is an elicitor required to activate SAR. Further these responses are not dependent on accumulation of SA but rather on increasing the sensitivity of tissue to SA (Van Loon and Bakker 2003).
15.4.2 NPR1 as a Common Component of ISR and SAR
Transmission of SAR to distal organs requires mediators. The transmitted SAR signal is chaperoned by one such mediator, Defective In Induced Resistance1 (DIR1) (Champigny et al. 2013) which assist Flavin-Dependent Monooxygenase 1 (FMO1) in receiving and amplifying signals for long distance SAR signaling (Mishina and Zeier 2007) (Fig. 15.1). The well-characterized transcriptional co-regulator, NPR1, plays a role in SA accumulation and in the SAR-signaling pathway (Dong 2004; Vlot et al. 2009; Pajerowska-Mukhtar et al. 2013). Pieterse et al. (1998) had implicated NPR1 in ISR through studies conducted on Arabidopsis. In studying the activation of ISR post treatment with P. fluorescens WCS417r and numerous other PGPR and PGPF , Pieterse et al. (1998) found a connection between NPR1 and the JA/ET-signaling pathways (Lavicoli et al. 2003; Ryu et al. 2003; Stein et al. 2008; Weller et al. 2012). This therefore demonstrates that SA signaling in response to either an avirulent pathogen or rhizobacteria can activate NPR1. While the role of NPR1 in SA signaling has been connected to nuclear function, recent studies have provided information that the NPR1 component of the JA/ET signaling is within the cytosol (Spoel et al. 2003; Stein et al. 2008; Ramirez et al. 2010). Both ISR and SAR defense mechanisms have additive effect within the host. At this juncture, it is difficult to ascertain the specific molecular mechanism involved in the NPR1 mediated JA/ET based ISR induction in host (Van Wees et al. 2000). Pieterse et al. (2014) reported that plant roots expressed high levels of NPR1, NPR3, and NPR4 suggesting a crucial role for these genes in belowground interactions. Both NPR3 and NPR4 together with Cullin 3 (CUL3) ubiquitin E3 ligase are involved in the degradation of NPR1. NPR3 degrades NPR1 when the levels of SA are high causing localized cell death during effector triggered immunity (ETI), while at lower SA levels, NPR4 maintains NPR1 during pathogen-associated molecular patterns triggered immunity (PTI) and thus results in PR expression (Fig. 15.1). In ISR, NPR1 itself acts to mediate the systemic response together with MYC and TFs and the JA/ET pathways. Though NPR1 is a shared component of ISR and SAR, the mechanism downstream of NPR1 perception is different as SAR results in PR accumulation while ISR does not. This could perhaps be due to lower levels of SA-induced ISR that perhaps was insufficient to result in PR production. Studies with the npr1 mutant plants that did not express ISR post cultivation with WCS417 indicated that the expression was dependent on regulation and sensitivity and not towards the SA levels within the host. However, further research is required to understand the role of NPR1 and the possible involvement of other regulatory factors in the SA-NPR1 interaction in ISR (Pieterse et al. 2012).
15.4.3 Other Pathways That Control ISR
As mentioned in the above sections, the JA/ET-signaling pathway is crucial in the induction of ISR in plants. Arabidopsis JA (jar1, coi1, jin1) and ET (ein2, etr1, eir1, ein3) mutants were utilized to establish the function of JA/ET in the plant immune system (Thomma et al. 2001). When these mutants were treated with PGPR (Pseudomonas CHA0, P. fluorescens WCS417r–ISR, P. syringae pv. maculicola, P. fluorescens Q2-87, S. marcescens 90–166) (Pieterse et al. 1998; Ryu et al. 2003; Pozo et al. 2008) and PGPF (P. indica, Penicillium sp. GP16-2, Trichoderma harzianum T39) they failed to induce ISR confirming the role of JA and ET in ISR (Ryu et al. 2004, Stein et al. 2008, Weller et al. 2012; Pieterse et al. 2014). Similar observations were also made in other plant systems, thus supporting the notion that in SA-independent ISR, JA/ET are the main regulators of plant immunity (Yan et al. 2002; De Vleesschauwer et al. 2008; Van der Ent et al. 2009). These pathways are also effective against necrotrophs and herbivores (Van Loon et al. 2008; Van Wees et al. 2008; De Vleesschauwer et al. 2009; Ding et al. 2015; Yezhang et al. 2015).
The Jar1 gene encodes JA-amino acid synthetase which activates the JA signaling. Treatment of wild-type plants with meJA and the ET precursor1-aminocyclopropane-1-carboxylate (ACC) did elicit a response similar to rhizobacterial colonization in plants. However, when treated with these beneficial organisms, endogenous JA levels did not increase which led to the conclusion that the signaling was dependent on JA responsiveness (Pieterse et al. 2000; Staswick and Tiryaki 2004; Van Loon and Bakker 2005). Further, Knoester et al. (1999) using ethylene insensitive mutants demonstrated impaired ISR implicating the requirement of complete and functional ET pathway for proper ISR function.
