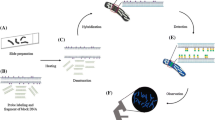
Abstract
Multicolored fluorescence-based chromosome biology or ‘molecular cytogenetics’ in common continue to flourish and make essential contributions to elucidate the plant gene regulation, genome architecture, and organization by revealing essential chromosomal landmarks. Fluorescence in situ hybridization (FISH) and its modifications, such as extended DNA fiber-FISH, bacterial artificial chromosome (BAC)-FISH, multicolor-FISH (McFISH), and super-stretched pachytene-FISH, allow the study of minute details of chromosome structure and subsequently permit sophisticated analyses of chromosomal behavior. Similarly, genomic in situ hybridization (GISH) facilitates genome-specific chromosome painting in hybrids and polyploids, analysis of recombination of partially homologous chromosomes in interspecific/generic natural hybrids, and detection of transgene and/or alien chromatin in synthetic hybrids. The global patterns of chromatin modification (e.g., DNA methylation and histone tail modifications) along with nuclear size and shape, relative content and distribution of hetero/euchromatin, and organization as well as structure of chromosomes (e.g., position and orientation) provide new insights into epigenomic evolution of the particular plant species. Molecular cytogenetics also provide information on gene pool diversity and relatedness of the plant to its wild relative that ultimately may serve as a baseline data for plant breeding programs. As more genomes become sequenced, such cytogenetic tools will play a greater role in investigating the function of those genomes. Attempts have been made to summarize the utility of molecular cytogenetic tools in exploration of important chromosomal landmarks in plants. The evolution of plant cytogenetic research from classical to molecular level and modern to next-generation era has been discussed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
6.1 Introduction
6.1.1 Classical Cytogenetics
The term cytogenetics is referred to the study of genetic consequences in terms of chromosome number, structure, and behavior vis-à-vis speciation and evolution. Cytogenetics has been proved to be an integral part of genome mapping projects owing to magnificent chromosomal dynamics during mitosis and meiosis. The field of plant cytogenetics was heavily induced by Barbara McClintock’s pioneering work on maize (Zea mays) (McClintock 1929, 1932, 1938, 1941a, b, 1984). McClintock used carmine for staining and uniquely identified all of the individual chromosomes from a single meiotic nucleus with a combination of two metrics, i.e., the relative lengths and arm ratios of the chromosomes. Her studies on unequivocal identification of individual chromosomes established a milestone in the scientific community, which allowed neo-discoveries regarding the dynamic structure and behavior of the maize genome (McClintock 1929, 1932, 1938, 1941a, b, 1984). Further, development of chromosome-banding techniques greatly improved the usefulness of chromosome biology to understand the basic genome architecture. In this context, Caspersson et al. (1968) proposed Q-banding pattern using the fluorescent dye quinacrine on plant chromosomes. Vosa and Marchi (1972) compared Giemsa C-banding to Q-banding on the chromosomes of bean (Vicia faba), keeled garlic (Allium carinatum), and maize. Further, Giemsa staining technique also showed its utility to identify individual rice prometaphase chromosomes (Kurata and Omura 1978), karyotype development for diploid rye (Secale cereale) (Gill and Kimber 1974), and barley (Hordeum vulgare) (Linde-Laursen 1975). With the advent of information on DNA and its characteristics, modifications of DNA staining dyes and banding techniques were adapted and optimized for cytogenetic characterization of different plant species. These classical approaches have proven invaluable for chromosome characterization, but the development of in situ hybridization, which allows for direct visualization of specific DNA sequences on chromosomes, forms a quantum leap forward for cytogenetics by combining cytology with molecular biology (Gill and Friebe 1998; Harper and Cande 2000).
6.1.2 Molecular Cytogenetics
A combination of ‘classical cytogenetics’ and ‘recombinant DNA technology’ gave birth to a versatile multicolored fluorescence engineering-based chromosome biology called ‘molecular cytogenetics.’ During initial development, such technique had been performed by using radioactive nucleic acid probes for the detection of specific DNA or RNA sequences in metaphase chromosomes or interphase. Subsequently, in the late nineties, methods for labeling nucleic acids with non-radioactive haptens such as biotin became available and adopted widely (Jiang and Gill 1994). The advantages of non-radioactive probes over radioactive probes include increased stability, safe handling, rapid, precise spatial localization, less back ground, and most importantly the ability to use multiple colors on a single chromosome preparation.
The development of in situ hybridization (ISH) techniques opened up opportunities for cytogenetic analysis of any species, regardless of its inherent chromosome morphology (Gall and Pardue 1969; Pardue and Gall 1975; John et al. 1969). In plants, the use of radioactive tagged or modified nucleotides (labeled with biotin, digoxigenin, or fluorescent haptens) and FISH probes also permits microscopic visualization and localization of complementary sequences in cells/nuclei and on individual chromosomes (Mukai et al. 1991; Fransz et al. 1996a; Mukai and Yamamoto 1998). Basic FISH makes use of green and red fluorochromes for probe detection and DAPI (4,6-diamidino-2-phenylindole) for counterstaining the chromosomal DNA. Although FISH is commonly used to map unique or low-copy-number sequences, however it also showed its potential to localize repetitive sequence in order to produce chromosome-specific landmarks or explore genome relations in polyploidy/closely related plant species (Lysak et al. 2001, 2003; Kato et al. 2004; Lamb and Birchler 2006). FISH has been found most successful in mapping the repetitive and single-copy DNA sequences on prometaphase chromosomes, interphase nuclei, pachytene complements, chromatin fibers, and naked DNA molecules. Accurate localization of repetitive and tandem arrays plays a major role in chromosome identification and karyotype analysis in plants (Mukai and Yamamoto 1998). The broad applications of FISH in structural, comparative, and functional genomics place plant cytogenetics in an important place to complement, accelerate, or guide plant genome research (Lamb et al. 2007; Danilova and Birchler 2008; Nagaki et al. 2012b). On the other hand, genomic in situ hybridization (GISH) (Le et al. 1989; Mukai and Gill 1991), a special type of FISH that uses genomic DNA of a donor species as a probe in combination with an excess amount of unlabeled blocking DNA, provides a powerful technique to monitor chromatin introgression during interspecific hybridization. In addition, the GISH technique allows the study of genome affinity between polyploid species and their progenitors (Mukai et al. 1993b; Raina et al. 1998; Raina and Mukai 1999). GISH is thus a valuable supplemental technique to traditional genome analysis such as conventional meiotic pairing analysis.
Molecular cytogenetics has now become an indispensible tool and a conceptual foundation for modern genome projects by providing significant information on individual chromosome portfolio of the organism under investigation.
6.2 Advances in Molecular Cytogenetic Techniques
Rapid developments in genetics, molecular genetics, molecular biology, and genomics, together with molecular cytogenetics , have driven major conceptual advances in mitotic, meiotic analysis, chromosome structure, and chromosome manipulation. Along such development although the principal steps of the FISH technique have remained same, various technical developments have been adapted in plant molecular cytogenetics . The basic development was the use of several colors for labeling the probes which provide holistic view of genome structure at a single glance, i.e., McFISH and McGISH (Mukai et al. 1993b; Mukai 1996). Some of the recent developments in the field of plant molecular cytogenetics in order to understand genome architecture and organization at ultra-resolution are described below.
6.2.1 Tyr-FISH
Tyr-FISH was developed to improve the detection sensitivity of FISH experiments. This method allows signal amplification by using a peroxidase-conjugated antibody as the first layer of signal detection. Fluorochrome-labeled tyramides as peroxidase substrate are used to generate and deposit many fluorochromes close to the in situ bound peroxidase (Raap et al. 1995). The sensitivity of the basic FISH technique can be increased by 10–100 times using such modification. DNA probes smaller than 1 kb were successfully visualized on plant chromosomes using Tyr-FISH (Khrustaleva and Kik 2001; Stephens et al. 2004).
6.2.2 DNA Fiber-FISH
The DNA fiber-FISH technology is applied to characterize complex genomic arrangements in plant nuclei by using decondensed chromatin and highly extended intact DNA fibers on microscopic slides (Fransz et al. 1996a). The method involves the release of DNA molecules from lysed nuclei followed by spreading them on the surface of a microscope slide and the hybridization of probes using a standard FISH method. Applying FISH probes to the stretched DNA molecules provides the higher spatial resolution with increased detection sensitivity. DNA prepared from BAC clones or plant tissues extends approximately 2.5–3.5 kb/μm on slides and provides fine-mapping resolution of up to a few kilobases. The drawback of the technique is that chromosome identification requires control DNA sequences, since there is no chromosome structure. In plants, Fransz et al. (1996b) demonstrated the utility of the extended DNA fiber-FISH (EDF-FISH) technology to characterize Arabidopsis thaliana and tomato genome. Later, this method was applied on other plants (e.g., rye, rice, and maize) in order to characterize complex genomic arrangements (Nagaki et al. 2004; Jin et al. 2004; Nakano et al. 2005; Yamamoto and Mukai 2007). Fiber-FISH is particularly informative when the exact position and ordering of DNA clones are needed. It can also evaluate the distances and overlaps between neighboring sequences (Ersfeld 2004; Suzuki et al. 2004; Yamamoto and Mukai 2007). The minimum target DNA size that can be distinguished unambiguously in plants is 10 kb (representing a ~3 μm fluorescent signal, de Jong et al. 1999); however, good flanking markers are crucial in order to differentiate and identify shorter DNA stretches.
6.2.3 Three-Dimensional (3D) FISH
The 3D-FISH technique had developed by Bass et al. (1997). Meiotic cells of maize were fixed in a buffer to preserve chromosome structure. Pollen mother cells were also gently extruded out of the fixed anthers and embedded in optically clear polyacrylamide for staining and imaging. Stacks of FISH images were taken and composed into a single 3D image. Individual chromosomes bearing the FISH signals were traced out and computationally straightened (Harper and Cande 2000). Since the chromosome structure can be preserved using this technique, it is advantageous for the identification of precise location of DNA probes on the chromosomes as well as within the nucleus.
6.2.4 FISH on Super-Stretched Chromosomes
Interphase nuclei, super-stretched mitotic metaphase chromosomes, and meiotic pachytene chromosome provide intermediate resolving power for FISH mapping. The relative positions of clone separated by <100 kb can be resolved on these cytological targets (Jiang et al. 1996; Wang et al. 2006). Pachytene chromosomes are particularly versatile targets for FISH mapping. Late pachytene chromosomes can be used to orient the telomere–centromere positions of the adjacent clones, whereas early pachytene chromosomes can be used to resolve even partially overlapped BAC clones. Nevertheless, pachytene chromosomes are not amenable for cytological analysis in many plant species.
On the other hand, flow-sorted plant chromosome at meiotic metaphase can be stretched to more than 100 times of their original size (Valarik et al. 2004). FISH on stretched chromosomes showed brighter signals than on the untreated control presumably as a result of better probe accessibility to the stretched chromatin. FISH on super-stretched metaphase chromosomes provides a mapping resolution of up to 70 kb (Valarik et al. 2004), similar to the resolution on meiotic pachytene chromosomes (Cheng et al. 2002). Thus, this modification of FISH provides an alternative mapping target for those plant species where meiotic pachytene chromosomes are not suitable for cytological analysis.
6.2.5 BAC-FISH
For the genome-wide sequencing project, a genomic library-holding large DNA fragments is an important tool for physical mapping or positional cloning of important chromosome landmarks. BAC-FISH, a unique tool of molecular cytogenetics , uses genomic DNA cloned in large-insert vectors such as bacterial artificial chromosomes (BACs) (Shizuya et al. 1992) in combination with FISH. This technique has shown its tremendous potential for physically mapping of specific DNA sequences and identifying individual chromosomes in plants (Suzuki and Mukai 2004). The BAC clones provide efficient resources for chromosome-specific FISH markers especially for plant species having small genomes such as rice, cotton, and sorghum. BAC-FISH favors the large clone as a probe for better resolution. The conventional FISH analysis on plant chromosomes employing probes containing over 10-kb insert DNA provides stable and distinct signals (Mukai and Yamamoto 1998; Suzuki et al. 2010). It is quite difficult to detect a single locus by using a plasmid clone of several kbs as the FISH probe. In this regard, the BAC clones containing around 50–100 kb fragments are suitable for probe of the FISH analysis.
6.3 Molecular Cytogenetics in Plant Genome Research
6.3.1 Physical Mapping and In Situ PCR
Plant genome are known for abundance of repeat sequences and cytogenetic or physical mapping of such repetitive DNA sequences decipher their genomic distribution and precisely identify the typical chromosome or set of chromosomes. These repeated rDNA gene clusters are being widely used and a common starting point for FISH-based mapping (Mukai et al. 1991; Yamamoto and Mukai 1991; Fransz et al. 1996a; Mukai and Yamamoto 1998; Sharma et al. 2012). The two types of ribosomal RNA genes (rDNA), 18S-5.8S-26S rDNA and 5S rDNA, have been extensively used as probes for physical mapping in higher plants due to their arrangement in tandem arrays (Mukai 1999). FISH mapping of rDNA clusters has provided a number of chromosomal markers that proved their efficacy in exploration of chromosome evolution and species interrelationships.