15.5 Elicitor Molecules in Rhizobacteria-ISR
The organisms that result in ISR do not cause any damage to host. Hence this has resulted in the early conclusion that the chemical compounds resulting in ISR and those resulting in SAR/HR are different. Unlike SAR, ISR is not dependent on localized cell death but rather on the production of elicitors/determinants that trigger the mechanism (Ebel and Mithöfer 1998). A host of chemical determinants have been identified as capable of inducing resistance either individually or in combination (Bakker et al. 2003). These determinants however seem to share similarities in the defense reaction elicited within the host (Gómez-Gómez 2004; Nürnberger et al. 2004). For instance, crude cell wall extracts and lipopolysaccharides (LPS) of P. fluorescens WCS358 resulted in the activation of defense-associated reactions in Arabidopsis (Raaijmakers et al. 1995; Van Wees et al. 1999; Meziane et al. 2005; Nadarajah 2016a) and reduced disease symptoms in pathogen challenged plants. Mutant analysis displayed a redundancy in elicitors as the lack of either O antigenic side chains in Lipopolysaccharide (LPS) or flagellin in these mutants still resulted in induced resistance, as the presence of either one elicitor compounds was sufficient to elicit a response. However, not all strains of P. fluorescence can elicit resistance in Arabidopsis or other plants (Van Wees et al. 1997). This variation may be due to differences in the chemical composition or structure of their determinants. For instance, it was reported that the O-antigenic side-chain of LPS differs from strain to strain probably resulting in perception specificity towards different plant species. Examples of specificity can be seen in application of LPS from Burkholderia cepacia against Phytophthora nicotianae strain ASP B 2D in tobacco (Coventry and Dubery 2001) and the efficient control of the nematode, Globodera pallida with LPS from Rhizobium etli strain G12 in potato (Reitz et al. 2002). Different species or strains of these rhizobacteria resulted in either induction or no response in the host.
Siderophore is another determinant that is involved in the induction of ISR. As there is a redundancy of determinants in rhizobacteria, ISR may be induced by different components in different plant species as exhibited by 7NSK2 in bean and tomato where SA and siderophores were implicated in the response (Audenaert et al. 2002). As siderophores are produced under iron-limiting conditions, it not only inhibits the pathogens within the soil but also induces systemic reaction within the host. However, while all siderophores are able to utilize iron, not every siderophore elicits ISR due to the differences in their chemical structure. Some siderophores produced by the rhizobacteria are pseudomonine, pyochelin, and pseudobactin (Nadarajah 2016a). Some examples of siderophore utilizing rhizobacteria are WCS374 (Leeman et al. 1996; Djavaheri et al. 2012), P. aeruginosa 7NSK2 (Audenaert et al. 2002), Serratia marcescens 90-166 (Press et al. 1997), and P. fluorescens CHA0 (Maurhofer et al. 1994; Weller et al. 2004).
Antibiotics play dual function in the rhizosphere as a microbial antagonist and a defense activator (Fernando et al. 2005). PGPR have been associated with producing more than one antibiotic which relates to its usefulness against phytopathogens (Glick et al. 2007). Diffusible (e.g., phenazines, pyoluteorin, pyrrolnitrin, cyclic lipopeptides (CLP)) and volatile (HCN) antimicrobial products are classified into six groups and interact effectively against microorganisms, nematodes, and plants (Haas and Défago 2005; Raaijmakers et al. 2010). The pyocyanin and pyochelin siderophore from 7NSK2 elicit ISR in conjunction with the 2,4-diacetylphloroglucinol (DAPG) antibiotic (Lavicoli et al. 2003) in tomato. DAPG likewise acts as an inducer in Q2-87 and CHA0 inducing resistance in tomato against Meloidogyne javanica (Siddiqui and Shaukat, 2003; Weller et al. 2004). These reports on DAPG suggest that there may be other rhizobacteria and antibiotics capable of eliciting ISR in plants. Pyrrolnitrin produced by the P. fluorescens (BL915) prevents damping-off by Rhizoctonia solani in cotton while phenazine producing pseudomonads possess redox potential with the ability to suppress various pathogens (Chin-A-Woeng et al. 2003). Phenazine-1-carboxamide that was isolated and studied from roots of tomato was able to mobilize iron from soil (P. chlororaphis PCL1391) (Haas and Défago 2005). A large number of Pseudomonads and Bacillus spp. have been reported to produce various antimicrobial compounds that have selective effect against various host and environments (Beneduzi et al. 2013).
Studies have shown that the interaction between these rhizobacteria and plant roots are dependent on plant variety, environmental conditions, and soil community (Ton et al. 1999; Nadarajah 2016b). While certain strains are perfect inducers of resistance in various plant species, most show tight specificity to root cell surface receptors (Van Loon et al. 1998). For example, WCS358 stimulates resistance in tomato, Arabidopsis, and bean (Raaijmakers et al. 1995; Meziane et al. 2005), and fails to do so in carnation or radish (Leeman et al. 1995). Other strains such as WCS374 induced a powerful response in radish (Leeman et al. 1995) while another, WCS417, could successfully elicit a response in all the above five species of plants (Leeman et al. 1995; Bakker et al. 2013, 2014). Over the course of the last two decades, many ISR determinants have been identified in certain rhizobacterial species. Some examples are provided in Table 15.1.
15.5.1 Key Early Root-Specific Regulator in ISR
Although signaling for ISR starts at the root–microbe interface, not much research has been done to investigate the signaling components involved at eliciting ISR at the root level. Knoester et al. (1999) in studying the root interaction in ISR used a root ET insensitive mutant (eir1) which exhibited the involvement of ET in the transmission of ISR, which was aided perhaps by some other regulatory elements. Further MYB72 was identified as a transcription factor that is involved in the signal transduction from the root as observed in response to P. fluorescens WCS417r in Arabidopsis (Verhagen et al. 2004; Pieterse et al. 2014). MYB72 shows high levels of expression in PGPR-induced roots and no expression was detected in the phloem of uninduced plants. PGPR (P. putida WCS358, P. fluorescens WCS417r) and PGPF (Trichoderma spp.) treated mutant MYB72 plants showed no ISR response hence indicating a significant role for this factor in ISR (Segarra et al. 2009). However, these studies showed that an overexpression of MYB72 did not result in enhanced ISR but rather is dependent on iron-limiting conditions making a connection between iron equilibrium and ISR induction (Van der Ent et al. 2008; Palmer et al. 2013). Treatment with ISR-inducing Pseudomonas strains resulted in the co-regulation of iron deficient marker genes (FRO2, IRT1) and MYB72 in Arabidopsis (Zamioudis et al. 2013). Transcriptome profiling of mutant myb72 and wild-type Arabidopsis provided evidence that iron deficiency response genes were the most dominant species found in roots. PGPR and PGPF however are known to produce siderophores which result in iron uptake from environment therefore resulting in the iron deficient environment. In order to establish if the siderophores are responsible for the deficiency, a siderophore mutant was utilized which exhibited normal MYB72, FRO2, and IRT1 gene activity confirming the role of these microbes in iron deficiency. This interaction requires further study for a better understanding of the connection between iron limitation and siderophore function in ISR (Zhang et al. 2007).