In hexaploid wheat, the six loci of 5S rRNA genes were identified on the short arm of the chromosomes of homoeologous group 1 and 5 (1A, 1B, 1D, 5A, 5B, and 5D) (Mukai et al. 1990), whereas 18S-5.8S-26S rDNA loci were mapped on the short arm of 1A, 1B, 6B, and 5D chromosomes and the long arm of 7D chromosome (Mukai et al. 1991) (Fig. 6.1). The rRNA genes are associated with the nucleolar organizing region (NOR), and the visualization of such repeat clusters at interphase represents the number of active rDNA loci. Multicolor FISH (McFISH) approach targeting repetitive DNA and rDNA probes also serves as chromosome identification markers in many plant species, for example, common wheat (Fig. 6.2). Similarly, Xu and Earle (1996) mapped the 45S rRNA DNA loci on to the tomato pachytene chromosomes, and Pedrosa et al. (2002) demonstrated the rDNA FISH for creating a karyotype of the model legume lotus. In addition, rDNA FISH in combination with other tandem repeats aids the generation of core cytogenetic maps, as demonstrated for maize, wheat (Jaing and Gill 1994), cotton (Hanson et al. 1996), tomato (Xu and Earle 1996), Pinus (Hizume et al. 2002), and Arabidopsis (Koornneef et al. 2003). The rDNA sequences are conserved across most plant species, but other tandem repeats exhibit variable degree of conservation.
Multicolor FISH mapping of 5S rRNA and 18S-5.8S-26S rRNA genes on the chromosomes of bread wheat (Triticum aestivum, 2n = 6x = 42, AABBDD genome), and fluorescence signals can be seen for 5S (red) and 18S-5.8S-26S (green) rRNA genes, respectively (Mukai 2004)
Seven-color FISH on a metaphase cell of common wheat. Seven DNA sequences-pSc119.2, pSc74, pAs1, telomere, 18S-26S rDNA, 5S rDNA, and gliadin were detected by red, bluish green, green, orange, pink, blue, and white fluorescence, respectively, and the photographs were taken by triple exposures (Mukai 1996)
Further, the chromosomal localization of rDNA has been widely used for comparative characterization of polyploid plant species. A comparison of FISH patterns of polyploid species with those of diploid progenitors of Aegilops revealed natural amphiplasty, in which the active rDNA sites either transformed to inactive or silent (deleted) during polyplodization event (Yamamoto 1994). Similarly, the U genome mostly suppresses the NOR activity of other genomes in tetraploids. On the other hand, the NOR activity of the d-genome chromosomes is completely suppressed by other genomes. In hexaploid species, all rDNA sites on the third genome remain active, reflecting time lapse after polyploid formation.
Simultaneously, Mukai and Apples (1996) invented the in situ polymerase chain reaction (in situ PCR)-FISH for mapping plant genes. This method uses the extreme temperature gradient sensitivity of PCR along with the cytological location of DNA sequences by means of in situ hybridization. The in situ locations of the rye-specific spacer region were determined on metaphase chromosomes. In such experiment, two pairs of primers for rye, i.e., Nor-R1 and rye 5S-Rrna-R1, were amplified in situ, which resulted in 386- and 107-bp amplified products, respectively. Rye NOR primers (45S) were localized on chromosome 1R and 4R, while 5S primers showed signals on the chromosome 1R and 3R. Interestingly, a previously described locus chromosome 5R did not show any signal in this experiment. It was concluded that the absence of a 5S site could be due to the sequence differences between the different 5S rDNA lineages. Several chromosome-specific sequences were also identified through primers specific to the chromosome. Thus, in situ PCR proved its utility in amplification of DNA sequences of specific plant chromosomes and for mapping low-copy genes of interest (Mukai and Yamamoto 1998).
Centromeric and telomeric sequences are also widely used in FISH mapping studies. Telomere repeats are highly conserved in plant species and occur in at least two major variants, i.e., (TTAGGG)n and (TTTAGGG)n (Lapitan et al. 1989; Adams et al. 1998; Fajkus et al. 2005). Similarly, the centromere associated 156-bp tandem repeat of maize, Cent C, was first discovered by Ananiev et al. (1998) and has become an invaluable cytogenetic milestone for maize and many related grass species. Cent O, a 155-bp centromere-specific satellite repeat sequence, the 180-bp satellite repeat, and CEN38, a 140-bp repeat sequence, have proven useful for labeling the primary constriction in rice, Arabidopsis, and sorghum, respectively (Heslop-Harrison et al. 1999; Cheng et al. 2002; Nagaki et al. 2003; Kim et al. 2005).
Further, employing BAC clones as a probes in FISH experiments become revolutionizing inventory in the field of molecular cytogenetic and extensively used in many plant species including cotton (Hanson et al. 1995), rice (Jiang et al. 1995), tomato (Zhong et al. 1996), Arabidopsis (Fransz et al. 1996b), onion (Suzuki et al. 2001), and sorghum (Kim et al. 2005). This approach can also be used to acquire insight for ongoing genome-sequencing projects worldwide.
King et al. (2002) demonstrated a GISH -based approach for physical mapping to distinguish recombination events between chromosomes of Festuca pratensis and Lolium perenne. A similar approach has also been used for the integration of genetic and physical maps of two Allium chromosomes (Khrustaleva et al. 2005). This GISH-based mapping strategy is similar to physical mapping using deletion and translocation stocks. This approach overcomes the major drawback of the tedious and time-consuming process of developing a large number of deletion and translocation stocks.
On the other hand, DNA clones were also used as probes for comparative FISH mapping in relative species. Several cytogenetics researchers reported FISH mapping of A. thaliana BACs on chromosomes of Brassica species. Comparative FISH mapping between Arabidopsis and Brassica provided a direct visualization of the genome duplication within Brassica species (Howell et al. 2005; Lysak et al. 2005). In addition, comparative chromosome painting with pooled BAC probes was used to investigate ancestral relationships among species that diverged within the Brassicaceae (Lysak et al. 2001, 2003, 2005). Recently, Koo and Jiang (2009) developed a technique by stretching maize pachytene chromosomes mechanically more than 20 times longer than their original size. Such super-stretched pachytene chromosomes can be directly used in conventional as well as molecular cytogenetic experiments. Super-stretching of the chromosomes coupled with immunofluorescence in situ detection of DNA methylation can lead to a new dimension and higher resolving power to modern molecular cytogenetics research. Collectively, these studies revealed that such FISH-based plant cytogenetical tools are uniquely informative and beneficial for genome analysis.
6.3.2 Chromosome Identification Subject to Parentage, Hybridity, and Ploidy
Fluorescence signal allows the identification of chromosomes, specific sequences, segments, or whole set of chromosome to gain a genome-wide view at a single glance in order to understand the plant genome organization and behavior. Fluorescence signals of either a single repetitive DNA probe or a mixture of several probes can be utilized for hybridization independently to identify individual chromosomes within a species. Chromosome identification through FISH method has advantage over the traditional chromosome-banding techniques due to availability of several probes for a particular species. Many repetitive DNA elements can also generate specific FISH signal pattern on individual chromosome within a single species (Mukai et al. 1991; Mukai and Yamamoto 1998; Koo et al. 2005). In this context, Pedersen and Langridge (1997) demonstrated the identification of all 21 chromosomes of hexaploid wheat through fluorescence signals derived from two different repetitive DNA probes. Later, similar approach has been adopted in several plant species for chromosome identification (Franz et al. 1998; Hizume et al. 2002; Kato et al. 2004; Koo et al. 2004).
On the other hand, GISH provides a direct visual method for distinguishing parental genomes and analyzing genome organization in intra-/interspecific hybrids, allopolyploid species, and introgression lines. This technique has an incredible prospective to identify application in identifying alien chromatin introgression and to study chromosomal pairing and recombination between divergent genomes. GISH has validated its utility in recognizing synthetic Hordeum chilense × Secale africanum (Schwarzacher et al. 1989) and Triticum aestivum (wheat) × S. cereale (rye) (Le et al. 1989). Mukai and Gill (1991) showed that GISH optimally detects barley chromosomes in a wheat background and further identified A-, B-, and D-genomes of the common wheat (Mukai et al. 1993b) using the same approach (Fig. 6.3). Similarly, Raina et al. (1998) and Raina and Mukai (1999) conclusively revealed that Coffea congensis and C. eugenioides, and Arachis villosa and A. ipaensis are the diploid wild progenitors of allotetraploid C. arabica (2n = 4x = 44) and A. hypogaea (2n = 4x = 40), respectively, using GISH as a tool. GISH has also been widely used to characterize the genome constitution of natural hybrids and to identify the parental origin of specific loci. By following the same approach, Takahashi et al. (1999) categorized the ancestral genome donors in maize and examined inter-genomic translocations and homeologous chromosome pairing (Zwierzykowski et al. 2008), as well as chromosomal areas with large species-specific sequences (alien chromatin introgression) or translocation break points (Qi et al. 2008). Such versatile approach of molecular cytogenetics also provides insight into somaclonal variation, the origin of B chromosomes, control of chromosome pairing, and other aspects of chromosome evolution (Kato et al. 2005).
Chromosome identification of Triticum aestivum (2n = 6x = 42). The AABBDD genome was simultaneously discriminated using GISH technique in which the diploid A genome progenitor Triticum urartu (yellow), diploid B genome progenitor Aegilops speltoides (brown), and diploid D genome progenitor Aegilops squarrosa (orange) have been identified precisely
6.3.3 Karyotype and Phylogenetic Analysis
FISH-based chromosome identification systems could lead to precise karyotyping and to understand the evolution of particular plant taxa by means of speciation from wild to cultivated ones. For example, several repetitive DNA probes generate specific hybridization pattern on chromosomes of wheat and related species (Mukai et al. 1993a; Pederson and Langridge 1997). The FISH karyotypes from some repetitive DNA probes are similar to karyotypes based on C- or N-banding analysis (Cuadrado et al. 1995; Pederson and Langridge 1997). FISH-based karyotyping also specifies the phylogenetic view of related plant species (Lim et al. 2000). A number of repetitive DNA probes had utilized to develop FISH karyotypes of several diploid and polyploid Triticum and Aegilops species by Badaeva et al. (1996a, b). Similarly, comparative FISH mapping using several repetitive DNA probes in Nicotiana species found N. tomentosiformis to be the T-genome donor (Lim et al. 2000). Comparison of such karyotypes evidently revealed chromosomal landmarks to understand the evolutionary relationship between these species. Karyotyping using repetitive DNA probes can also visualize inter-genomic chromosome translocations in polyploid species. Since molecular cytogenetic techniques are often used to compare the ability of different genomes to hybridize (homology of genomes), together with the use of interspecific hybrids and allopolyploids, there by can serve as a powerful tool to understand phylogenetic relationships between species that is independent of nucleotide sequence-based approaches.
6.3.4 Chromosome Painting
The basic principle of FISH was further exploited to ‘paint’ individual plant chromosomes. The ‘chromosome painting’ is one of the most powerful molecular cytogenetic techniques to analyze nuclear organization and genome structure through visualization of specific cytogenetic target regions or entire chromosomes using this technique (Pinkel et al. 1986). Such technique involves the hybridization of fluorescence-tagged chromosome-specific composite probe pools (generally BAC clones) to various cytological preparations. Lysak et al. (2001) painted the chromosome of dicotyledonous model organism A. thaliana for the first time by employing selected BACs as differential labeled probes. However, in plants, mainly due to the presence of large amounts of repetitive DNA sequences such technique is remained limited (Jiang and Gill 2006). Such technique was found to be useful to identify individual chromosome in the interphase nuclei and could reveal the spatial arrangement and functional properties of individual chromatin domains. Further, Han et al. (2003, 2004) modified the McGISH to identify closely related wheat-Thinopyrum intermediates. Such chromosome painting provides insight into genome duplication/multiplication and karyotype evolution in closely related taxa. Arabidopsis chromosome and/or segment-specific probes were hybridized to ‘paint’ the chromosomes from species related to A. thaliana (Lysak et al. 2005). In later studies, the chromosome painting technique was applied successfully in related Brassica species (Lysak et al. 2010). These experiments proved that the technique is feasible for the detailed investigation of the pairing behavior of homologous chromosomes during early prophase I. Painting by this method is found to be feasible on small B chromosomes as well as alien chromosomes that possess chromosome-specific repeats (Houben et al. 1996). Comparative chromosome painting is an efficient and powerful approach to study the partial genome duplications and karyotype evolution. This advantage of the technique has been used to investigate the mechanisms of chromosome number reduction in A. thaliana and related Brassicaceae species.
Successful interspecific chromosome painting experiments were carried out between sorghum and maize (Koumbaris and Bass 2003). Ma et al. (2010) used Brachypodium distachyon BAC-clone to map the barley genome. Recently, the evolution and taxonomic split of the model grass B. distachyon were analyzed, and substantial phenotypic, cytogenetic, and molecular differences were detected between three cytotypes with the help of chromosome painting (Catalán et al. 2012). The development of comparative chromosome painting paves the way toward comparative chromosome mapping in several crop taxa including Triticeae hexaploid wheat, thereby facilitating the formulation of meaningful breeding program in light of the gene pool diversity.
6.3.5 Alien Chromatin and Transgene Detection
Schwarzacher et al. (1992) ascertained the alien chromatin incorporated from Leymus, Thinopyrum, Hordeum, or Secale in five bread wheat lines by GISH analysis. Friebe et al. (1991) also used GISH to locate the translocation chromosomes in different leaf rust-resistant wheat using GISH technology. Mukai et al. (1993a) also noticed the rye chromatin transfer in wheat. This technique has been effectively applied to detect genome donors in Brassica allopolyploids (Snowdon et al. 1997).