15.6 Expression of ISR
The consequence of ISR expression leads to reduced disease incidence as well as severity post treatment. While ISR executes its defense from belowground, SAR spreads its defense to distal organs from site of pathogenic infection. While both share some overlap in the mechanism of defense moderation, their signals differ. Studies have also shown that due to these differences in signals and moderation, SAR is more effective against biotrophic pathogens while ISR are active against necrotrophs (Bakker et al. 2013). Therefore through the activation of JA and SA-dependent pathways, plants defends themselves against different pathogens in different plant species. This preparatory state of the plant to defend against invasion is called “priming” where there is enhanced level of cellular defense resulting in improved resistance (Van Wees et al. 2008). Various studies conducted on PGPR and PGPF have shown a role for priming in ISR defense (Van Loon and Bakker 2005; Wang et al. 2005). Priming is an important biological and chemical process that is fit and cost effective in adapting plants to its hostile environment (Pozo et al. 2008; Conrath 2011). In addition to the chemical changes observed within the plants in defense, there are structural changes such as callose deposition observed at the site of pathogen entry as seen in P. fluorescens WCS417r treated Arabidopsis (Van der Ent et al. 2009). Abscisic acid (ABA) has been indicated as essential in primed response against insects and pathogens in ISR (Corné et al. 2013; Vos et al. 2013). Besides callose deposition, Bacillus subtilis FB17 was observed to aggregate around the roots of plants infected by P. syringae. The presence of this organism induced stomatal closure and thence reduced the potential of invasion by foliar pathogens through the stomata (Walters et al. 2008). Transcription factors have a responsibility in signaling and regulating the primed state. These factors remain inactive in a non-induced stage but are rapidly activated when the host is affected by pathogens or insects. One transcriptional factor that has been linked to regulation and signaling of ISR is a member of the AP2/ERF family (Van der Ent et al. 2009). These factors have been linked with JA/ET regulated genes and are directly linked to ISR expression (Verhagen et al. 2004). Promoter region analysis of these ISR-related genes revealed the presence of cis-acting G box motif. These motifs are linked to a regulator of JA dependent response, MYC2 (Pozo et al. 2008; Stein et al. 2008) (Fig. 15.1) which is required for proper execution of this pathway. Out of the genes expressed in Arabidopsis, only ~1% of these genes are expressed at the root level and there is no constancy in the expression level observed in the distal leaves (Verhagen et al. 2004).
15.7 Disease-Suppressed Soil
Disease suppressive soil has been described as soil that shows suppression of pathogen through competition for nutrient, antagonism, lytic enzymes, quorum sensing, and various other means by which a non-beneficial microbial population is kept at check (Weller et al. 2002; Loper et al. 2012; Philippot et al. 2013). Through these belowground activities, damage is reduced significantly to the host or the establishment of disease becomes less important over time in a particular soil (Mazzola 2002). Beneficial microbes have been used to control agriculturally significant organisms such as Gaeumannomyces graminis var. tritici through the production of DAPG on Take All Disease. Over successive events of take all in a particular location, the soil eventually became suppressive towards the pathogen. This has been seen in events where monocultures were grown over a period of time resulting in inhibition of the pathogen due to eventual multiplication and high dosage levels of the beneficial microbes (Pseudomonas fluorescens) within the soil (Weller et al. 2002). This disease suppressive soil can also be used in amending condusive soil to reduce disease incidences (Raaijmakers and Weller 1998). Another example in disease suppresiveness is against Fusarium wilt (Alabouvette 1999), and Rhizoctonia solani infestations (Mendes et al. 2011). The competition for nutrient and the production of phenazines appear to reduce the wilt symptom in infected soil (Mazurier et al. 2009). In each incident, it has been reported that while there may be a dominant microbe facilitating this suppression, in most cases it will involve a consortium. This consortium may be made up of microbes from the groups: Azospirillum, Bulkholderia, Comamonas, Gluconacetobacter, Pseudomonas, and Sphingomonadaceae genus (Kyselkova et al. 2009). Through Chip technology, 17 taxa of β and γ-proteobacteria and firmicutes were linked to disease suppressiveness (Mendes et al. 2011). Most often, disease suppressiveness has been linked to antibiosis (Raaijmakers et al. 2002), siderophore producing Pseudomonaceae (Duijff et al. 1998; Zhang et al. 2007) and ISR (Bakker et al. 2007) subsequently resulting in reduced disease incidence and severity (Pieterse et al. 2013).
15.8 Concluding Remarks and Future Prospects
Much research has been devoted towards understanding the role of beneficial microbes in the elicitation of plant immunity and its specific role in ISR since its discovery more than two decades ago. The plant immune system is unique in a way that it is activated to fend off enemies while it remains suppressed to support beneficial interactions. Both these interactions of the plant immune system are in play in ISR to benefit the host. It remains to be deciphered how a phenomenon that enhances plant immunity towards both biotic and abiotic stresses can at the same time contribute towards improved growth and development in the host. One would expect that the initial approach would be to try and determine or understand the “message” transmitted at the point of contact and how this message is then amplified and transmitted to the rest of the plant. We should also look at how both the ISR and SAR mechanisms overlap and what are the shared or different points of regulation between these mechanisms in incurring an effective immune response in host. The perception of the signal by the receptors as well as the regulators involved in the long distance signaling and perception of the signal in both ISR and SAR is still not completely understood. We believe through the use of the “omics” platforms, a wealth of information will surface not only to enable us to better understand the key players in these processes but to add on and gap fill on issues such as regulators, receptors, signal molecules, pathways, and other participants of the complex system of plant–microbe interactions in ISR and SAR. While we may have acquired sufficient knowledge on how the soil microbiomes improve plant heath and development, we are still vague on how the host is able to shape the microbial community surrounding the roots to best benefit it. Likewise, while we may know the key processes involved in the perception and signaling of SAR in plant, there are still gaps in our knowledge with regards to the signals , the regulation, the perception and the mode of transmission of signals long distance.
As a major societal challenge is producing sufficient agricultural produce to meet the market and population demands, any development in science that lends towards this is a positive contribution. In this chapter, we see how both SAR and ISR are two main contributors of the plant defense mechanism and how a better knowledge and understanding of these can assist us with the challenge. ISR has been used in the past decades as biocontrols and in soil amendments all with the hope of reducing disease incidence and severity and at the same time contributing towards better yield and development. A better understanding of SAR on the other hand will likewise contribute towards the knowledge to enhance plant immunity through external stimuli, breeding and the utilization of transgenics towards generating crops with heightened defense mechanisms.
Some answers that may assist with obtaining a clearer and more well-defined picture of SAR and ISR in plants may arise from addressing the following questions:
-
1.
How does the plant facilitate the colonization of a suitable community to enhance growth and immunity? How do these communities play a dual role in growth and immunity?
-
2.
What are the long distance signal(s) involved in both ISR and SAR? How are they transmitted and how do they trigger ISR/SAR?
-
3.
Are there any other regulators than MYB72 in the root for ISR?
-
4.
Are there any other transcription factors or regulators that are involved in ISR and SAR? What are their function and contribution in these processes?
-
5.
Is there a role for autoregulation of mutualism in ISR?
-
6.
How exactly does NPR1 regulate ISR?
-
7.
What are the differences and similarities in genes triggered by ISR and SAR in plants?
While the above questions are not the only areas left with gaps to fill, the constant inquisition on the above mechanisms will only increase our knowledge. However there is always a possibility that with new knowledge comes new questions and new issues to address.
References
Alabouvette C (1999) Fusarium wilt suppressive soils: an example of disease-suppressive soils. Australasian Plant Pathol 28:57–64
Audenaert K, Pattery T, Cornelis P, Hofte M (2002) Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7NSK2: role of salicylic acid, pyochelin, and pyocyanin. Mol Plant-Microbe Interact 15:1147–1156
Bakker PAHM, Ran LX, Pieterse CMJ, Van Loon LC (2003) Understanding the involvement of rhizobacteria-mediated induction of systemic resistance in biocontrol of plant diseases. Can J Plant Pathol 25:5–9
Bakker PAHM, Pieterse CMJ, Van Loon LC (2007) Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology 97:239–243
Bakker PAHM, Berendsen RL, Doornbos RF, Wintermans PCA, Pieterse CMJ (2013) The rhizospere revisited: root microbiomics. Front Plant Sci 4:165
Bakker PAHM, Ran LX, Mercado-Blanco J (2014) Rhizobacterial salicylate production provokes headaches. Plant Soil 382:1–16
Beardon E, Scholes J, Ton J (2014) How do beneficial microbes induce systemic resistance? In: Walters RD, Newton AC, Gary DL (eds) Induced resistance for plant defense: a sustainable approach to crop protection. Wiley, Chichester, pp 232–248
Beneduzi A, Moreira F, Costa PB, Vargas LK, Lisboa BB, Favreto R et al (2013) Diversity and plant growth promoting evaluation abilities of bacteria isolated from sugarcane cultivated in the South of Brazil. Appl Soil Ecol 63:94–104
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88:57–63
Champigny MJ, Isaacs M, Carella P, Faubert J, Fobert PR, Cameron RK (2013) Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front Plant Sci 4:230
Chin-A-Woeng TFC, Bloemberg GV, Lugtenberg BJJ (2003) Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol 157:503–523
Choudhary DK, Prakash A, Johri BN (2007) Induced systemic resistance (ISR) in plants: mechanism of action. Indian J Microbiol 47:289–297
Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12:2175–2190
Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK Jr, Bent AF (2000) The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA 97:9323–9328
Conrath U (2011) Molecular aspects of defence priming. Trends Plant Sci 16:524–531
Conrath U, Pieterse CMJ, Mauch-Mani B (2002) Priming in plant-pathogen interactions. Trends Plant Sci 7:210–216
Conrath U, Beckers GJM, Flors V, García-Agustín P, Jakab G, Mauch F, Newman MA, Pieterse CM, Poinssot B, Pozo MJ, Pugin A, Schaffrath U, Ton J, Wendehenne D, Zimmerly L, Mouch-Mani B (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19:1062–1071
Corné MJP, Poelman EH, Van Wees SCM, Marcel D (2013) Induced plant responses to microbes and insects. Front Plant Sci 4:475
Coventry HS, Dubery IA (2001) Lipopolysaccharides from Burkholderia cepacia contribute to an enhanced defensive capacity and the induction of pathogenesis-related proteins in Nicotiana tabacum. Physiol Mol Plant Pathol 58:149–158
De Meyer G, Höfte M (1997) Salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 induces resistance to leaf infection by Botrytis cinerea on bean. Phytopathology 87:588–593
De Meyer G, Audenaert K, Höfte M (1999a) Pseudomonas aeruginosa 7NSK2-induced systemic resistance in tobacco depends on in planta salicylic acid accumulation but is not associated with PR1a expression. Eur J Plant Pathol 105:513–517
De Meyer G, Capieau K, Audenaert K, Buchala A, Métraux JP, Höfte M (1999b) Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 activate the systemic acquired resistance pathway in bean. Mol Plant Microbe Interact 12:450–458