FISH has also analyzed the structure of the transgene loci on interphase nuclei, metaphase chromosomes, and on extended DNA fibers (Forsbach et al. 2003; Chen et al. 2003). Particle bombardment often generates very large, high-copy-number transgenic arrays that can extend for megabases. Interestingly, earlier studies showed that dispersed metaphase FISH signals come together at interphase. By contrast, Agrobacterium transformation gives rise to lower transgene copy numbers and is usually characterized by single discrete FISH signals. Employing molecular cytogenetic approaches, transgenes have been identified in Arabidopsis, barley, and rice, respectively (Forsbach et al. 2003; Chen et al. 2003).
6.4 Modern Molecular Cytogenetics
A biological question may not be solved by a simple localization of DNA sequences in interphase nuclei or on chromosomes. However, physical localization of a DNA sequence together with its associated protein may dramatically enhance the power of FISH. The global patterns of chromatin modification (e.g., DNA methylation and histone tail modifications) along with nuclear size and shape, relative content and distribution of heterochromatin/euchromatin , and organization and structure of chromosomes (e.g., position and orientation) provides new insight into evolution of the particular plant species at chromosomal level. Therefore, it has acquired am important share in this newly developing research field of studying chromatin dynamics through localization of epigenetic signatures of histone/DNA modifications and methylation. It has also been emphasized that amino-terminal tails of histone proteins are targets for a series of posttranslational modifications (PTMs), including acetylation, phosphorylation, and methylation. These modifications regulate chromatin structure and gene expression (Jenuwein and Allis 2001).
6.4.1 Immuno-FISH
Several plant laboratories have developed techniques that combine FISH with immunoassay methods (Jasencakova et al. 2001; Zhong et al. 2002; Nagaki et al. 2005, 2012a, b; Lavania et al. 2012). Such modernization of cytogenetic technique involves an immunoassay of specific antibodies and cytological preparations followed by standard FISH procedure. Immuno-FISH has been used to reveal DNA methylation and histone modifications with specific genomic region. A number of antibodies are available for studying 5mC and histone modifications vis-à-vis chromatin status. Recently, several studies have been conducted on plants using immunohistochemical staining to elucidate chromosomal distribution pattern of the epigenetic marks including Arabidopsis (Zhang et al. 2008), Allium (Suzuki et al. 2010; Nagaki et al. 2012b), maize (Jin et al. 2008; Koo and Jiang 2009; Koo et al. 2011), rice (Yan et al. 2010), brassica (Wang et al. 2011), Barley (Sanei et al. 2011), tobacco (Nagaki et al. 2009), sugarcane (Nagaki et al. 2005), and other taxa (Lavania et al. 2012). Most of the studies suggest that H3K4me1,2,3 mostly mark euchromatin, while H3K9me1 and H3K27me1 mostly target heterochromatin (Fuchs et al. 2006). While H3K9me2 and H3K27me2,3 showed diverse distribution pattern among angiosperms (Fuchs et al. 2006). On the other hand, centromere-specific histone H3 (CENH3) is one of the most fundamental centromeric proteins known to be involved in recruiting other centromeric proteins. CENH3 was first identified as CENP-A in humans (Earnshaw and Rothfield 1985) and subsequently found in a large number of plant species including Brassicaceae, Solanaceae, Leguminosae, Poaceae, and Juncaceae species (Zhong et al. 2002; Telbert et al. 2002; Nagaki et al. 2004, 2005, 2009, 2012a; Sanei et al. 2011; Tek et al. 2011; Wang et al. 2011; Neumann et al. 2012). Since CENH3 comprises part of the core histone that binds directly to DNA at centromeres, centromeric DNA has been isolated from several plant species using antibodies against CENH3 (Nagaki et al. 2003, 2004, 2009, 2011, 2012b; Nagaki and Murata 2005; Tek et al. 2011; Zhong et al. 2002; Neumann et al. 2012; Houben et al. 2007). Immunostaining of chromosomes of Allium species using anti-AfiCENH3 antibody has been shown in Fig. 6.4. Such studies suggest that these histone variants have immense potential to generate extensive information about chromosomal distribution pattern of the epigenetic marks in a wide range of plant species (Sharma et al. 2015).
6.5 Future Prospects
Exciting advances in plant molecular cytogenetic tools and array-based techniques are changing the nature of chromosome biology, in both basic research and at molecular diagnostic levels. Cytogenetic analysis now extends beyond the simple description of the chromosomal status of a genome and allows the study of fundamental biological questions of chromosomal evolution underlying speciation and adaptation. One of the major challenges in plant cytogenetics includes the increment of the resolution power of in situ hybridization and immunostaining techniques to detect shorter nucleotide stretches or single antigen molecules reliably on metaphase chromosomes, extended chromatin fibers and/or in interphase nuclei. Further, improvement of efficient and effective fluorescent chromatin tags for in vivo studies is also needed. FISH may play a powerful role to delineate the structure and DNA composition of long track of highly repeated regions, for example, centromere as well as telomeric ends that are difficult to clone.
As discussed earlier in this article, DNA methylation, nucleosome remodeling (including histone modification and histone variants), and noncoding RNAs can organize chromatin into accessible (euchromatic) and inaccessible (heterochromatic) sub-domains. This extends the information potential of the genetic code, and one genome can generate many ‘epigenomes’ in time and space, during the life span of an organism. The implications of epigenetic research seek attention and efforts that should be targeted to epigenome in a variety of plant systems especially at chromosome inheritance level. In a recent study, it was shown that these epigenetic modifications are not as conserved as was once thought. Further, very little is known about histone/DNA methylation/modification in large genome plants (Houben et al. 2003), which make up the bulk of the angiosperms (Arumuganthan and Earle 1991). Immuno-FISH should be practiced worldwide that has potential to significantly increase the resolving power to reveal fine interaction between DNA and proteins.
6.6 Next-Generation FISH
Next-generation sequencing (NGS) technologies continue to develop at a fast pace, and whole genome sequence of several plants have either been released or to be released soon. NGS technologies of third-generation platforms could produce reads reaching up to a few kilobases, whereas read lengths presently range from 30 to 400 bp depending on the platform. NGS may also facilitate probe development for studies of chromosome using FISH . These genomic regions can be mapped on the chromosome for precise location information with reference to chromosome rearrangements and translocation events and to identify chromosome with/without physical gaps, if any. Further, transcriptome sequencing has also been engaged in construction of large datasets of nuclear genes. NGS is also making the rapid sequencing of complete nuclear genomes routine, thus transforming the genomics research field and opening up new avenues of systematic endeavor in comparative genomics. Further, research should be aimed at understanding the distribution, location, and copy number of the epigenetically inherited gene/genic regions identified through NGS data in several crop/plant species/families in order to shed light on the role of chromatin dynamics in speciation and evolution.