De Vleesschauwer D, Cornelis P, Höfte M (2006) Redox-active pyocyanin secreted by Pseudomonas aeruginosa 7NSK2 triggers systemic resistance to Magnaporte grisea but enhances Rhizoctonia solani susceptibility in rice. Mol Plant Microbe Interact 19:1406–1419
De Vleesschauwer D, Djavaheri M, Bakker PAHM, Hofte M (2008) Pseudomonas fluorescens WCS374r-induced systemic resistance in rice against Magnaporthe oryzae is based on pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response. Plant Physiol 148:1996–2012
De Vleesschauwer D, Chernin L, Höfte MM (2009) Differential effectiveness of Serratia plymuthica IC1270-induced systemic resistance against hemibiotrophic and necrotrophic leaf pathogens in rice. BMC Plant Biol 9:9
Despres C, Delong C, Glaze S, Liu E, Fobert PR (2000) The Arabidopsis NPR1/Nim 1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12:279–290
Ding Y, Shaholli D, Mou Z (2015) A large-scale genetic screen for mutants with altered salicylic acid accumulation in Arabidopsis. Front Plant Sci 5:763
Djavaheri M, Mercado-Blanco J, Versluis C, Meyer J-M, Van Loon LC, Bakker PAHM (2012) Iron regulated metabolites produced by Pseudomonas fluorescens WCS374r are not required for eliciting induced systemic resistance (ISR) against Pseudomonas syringae pv. tomato in Arabidopsis. Microbiol Open 1:311–325
Dong X (2004) NPR1, all things considered. Curr Opin Plant Biol 7:547–552
Duijff BJ, Pouhair D, Olivain C, Alabouvette C, Lemanceau P (1998) Implication of systemic induced resistance in the suppression of fusarium wilt of tomato by Pseudomonas fluorescens WCS417r and by nonpathogenic Fusarium oxysporum Fo47. Eur J Plant Pathol 104:903–910
Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42:185–209
Ebel J, Mithöfer A (1998) Early events in the elicitation of plant defence. Planta 206:335–348
Ellis C, Karafyllidis I, Turner JG (2002) Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol Plant Microbe Interact 15:1025–1030
Farmer EE, Mueller MJ (2013) ROS-mediated lipid peroxidation and RES-activated signaling. Annu Rev Plant Biol 64:1–22
Fernando WG, Ramarathnam R, Krishnamoorthy AS, Savchuk SC (2005) Identification and use of potential bacterial organic antifungal volatiles in biocontol. Soil Biol Biochem 37:955–964
Glazebrook J (1999) Genes controlling expression of defense responses in Arabidopsis. Curr Opin Plant Biol 2:280–286
Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis. Curr Opin Plant Biol 4:301–308
Glick BR, Cheng Z, Czarny J, Duan J (2007) Promotion of plant growth by ACC deaminase-containing soil bacteria. Eur J Plant Pathol 119:329–339
Gómez-Gómez L (2004) Plant perception systems for pathogen recognition and defence. Mol Immunol 41:1055–1062
Haas D, Défago G (2005) Biological control of soilborne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 4:1–13
Hammond-Kosack KE, Jones JD (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607
Hartmann A, Schmid M, van Tuinen D, Berg G (2009) Plant-driven selection of microbes. Plant and Soil 321:235–257
Heil M, Bostock RM (2002) Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Ann Bot 89:503–512
Hoffland E, Pieterse CMJ, Bik L, Van Pelt JA (1995) Induced systemic resistance in radish is not associated with accumulation of pathogenesis-related proteins. Physiol Mol Plant Pathol 46:309–320
Hunt M, Ryals J (1996) Systemic acquired resistance signal transduction. Crit Rev Plant Sci 15:583–606
Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci U S A 96:13583–13588
Katz VA, Oliver U, Thulke CU (1998) Benzothiadiazole primes parsley cells for augmented elicitation of defense responses. Plant Physiol 117:1333–1339
Kauss H, Fauth M, Merten A, Jeblick W (1999) Cucumber hypocotyls respond to cutin monomers via both an inducible and a constitutive H2O2-generating system. Plant Physiol 120:1175–1182
Kessmann H, Staub T, Ligon J, Oostendorp M, Ryals J (1994) Activation of systemic acquired disease resistance in plants. Eur J Plant Pathol 100:359–369
Khan MS, Zaidi A, Wani PA, Oves M (2009) Role of plant growth promoting rhizobacteria in the remediation of metal contaminated soils. Environ Chem Lett 7:1–19
Kloepper JW, Ryu C-M, Zhang SA (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266
Knoester M, Pieterse CMJ, Bol JF, Van Loon LC (1999) Systemic resistance in Arabidopsis induced by rhizobacteria requires ethylene-dependent signaling at the site of application. Mol Plant Microbe Interact 12:720–727
Kraiser T, Gras DE, Gutiérrez AG, González B, Gutiérrez RA (2011) A holistic view of nitrogen acquisition in plants. J Exp Bot 62:1455–1466
Kyselkova M, Kopecky J, Frapolli M, Defago G, Sagova-Mareckova M, Grundmann GL, Moënne-Loccoz Y (2009) Comparison of rhizobacterial community composition in soil suppressive or conducive to tobacco black root rot disease. ISME J 3:1127–1138
Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48:251–275
Lavicoli A, Boutet E, Buchala A, Métraux J-P (2003) Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Mol Plant Microbe Interact 16:851–858
Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E (1998) Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J 16:223–233
Leeman M, Van Pelt JA, Den Ouden FM, Heinsbroek M, Bakker PAHM, Schippers B (1995) Induction of systemic resistance by Pseudomonas fluorescens in radish cultivars differing in susceptibility to fusarium wilt, using a novel bioassay. Eur J Plant Pathol 101:655–664
Leeman M, Den OFM, Van Pelt JA, Dirkx FPM, Steijl H, Bakker PAHM, Schippers B (1996) Iron availability affects induction of systemic resistance against fusarium wilt of radish by Pseudomonas fluorescens. Phytopathology 86:149–155
Li X, Zhang Y, Clarke JD, Li Y, Dong X (1999) Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98:329–339
Loper JE, Hassan KA, Mavrodi DV, Davis EW, Lim CK, Shaffer BT, Elbourne LD, Stockwell VO, Hartney SL, Breakwell K, Henkels MD, Tetu SG, Rengel LI, Kidarsa TA, Wilson NL, van de Mortel JE, Song C, Blumhagen R, Radune D, Hostetler JB, Brinkac LM, Durkin AS, Kluepfel DA, Wechter WP, Anderson AJ, Kim YC, Pierson LS 3rd, Pierson EA, Lindow SE, Kobayashi D, Raajimakers JM, Weller DM, Thomashow LS, Allen AE, Paulsen IT (2012) Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8:e1002784
Malamy J, Carr JP, Klessig DF, Raskin I (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250:1002–1004
Marek G, Carver R, Ding Y, Sathyanarayan D, Zhang X, Mou Z (2010) A high-throughput method for isolation of salicylic acid metabolic mutants. Plant Methods 6:21
Martínez-Medina A, Fernández I, Sánchez-Guzmán MJ, Jung SC, Pascual JA, Pozo MJ (2013) Deciphering the hormonal signalling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front Plant Sci 4:206
Mathys J, De Cremer K, Timmermans P, Van KS, Lievens B, Vanhaecke M, Cammue BP, De Connick B (2012) Genome-wide characterization of ISR induced in Arabidopsis thaliana by Trichoderma hamatum T382 against Botrytis cinerea infection. Front Plant Sci 3:108
Maurhofer M, Hase C, Meuwly P, Métraux JP, Défago G (1994) Induction of systemic resistance of tobacco to tobacco necrosis virus by the root-colonizing Pseudomonas fluorescens strain CHA0: influence of the gacA gene and of pyoverdine production. Phytopathology 84:139–146
Maurhofer M, Reimmann C, Schmidli-Sacherer P, Heeb SD, Défago G (1998) Salicylic acid biosynthesis genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology 88:678–684
Mazurier S, Corberand T, Lemanceau P, Raaijmakers JM (2009) Phenazine antibiotics produced by fluorescent pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J 3:977–991
Mazzola M (2002) Mechanisms of natural soil suppressiveness to soilborne diseases. Antonie Van Leeuwenhoek 81:557–564
Mendes R, Kruijt M, De Bruijn I, Dekkers E, Van der Voort M, Schneider JH, Piceno YM, DeSantis TZ, Andersen GL, Bakker PA, Raaijmakers JM (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100
Métraux JP (2002) Recent breakthroughs in the study of salicylic acid biosynthesis. Trends Plant Sci 7:332–334
Métraux JP, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B (1990) Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250:1004–1006
Meziane H, Van der SI, Van Loon LC, Hofte M, Bakker PAHM (2005) Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol Plant Pathol 6:177–185
Mishina TE, Zeier J (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50:500–513
Mur LA, Brown IR, Darby RM, Bestwick CS, Bi YM, Mansfield JW, Draper J (2000) A loss of resistance to avirulent bacterial pathogens in tobacco is associated with the attenuation of a salicylic acid-potentiated oxidative burst. Plant J 23:609–621
Nadarajah K (2016a) Induced systemic resistance in rice. In: Choudhary KD, Varma A (eds) Microbial-mediated induced systemic resistance in plants. Springer Singapore, Singapore, pp 103–124. doi:10.1007/978-981-10-0388-2_7
Nadarajah K (2016b) Rhizosphere interactions: life below ground. In: Choudhary KD, Varma S, Tuteja N (eds) Plant microbe interactions: an approach to sustainable agriculture. Springer Singapore, Singapore
Nawrath C, Metraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11:1393–1404
Nawrath C, Heck S, Parinthawong N, Metraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14:275–286
Nürnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198:249–266
Pajerowska-Mukhtar KM, Emerine DK, Mukhtar MS (2013) Tell me more: roles of NPRs in plant immunity. Trends Plant Sci 18:402–411
Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML (2013) MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet 9:e1003953
Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799
Pieterse CMJ, Van Wees SCM, Hoffland E, Van Pelt JA, Van Loon LC (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis related gene expression. Plant Cell 8:1225–1237
Pieterse CMJ, Van Wees SCM, Van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, Van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10:1571–1580
Pieterse CMJ, Van Pelt JA, Ton J, Parchmann S, Mueller MJ, Buchala AJ, Métraux JP, Van Loon LC (2000) Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol Mol Plant Pathol 57:123–134
Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521
Pieterse CMJ, Poelman EH, Van Wees SC, Dicke M (2013) Induced plant responses to microbes and insects. Front Plant Sci 4:475
Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52:347–375
Pozo MJ, Azcon-Aguilar C (2007) Unraveling mycorrhiza-induced resistance. Curr Opion Plant Biol 10:393–398
Pozo MJ, Van der Ent S, Van Loon LC, Pieterse CMJ (2008) Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol 180:511–523
Press CM, Wilson M, Tuzun S, Kloepper JW (1997) Salicylic acid produced by Serratia marcescens 91-166 is not the primary determinant of induced systemic resistance in cucumber or tobacco. Mol Plant Microbe Interact 10:761–768
Raaijmakers JM, Weller DM (1998) Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol Plant Microbe Interact 11:144–152
Raaijmakers JM, Leeman M, Van Oorschot MMP, Van der Sluis I, Schippers B, Bakker PAHM (1995) Dose-response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathology 85:1075–1081
Raaijmakers JM, Vlami M, de Souza JT (2002) Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 81:537–547
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y (2009) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant and Soil 321:341–361