References
Adams Ananiev EV, Phillips RL, Rines HW (1998) Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc Natl Acad Sci USA 95:13073–13078
Arumuganthan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Badaeva ED, Friebe B, Gill BS (1996a) Genome differentiation in Aegilops. 1. Distribution of highly repetitive DNA sequences on chromosomes of diploid species. Genome 39:293–306
Badaeva ED, Friebe B, Gill BS (1996b) Genome differentiation in Aegilops. 1. Physical mapping of 5S and 18S-26S ribosomal RNA gene families in diploid species. Genome 39:1150–1158
Bass HW, Marshall WF, Sedat JW et al (1997) Telomeres cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol 137:5–18
Caspersson T, Farber S, Foley GE et al (1968) Chemical differentiation along metaphase chromosomes. Exp Cell Res 49:219–222
Catalán P, Müller J, Hasterok R et al (2012) Evolution and taxonomic split of the model grass Brachypodium distachyon. Ann Bot 109:385–405
Chen S, Jin W, Wang M et al (2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J 36:105–113
Cheng Z, Buell CR, Wing RA, Jiang J (2002) Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes and extended DNA fibers. Chromosome Res 10:379–387
Cuadrado A, Ceoloni C, Jouve N (1995) Variation in highly repetitive DNA composition of heterochromatin in rye studied by fluorescence in situ hybridization. Genome 38:1061–1069
Danilova TV, Birchler JA (2008) Integrated cytogenetic map of mitotic metaphase chromosome 9 of maize: resolution, sensitivity, and banding paint development. Chromosoma 117:345–356
de Jong J, Fransz P, Zabel P (1999) High resolution FISH in plants—techniques and applications. Trends Plant Sci 4:258–263
Earnshaw WC, Rothfield N (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313–321
Ersfeld K (2004) Fiber-FISH: fluorescence in situ hybridization on stretched DNA. Methods Mol Biol 270:395–402
Fajkus J, Sykorova E, Leitch AR (2005) Telomeres in evolution and evolution in telomeres. Chromosome Res 13:469–479
Forsbach A, Schubert D, Lechtenberg B et al (2003) A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol Biol 52:161–176
Fransz PF, Alonso-Blanco C, Liharska TB et al (1996a) High resolution physical mapping in Arabidopsis thaliana and tomato by fluorescence in situ hybridization to extended DNA fibers. Plant J 9:421–430
Fransz PF, Stam M, Montijn B et al (1996b) Detection of single-copy genes and chromosome rearrangements in Petunia hybrida by fluorescence in situ hybridization. Plant J 9:767–774
Fransz PF, Armstrong S, Alonso-Blanco C et al (1998) Cytogenetics for the model system Arabidopsis thaliana. Plant J 13:867–876
Friebe B, Mukai Y, Dhaliwal HS et al (1991) Identification of alien chromatin specifying resistance to wheat streak mosaic virus and greenbug in wheat germplasm by C-banding and in situ hybridization. Theor Appl Genet 81:381–389
Fuchs J, Demidov D, Houben A, Schubert I (2006) Chromosomal histone modification patterns—from conservation to diversity. Trends Plant Sci 11:199–208
Gall JG, Pardue ML (1969) Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc Natl Acad Sci USA 63:378–383
Gill BS, Friebe B (1998) Plant cytogenetics at the dawn of the 21st century. Curr Opin Plant Biol 1:109–115
Gill BS, Kimber G (1974) Giemsa C-banding and the evolution of wheat. Proc Natl Acad Sci USA 71:4086–4090
Han FP, Fedak G, Benabdelmouna A et al (2003) Characterization of six wheat × Thinopyrum intermedium derivatives by GISH, RFLP and multicolor GISH. Genome 46:490–495
Han F, Liu B, Fedak G, Liu Z (2004) Genomic constitution and variation in five partial amphiploids of wheat-Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor Appl Genet 109:1070–1076
Hanson RE, Zwick MS, Choi S et al (1995) Fluorescent in situ hybridization of a bacterial artificial chromosome. Genome 38:646–651
Hanson RE, Islam-Faridi MN, Percival EA (1996) Distribution of 5S and 18S–28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 105:55–61
Harper LC, Cande WZ (2000) Mapping a new frontier; development of integrated cytogenetic maps in plants. Funct Integr Genomics 1:89–98
Heslop-Harrison JS, Murata M, Ogura Y (1999) Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis thaliana chromosomes. Plant Cell 11:31–42
Hizume M, Shibata F, Matsusaki Y, Garajova Z (2002) Chromosome identification and comparative karyotypic analyses of four Pinus species. Theor Appl Genet 105:491–497
Houben A, Kynast RG, Heim U et al (1996) Molecular cytogenetic characterization of the terminal heterochromatic segment of the B-chromosome of rye (Secale cereale). Chromosoma 105:97–103
Houben A, Demidov D, Gernand D et al (2003) Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J 33:967–973
Houben A, Demidov D, Caperta AD et al (2007) Phosphorylation of histone H3 in plants—a dynamic affair. Biochim Biophys Acta 1769:308–315
Howell EC, Armstrong SJ, Barker GC et al (2005) Physical organization of the major duplication on Brassica oleracea chromosome O6 revealed through fluorescence in situ hybridization with Arabidopsis and Brassica BAC probes. Genome 48:1093–1103
Jasencakova Z, Meister A, Schubert I (2001) Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma 110:83–92
Jenuwein T, Allis C (2001) Translating the histone code. Science 293:1074–1080
Jiang J, Gill BS (1994) Non-isotopic in situ hybridization and plant genome mapping: the first 10 years. Genome 37:717–725
Jiang J, Gill BS (2006) Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49:1057–1068
Jiang J, Gill BS, Wang GL et al (1995) Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA 92:4487–4491
Jiang J, Hulbert SH, Gill BS, Ward DC (1996) Interphase fluorescence in situ hybridization mapping: a physical mapping strategy for plant species with large complex genomes. Mol Gen Genet 252:497–502
Jin WW, Melo JR, Nagaki K et al (2004) Maize centromeres: organization and functional adaptation in the genetic background of Oat. Plant Cell 16:571–581
Jin W, Lamb JC, Zhang W (2008) Histone modifications associated with both A and B chromosomes of maize. Chromosome Res 16:1203–1214
John HA, Birnstiel ML, Jones KW (1969) RNA-DNA hybrids at the cytological level. Nature 223:582–587
Kato A, Lamb JC, Birchler JA (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA 101:13554–13559
Kato A, Vega JM, Han F et al (2005) Advances in plant chromosome identification and cytogenetic techniques. Curr Opin Plant Biol 8:148–154
Khrustaleva LI, Kik C (2001) Localization of single-copy T-DNA insertion in transgenic shallots (Allium cepa) by using ultra-sensitive FISH with tyramide signal amplification. Plant J 25:699–707
Khrustaleva LI, de Melo PE, van Heusden AW, Kik C (2005) The integration of recombination and physical maps in a large-genome monocots using haploid genome analysis in a trihybrid Allium population. Genetics 169:1673–1685
Kim JS, Klein PE, Klein RR (2005) Molecular cytogenetic maps of sorghum linkage groups 2 and 8. Genetics 169:955–965
King J, Armstead IP, Donnison IS et al (2002) Physical and genetic mapping in the grasses Lolium perenne and Festuca pratensis. Genetics 161:315–324
Koo DH, Jiang JM (2009) Super-stretched pachytene chromosomes for fluorescence in situ hybridization mapping and immunodetection of DNA methylation. Plant J 59:509–516
Koo DH, Plaha P, Lim YP et al (2004) A high-resolution karyotype of Brassica rapa ssp. pekinensis revealed by pachytene analysis and multicolor fluorescence in situ hybridization. Theor Appl Genet 109:1346–1352
Koo DH, Choi HW, Cho J et al (2005) A high-resolution karyotype of cucumber (Cucumis sativus L ‘Winter Long’) revealed by C-banding, pachytene analysis, and RAPD-aided fluorescence in situ hybridization. Genome 48:534–540
Koo DH, Han F, Birchler JA, Jiang J (2011) Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res 21:908–914
Koornneef M, Fransz P, De Jong H (2003) Cytogenetic tools for Arabidopsis thaliana. Chromosome Res 11:183–194
Koumbaris GL, Bass HW (2003) A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J 35:647–659
Kurata N, Omura T (1978) Karyotype analysis in rice, I. A new method for identifying all chromosome pairs. Jpn J Genet 53:251–255
Lamb JC, Birchler JA (2006) Retroelement genome painting: cytological visualization of retroelement expansions in the genera Zea and Tripsacum. Genetics 173:1007–1021
Lamb JC, Meyer JM, Corcoran B et al (2007) Distinct chromosomal distributions of highly repetitive sequences in maize. Chromosome Res 15:33–49
Lapitan NL, Ganal MW, Tanksley SD (1989) Somatic chromosome karyotype of tomato based on in situ hybridization of the TGR I satellite repeat. Genome 32:992–998
Lavania UC, Srivastava S, Lavania S (2012) Autopolyploidy differentially influences body size in plants, but facilitates enhanced accumulation of secondary metabolites, causing increased cytosine methylation. Plant J 71:539–549
Le HT, Armstrong KC, Miki B (1989) Detection of rye DNA in wheat-rye hybrids and wheat translocation stocks using total genomic DNA as a probe. Mol Biol Rep 7:150–158
Lim KB, Chung JD, Van Kronenburg BCE et al (2000) Introgression of Lilium rubellum Baker chromosomes into L. Longiflorum Thunb.: a genome painting study of the F1 hybrid, BC1 and BC2 progenies. Chromosome Res 8:119–125
Linde-Laursen I (1975) Giemsa C-banding of the chromosomes of ‘Emir’ barley. Hereditas 81:285–289
Lysak MA, Fransz PF, Ali HBM, Schubert I (2001) Chromosome painting in Arabidopsis thaliana. Plant J 28:689–697
Lysak MA, Pecinka A, Schubert I (2003) Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Res 11:195–204
Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassicaceae. Genome Res 15:516–525
Lysak MA, Mandakova T, Lacombe E (2010) Reciprocal and multi-species chromosome BAC painting in crucifers (Brassicaceae). Cytogenet Genome Res 129:184–189
Ma L, Vu GT, Schubert V et al (2010) Synteny between Brachypodium distachyon and Hordeum vulgare as revealed by FISH. Chromosome Res 18:841–850
McClintock B (1929) A cytological and genetical study of triploid maize. Genetics 14:180–222
McClintock B (1932) A correlation of ring-shaped chromosomes with variation in Zea mays. Proc Natl Acad Sci 18:677–681
McClintock B (1938) The fusion of broken ends of sister half-chromatids following breakage at meiotic anaphase. Mo Agric Exp Stat Res Bull 290:1–48
McClintock B (1941a) The association of mutants with homozygous deficiencies in Zea mays. Genetics 26:542–571
McClintock B (1941b) The stability of broken ends of chromosomes in Zea mays. Genetics 26:234–282
McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801
Mukai Y (1996) Multicolor fluorescence in situ hybridization: a new tool for genome analysis. In: Jauhar PP (ed) Methods of genome analysis in plants. CRC Press, Boca Raton, pp 181–192
Mukai Y (1999) Molecular cytogenetic studies on chromosomes of useful plants. Breed Res 1:165–172 (Japanese)
Mukai Y (2004) Fluorescence in situ hybridization. In: Goodman RM (ed) Encyclopedia of plant and crop science. Marcel Dekker, New York, pp 468–471
Mukai Y, Appels R (1996) Direct chromosome mapping of plant genes by in situ polymerase chain reaction (in situ PCR). Chromosome Res 4:401–404
Mukai Y, Gill BS (1991) Detection of barley chromatin added to wheat by genomic in situ hybridization. Genome 34:448–452
Mukai Y, Yamamoto M (1998) Application of multicolor fluorescence in situ hybridization to plant genome analysis. In: Gupta PK (ed) Genetics and biotechnology in crop improvement. Rastogi Publication, Meerut, India, pp 14–23
Mukai Y, Endo TR, Gill BS (1990) Physical mapping of the 5S rRNA multigene family in common wheat. J Heredity 81:290–295
Mukai Y, Endo TR, Gill BS (1991) Physical mapping of the 18S-26S rRNA multigene family in common wheat: identification of a new locus. Chromosoma 100:71–78
Mukai Y, Friebe B, Hatchett JH, Yamamoto M, Gill BS (1993a) Molecular cytogenetic analysis of radiation-induced wheat-rye terminal and intercalary chromosomal translocations and the detection of rye chromatin specifying resistance to Hessian fly. Chromosoma 102:88–95
Mukai Y, Nakahara Y, Yamamoto M (1993b) Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 36:489–494
Nagaki K, Murata M (2005) Characterization of CENH3 and centromere—associated DNA sequences in sugarcane. Chromosome Res 13:195–203
Nagaki K, Talbert PB, Zhong CX (2003) Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163:1221–1225
Nagaki K, Cheng Z, Ouyang S et al (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Nagaki K, Kashihara K, Murata M (2005) Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell 17:1886–1893
Nagaki K, Kashihara K, Murata M (2009) A centromeric DNA sequence colocalized with a centromere-specific histone H3 in tobacco. Chromosoma 118:249–257
Nagaki K, Shibata F, Suzuki G et al (2011) Coexistence of NtCENH3 and two retrotransposons in tobacco centromeres. Chromosome Res 19:591–605
Nagaki K, Shibata F, Kanatani A et al (2012a) Isolation of centromeric-tandem repetitive DNA sequences by chromatin affinity purification using a HaloTag7-fused centromere-specific histone H3 in tobacco. Plant Cell Rep 31:771–779
Nagaki K, Yamamoto M, Yamaji N et al (2012b) Chromosome dynamics visualized with an anti-centromeric histone H3 antibody in Allium. PLoS ONE 7(2):e51315