Raaijmakers JM, de Bruijn I, Nybroe O, Ongena M (2010) Natural functions of lipopeptides from bacillus and pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev 34:1037–1062
Ramirez V, Van der Ent S, Garcia-Andrade J, Coego A, Pieterse CMJ, Vera P (2010) OCP3 is an important modulator of NPR1-mediated jasmonic acid-dependent induced defenses in Arabidopsis. BMC Plant Biol 10:199
Ran LX, Van Loon LC, Bakker PAHM (2005) No role for bacterially produced salicylic acid in rhizobacterial induction of systemic resistance in Arabidopsis. Phytopathology 95:1349–1355
Reitz M, Oger P, Meyer A, Niehaus K, Farrand SK, Hallmann J, Sikora RA (2002) Importance of the O-antigen, core-region and lipid a of rhizobial lipopolysaccharides for the induction of systemic resistance in potato to Globodera pallida. Nematol 4:73–79
Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner H-Y, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8:1808–1819
Ryals JA, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner H-Y, Johnson J, Delaney TP, Jesse T, Vos P, Uknes S (1997) The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell 9:425–439
Ryu C-M, Hu C-H, Reddy MS, Kloepper JW (2003) Different signaling pathways of induced resistance by rhizobacteria in Arabidopsis thaliana against two pathovars of Pseudomonas syringae. New Phytol 160:413–420
Ryu C-M, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW (2004) Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134:1017–1026
Segarra G, Van der Ent S, Trillas I, Pieterse CMJ (2009) MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol 11:90–96
Seskar M, Shulaev V, Raskin I (1998) Endogenous methyl salicylate in pathogen-inoculated tobacco plants. Plant Physiol 116:387–392
Shulaev V, Leon J, Raskin I (1995) Is salicylic acid a transported signal of systemic acquired resistance in tobacco? Plant Cell 7:1691–1701
Siddiqui IA, Shaukat SS (2003) Suppression of root-knot disease by Pseudomonas fluorescens CHA0 in tomato: importance of bacterial secondary metabolite, 2,4-diacetylpholoroglucinol. Soil Biol Biochem 35:1615–1623
Somers E, Vanderleyden J, Srinivasan M (2004) Rhizosphere bacterial signalling, a love parade beneath our feet. Crit Rev Microbiol 30:205–240
Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt MJ, Mueller AJ, Buchala J-P, Métraux R, Brown K, Kazan K (2003) NPR1 modulates crosstalk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15:760–770
Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16:2117–2127
Stein E, Molitor A, Kogel KH, Waller F (2008) Systemic resistance in Arabidopsis conferred by the mycorrhizal fungus Piriformospora indica requires jasmonic acid signaling and the cytoplasmic function of NPR1. Plant Cell Physiol 49:1747–1751
Sticher L, Mauch-Mani B, Métraux JP (1997) Sistemathic acquired resistance. Annu Rev Phytopathol 35:235–270
Stone JK, Bacon CW, White JR (2000) An overview of endophytic microbe: endophytism defined. In: Bacon CW, White JF Jr (eds) Microbial endophytes. Marcel Dekker, New York, pp 3–30
Thomma BPHJ, Penninckx IAMA, Broekaert WF, Cammue BPA (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13:63–68
Thulke OU, Conrath U (1998) Salicylic acid has a dual role in the activation of defense-related genes in parsley. Plant J 14:35–42
Tjamos SE, Flemetakis E, Paplomatas EJ, Katinakis P (2005) Induction of resistance to Verticillium dahlia in Arabidopsis thaliana by the biocontrol agent K-165 and pathogenesis-related proteins gene expression. Mol Plant Microbe Interact 18:555–561
Ton J, Pieterse CMJ, Van Loon LC (1999) Identification of a locus in Arabidopsis controlling both the expression of rhizobacteria-mediated induced systemic resistance (ISR) and basal resistance against Pseudomonas syringae pv. Tomato. Mol Plant Microbe Interact 12:911–918
Vallad GE, Goodman RM (2004) Systematic acquired resistance and induced systematic resistance in conventional agriculture. Crop Sci 44:1920–1934
Van de Mortel JE, De Vos RCH, Dekkers E, Pineda A, Guillod L, Bouwmeester K, van Loon JJ, Dicke M, Raaijmakers JM (2012) Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol 160:2173–2188
Van der Ent S, Verhagen BW, Van Doorn R, Bakker D, Verlaan MG, Pel MJ, Joosten RG, Proveniers MC, Van Loon LC, Ton J, Pieterse CM (2008) MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol 146:1293–1304
Van der Ent S, Van Hulten MHA, Pozo MJ, Czechowski T, Udvardi MK, Pieterse CM, Ton J (2009) Priming of plant innate immunity by rhizobacteria and β-aminobutyric acid: differences and similarities in regulation. New Phytol 183:419–431
Van der Heijden MGA, Bardgett RD, Van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310
Van Loon LC (1997) Induced resistance in plants and the role of pathogenesis-related proteins. Eur J Plant Pathol 103:753–765
Van Loon LC, Antoniw JF (1982) Comparison of the effects of salicylic acid and ethephon with virus-induced hypersensitivity and acquired resistance in tobacco. Neth J Plant Pathol 88:237–256
Van Loon LC, Bakker PAHM (2003) Signalling in rhizobacteria-plant interactions. In: De Kroon H, Visser EJW (eds) Root ecology. Springer, Berlin, pp 297–330
Van Loon LC, Bakker PAHM (2005) Induced systemic resistance as a mechanism of disease suppression by rhizobacteria. In: Siddiqui ZA (ed) PGPR: biocontrol and biofertilization. Springer, Dordrecht, pp 39–66
Van Loon LC, Bakker PAHM (2006) Root-associated bacteria inducing systemic resistance. In: Gnanamanickam SS (ed) Plant-associated bacteria. Springer, Dordrecht, pp 269–316