Nakano A, Suzuki G, Yamamoto M et al (2005) Rearrangements of large-insert T-DNA in transgenic rice. Mol Genet Genomics 273:123–129
Neumann P, Navratilova A, Schroeder-Reiter E et al (2012) Stretching the rules: monocentric chromosomes with multiple centromere domains. PLoS Genet 8(6):e1002777
Pardue ML, Gall JG (1975) Nucleic acid hybridization to the DNA of cytological preparations. Methods Cell Biol 10:1–16
Pederson C, Langridge P (1997) Identification of the entire chromosome complement of bread wheat by two-color FISH. Genome 40:589–593
Pedrosa A, Sandal N, Stougaard J et al (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161:1661–1672
Pinkel DT, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 85:2934–2938
Qi LL, Pumphrey MO, Friebe B et al (2008) Molecular cytogenetic characterization of alien introgressions with gene Fhb3 for resistance to Fusarium head blight disease of wheat. Theor Appl Genet 117:1155–1166
Raap A, van de Corput M, Vervenne R et al (1995) Ultra-sensitive FISH using peroxidase-mediated deposition of biotin- or fluorochrome tyramides. Hum Mol Genet 4:529–534
Raina SN, Mukai Y (1999) Genomic in situ hybridization identifies the diploid wild progenitors of cultivated (Arachis hypogaea) and related wild (A. monticola) peanut species. Pl Syst Evol 214:251–262
Raina SN, Mukai Y, Yamamoto M (1998) In situ hybridization identifies the diploid progenitor species of Coffea arabica (Rubiaceae). Theor Appl Genet 97:1204–1209
Sanei M, Pickering R, Kumke K et al (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci USA 108:E498–E505
Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS (1989) In situ localization of parental genomes in a wide hybrid. Ann Bot 63:315–324
Schwarzacher T, Anamthawat-Jónsson K, Harrison GE et al (1992) Genomic in situ hybridization to identify alien chromosomes and chromosome segments in wheat. Theor Appl Genet 84:778–786
Sharma SK, Mehra P, Kumaria S et al (2012) Physical localization and probable transcriptional activity of 18S–5.8 S–26S rRNA gene loci in some Asiatic Cymbidiums (Orchidaceae) from north-east India. Gene 499:362–366
Sharma SK, Yamamoto M, Mukai Y (2015) Immuno-cytogenetic manifestation of epigenetic chromatin modification marks in plants. Planta 241:291–301
Shizuya H, Birren B, Kim UJ (1992) Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci 89:8794–8797
Snowdon RJ, Kohler W, Kohler A (1997) Chromosomal localization and characterization of rDNA loci in the Brassica A and C genomes. Genome 40:582–587
Stephens JL, Brown SE, Lapitan NL, Knudson DL (2004) Physical mapping of barley genes using an ultrasensitive fluorescence in situ hybridization technique. Genome 47:179–189
Suzuki G, Mukai Y (2004) Plant BAC libraries as tools for molecular cytogenetics. In: Williams CR (ed) Focus on genome research. Nova Science Publishers, New York, pp 205–220
Suzuki G, Ura A, Saito N, Do GS, Seo BB, Yamamoto M, Mukai Y (2001) BAC FISH analysis in Allium cepa. Genes Genet Syst 76:251–255
Suzuki G, Tanaka S, Yamamoto M (2004) Visualization of the S-locus region in Ipomoea trifida: toward positional cloning of self-incompatibility genes. Chromosome Res 12:475–481
Suzuki G, Shiomi M, Morihana S (2010) DNA methylation and histone modification in onion chromosomes. Genes Genet Syst 85:377–382
Takahashi C, Marshall JA, Bennett MD, Leitch IJ (1999) Genomic relationships between maize and its wild relatives. Genome 42:1201–1207
Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S (2002) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14:1053–1066
Tek AL, Kashihara K, Murata M, Nagaki K (2011) Functional centromeres in Astragalus sinicus include a compact centromere-specific histone H3 and a 20-bp tandem repeat. Chromosome Res 19:969–978
Valarik M, Bartos J, Kovarova P et al (2004) High-resolution FISH on super-stretched flow-sorted plant chromosomes. Plant J 37:940–950
Vosa CG, Marchi P (1972) Quinacrine fluorescence and giemsa staining in plants. Nature New Biol 237:191–192
Wang CJ, Harper L, Cande WZ (2006) High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 18:529–544
Wang G, He Q, Cheng Z (2011) Characterization of CENH3 proteins and centromere-associated DNA sequences in diploid and allotetraploid Brassica species. Chromosoma 120:353–365
Xu J, Earle ED (1996) High resolution physical mapping of 45S (5.8S, 18S and 25S) rDNA gene loci in the tomato genome using a combination of karyotyping and FISH of pachytene chromosomes. Chromosoma 104:545–550
Yamamoto M (1994) Molecular-cytogenetic analysis of the rDNA region in Triticum and Aegilops. Ph.D. thesis, Kyoto University, Japan, p 248
Yamamoto M, Mukai Y (1991) Fluorescence in situ hybridization: a new approach to study chromosome structure. In: Proceedings of 2nd international symposium on chromosome engineering in plants, Columbia, MO, Aug 13–15, pp 296–301
Yamamoto M, Mukai Y (2007) Extended DNA fiber FISH in plants: visible message from cell nuclei. The Nucleus 50:439–452
Yan H, Kikuchi S, Neumann P et al (2010) Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J 63:353–365
Zhang WL, Lee HR, Koo DH, Jiang JM (2008) Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell 20:25–34
Zhong XB, Fransz PF, Wennekes-van E (1996) High-resolution mapping on pachytene chromosomes and extended DNA fibres by fluorescence in-situ hybridization. Plant Mol Biol Rep 14:232–242
Zhong CX, Marshall JB, Topp C et al (2002) Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14:2825–2836
Zwierzykowski Z, Zwierzykowska E, Taciak M (2008) Chromosome pairing in allotetraploid hybrids of Festuca pratensis × Lolium perenne revealed by genomic in situ hybridization (GISH). Chromosome Res 16:575–585
Acknowledgement
We thank ‘Japan Society for the Promotion of Science (JSPS)’ and ‘Osaka Kyoiku University,’ Osaka, Japan, for providing research funds (SKS: P13399, 2013) and facilities for plant molecular cytogenetic research, respectively. Sincere thanks to Dr. Go Suzuki and all members of Plant Molecular Genetics Laboratory, Osaka Kyoiku University, Osaka, Japan, for their constant encouragement and help.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Sharma, S.K., Yamamoto, M., Mukai, Y. (2016). Molecular Cytogenetic Approaches in Exploration of Important Chromosomal Landmarks in Plants. In: Rajpal, V., Rao, S., Raina, S. (eds) Molecular Breeding for Sustainable Crop Improvement. Sustainable Development and Biodiversity, vol 11. Springer, Cham. https://doi.org/10.1007/978-3-319-27090-6_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-27090-6_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-27088-3
Online ISBN: 978-3-319-27090-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)