Van Loon LC, Glick BR (2004) Increased plant fitness by rhizobacteria. In: Sandermann H (ed) Molecular ecotoxicology of plants. Springer, Berlin, pp 177–205
Van Loon LC, Van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55:85–97
Van Loon LC, Bakker PAHM, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483
Van Loon LC, Bakker PA, van der Heijdt WH, Wendehenne D, Pugin A (2008) Early responses of tobacco suspension cells to rhizobacterial elicitors of induced systemic resistance. Mol Plant Microbe Interact 21:1609–1621
Van Wees SCM, Pieterse CMJ, Trijssenaar A, Van’t Westende Y, Hartog F, Van Loon LC (1997) Differential induction of systemic resistance in Arabidopsis by biocontrol bacteria. Mol Plant Microbe Interact 10:716–724
Van Wees SCM, Luijendijk M, Smoorenburg I, Van Loon LC, Pieterse CMJ (1999) Rhizobacteria mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol Biol 41:537–549
Van Wees SCM, De Swart EAM, Van Pelt JA, Van Loon LC, Pieterse CMJ (2000) Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc Natl Acad Sci U S A 97:8711–8716
Van Wees SCM, Van der Ent S, Pieterse CMJ (2008) Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol 11:443–448
Verberne MC, Hoekstra J, Bol JF, Linthorst HJM (2003) Signaling of systemic acquired resistance in tobacco depends on ethylene perception. Plant J 35:27–32
Verhagen BWM, Glazebrook J, Zhu T, Chang H-S, Van Loon LC, Pieterse CMJ (2004) The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol Plant Microbe Interact 17:895–908
Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47:177–206
Vos IA, Pieterse CMJ, Van Wees SCM (2013) Costs and benefits of hormone-regulated plant defences. Plant Pathol 62:43–55
Walters DR, Paterson L, Walsh DJ, Havis ND (2008) Priming for plant defense in barley provides benefits only under high disease pressure. Physiol Mol Plant Pathol 73:95–100
Walters DR, Ratsep J, Havis ND (2013) Controlling crop diseases using induced resistance: challenges for the future. J Exp Bot 64:1263–1280
Wang YQ, Ohara Y, Nakayashiki H, Tosa Y, Mayama S (2005) Microarray analysis of the gene expression profile induced by the endophytic plant growth–promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. Mol Plant Microbe Interact 18:385–396
Wang L, Tsuda K, Truman W, Sato M, Nguyen le V, Katagiri F, Glazebrook J (2011) CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J 67:1029–1041
Wei G, Kloepper JW, Tuzun S (1991) Induction of systemic resistance of cucumber to Colletotrichum orbiculare by select strains of plant-growth promoting rhizobacteria. Phytopathology 81:1508–1512
Weller DM, Raaijmakers JM, McSpadden Gardener BB, Thomashow LS (2002) Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu Rev Phytopathol 40:309–348
Weller DM, Van Pelt JA, Mavrodi DV, Pieterse CMJ, Bakker PAHM, Van Loon LC (2004) Induced systemic resistance (ISR) in Arabidopsis against Pseudomonas syringae pv. tomato by 2,4-diacetylphloroglucinol (DAPG)-producing Pseudomonas fluorescens. Phytopathology 94:S108
Weller DM, Mavrodi DV, Van Pelt JA, Pieterse CMJ, Van Loon LC, Bakker PAHM (2012) Induced systemic resistance (ISR) in Arabidopsis thaliana against Pseudomonas syringae pv. tomato by 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology 102:403–412
Yan Z, Reddy MS, Ryu C-M, McInroy JA, Wilson M, Kloepper JW (2002) Induced systemic protection against tomato late blight elicited by plant growth–promoting rhizobacteria. Phytopathology 92:1329–1333
Yang P, Chen C, Wang Z, Fan B, Chen Z (1999) A pathogen- and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of the tobacco class I chitinase gene promoter. Plant J 18:141–149
Yang YN, Qi M, Mei CS (2004) Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J 40:909–919
Yezhang D, Danjela S, Zhonglin M (2015) A large-scale genetic screen for mutants with altered salicylic acid accumulation in Arabidopsis. Front Plant Sci 5:763. doi:10.3389/fpls.2014.00763
Zamioudis C, Mastranesti P, Dhonukshe P, Blilou I, Pieterse CMJ (2013) Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiol 162:304–318
Zhang S, Klessig DF (1997) Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9:809–824
Zhang Y, Fan W, Kinkema M, Li X, Dong X (1999) Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci U S A 96:6523–6528
Zhang S, Moyne AL, Reddy MS, Kloepper JW (2002) The role of salicylic acid in induced systemic resistance elicited by plant growth-promoting rhizobacteria against blue mold of tobacco. Biol Control 25:288–296
Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, He Z (2007) Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol 145:450–464
Zhang Y, Xu S, Ding P, Wang D, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, Li X, Zhang Y (2010) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci U S A 107:18220–18225. doi:10.1073/pnas.1005225107
Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF (2000) NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe Interact 13:191–202. doi:10.1094/MPMI.2000.13.2.191
Zimmerli L, Jakab G, Métraux J-P, Mauch-Mani B (2000) Potentiation of pathogen-specific defense mechanisms in Arabidopsis by β-aminobutyric acid. Proc Natl Acad Sci U S A 97:12920–12925
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Nadarajah, K.K. (2017). Induction of Systemic Resistance for Disease Suppression. In: Abdullah, S., Chai-Ling, H., Wagstaff, C. (eds) Crop Improvement. Springer, Cham. https://doi.org/10.1007/978-3-319-65079-1_15
Download citation
DOI: https://doi.org/10.1007/978-3-319-65079-1_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-65078-4
Online ISBN: 978-3-319-65079-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)