Abstract
The kidney shows many pathophysiological changes during aging, such as glomerulosclerosis, tubular atrophy, interstitial fibrosis and arterial intimal fibrosis. Recent work has begun to reveal the genes and pathways that are responsible for some of these age-related changes in the kidney. Understanding the genetic and molecular basis for renal aging is of clinical interest because donor age is a major criterion for success in renal transplantation. Kidneys from cadaveric donors over age 60 are considered less desirable and show shorter median survival times. There is a surplus of patients in need of kidney transplantations resulting in many premature deaths, and yet many potential donor kidneys are discarded on the basis of donor age alone. Insight into the mechanism of renal aging could lead to better criteria to identify a subset of kidneys from elderly donors that are suitable for transplantation. Further, it may soon be possible to slow down or rejuvenate renal aging. Better understanding of the renal aging process could help alleviate the critical shortage of kidneys for transplants.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The kidney plays an important role in human aging because it shows stereotyped changes in morphology and physiology beginning around age 50. Kidneys show clear changes in structure and morphology with age. Starting at about age 50, the weight and volume of the kidney shrinks by about one third [1]. The glomerulus is a network of capillaries that is located at the beginning of the nephron that filters blood to form urine. The number of glomeruli declines by one third to one half in old age through obsolescence or glomerulosclerosis. The tubules associated with the sclerosed glomeruli cease to function and the filtration capacity of each kidney declines. As the tubules atrophy, the tubular epithelium shrinks, the tubules contract and the basement membranes of the tubules thicken. Interstitial fibrosis increases with age, and refers to an increase in connective tissue in the space between the tubules. With age, the walls of arterioles become thick, caused by a deposition of hyaline. Hyaline is composed of plasma protein (for example C3 and IgM) that has leaked across the endothelium and accumulated in the wall of the arterioles. In Fig. 1, the kidney section on the right is from a 70 yo and shows many of the morphological hallmarks of aging: atrophied tubules, glomerulosclerosis and interstitial fibrosis.
Physiological kidney aging. Shown are two kidneys from elderly donors with a similar chronological age. The left kidney is physiologically younger than the right kidney. The kidney on the right shows classic signs of structural and morphological changes associated with renal aging, decreased kidney function, and poor renal transplant outcome. The glomeruli show signs of scarring (glomerulosclerosis). The cells in the interstitial space have thickened extracellular membranes indicative of fibrosis (interstitial fibrosis). The tubules are smaller, have thickened walls and have atrophied (tubular atrophy). Genetic and molecular biomarkers for physiological aging could be used to distinguish kidney donors based on physiological age. This could rescue renal organs such as the one on the left from exclusion, possibly making them eligible for renal transplant. The net effect would be to expand the pool of renal donors available for patients with end stage renal disease
The rate of filtration of the blood through the glomerulus (i.e. the glomerular filtration rate; GFR) is one of the primary indicators of renal function. On average, the glomerular filtration rate begins to decline at age 40, although the rate of decline is different in different individuals [1–3]. The loss of renal function due to advancing age may become clinically significant over a normal human life span. In the elderly, glomerular filtration rate often reaches levels low enough to indicate chronic kidney disease. By age 70, 35 % of people have moderate chronic kidney disease (stage 3) according to the National Health and Nutrition Examination Survey [4]. A healthy GFR is ≥90 ml/min for an adult, but when the GFR falls to <15 ml/min the patient is considered to have end stage renal disease. Patients with end stage renal disease require dialysis in order to survive as the blood no longer receives adequate renal filtration, but simply going on dialysis doubles the 5-year risk for mortality. Renal transplantation is preferable to dialysis for end stage renal disease because the donated kidney can function at a relatively normal level and restore glomerular filtration rate. Both quality of life and survival are greatly improved by transplantation compared to dialysis [6]. The decline in glomerular filtration rate is likely caused by structural changes to the glomerulus, the interstitium and the arterioles [3, 7].
Understanding the genetic and molecular mechanisms that contribute to kidney aging will advance our basic understanding of the aging process in humans. Furthermore, aging research on the kidney could have important clinical applications. In the long run, a better understanding of renal aging could lead to strategies or treatments to delay the aging process. This could delay or prevent chronic kidney disease and reduce the number of people suffering from end stage renal disease.
In the short run, one promising opportunity is to use knowledge of aging to develop biomarkers in order to measure physiological age, as opposed to chronological age. For instance, from a cohort of elderly, it would be desirable to be able to identify those that have physiologically young kidneys. Figure 1 illustrates two kidneys from donors of similar chronological age of about 70 years. The kidney on the left retains a youthful morphological appearance, equivalent to the appearance of kidneys from middle-aged donors, suggesting that this kidney is physiologically young. The kidney on the right shows classic signs of aging. Figure 1 illustrates the concept that adding information from histopathological and molecular biomarkers to chronological age can improve our knowledge of the true age of an organ better than chronological age alone
Individuals with kidneys that are physiologically younger are likely to show a lower incidence of renal disease as they grow older. Furthermore, donor age is the major criterion for success of a kidney in renal transplantation [8, 9], which means that individuals with kidneys that are physiologically young are likely to be better renal transplantation donors than individuals with kidneys that are physiologically old irrespective of their chronological age. Instead of categorically discarding all of the organs from donors above a certain age, it may be possible to select a subset of organs that are physiologically young and suitable for transplantation (Fig. 2). Renal transplant outcome declines gradually with age, and the difference between youthful and elderly kidney donors is relative but not absolute. With elderly renal donors, the fraction of renal transplants that are successful (as measured by graft survival after 1 and 5 years) is lower than the fraction of successful transplants from youthful donors. Still, some of the transplants from elderly donors are successful. Figure 2 illustrates the concept of using physiological age to help increase the pool of renal transplant donors. Exclusion criteria based on chronological age alone become increasingly strong as the donor ages (gradient arrow). Aging biomarkers could be used to provide information about the physiological age of the tissue, which might permit certain prospective donors (dots shown in red) by expanding the criteria to include physiological in addition to chronological age. This strategy would expand the pool of kidney organs suitable for transplantation, and thereby allow patients with end stage renal disease to receive a transplant and end their time on dialysis treatment.
Kidney aging and renal transplantation. Figure depicts how some kidneys from elderly donors may be suitable for renal transplantation. With increasing age of the donor, there is a steady decline in the percent of renal transplants that survive 1 and 5 years after transplantation. Nevertheless, there are many renal transplants from elderly donors that last for a suitable length of time. In principle, aging biomarkers could be used to identify kidneys that are physiologically young, and perhaps could be used to rescue organs that are currently discarded due to old age. The gradient arrow indicates how donor age becomes a stronger criterion for exclusion with increasing chronological age. The red triangles indicate kidney donors that may still be suitable for renal transplantation, even though their chronological age may have exceeded an exclusion criteria cut-off that is currently used. The black dots indicate individual donor kidneys
2 Renal Transplantation
Renal transplantation is the best option for patients with end stage renal disease. The survival rates for kidneys in recipients following renal transplantation are 80 % after 1 year and about 60 % after 5 years [6, 10, 11]. Renal transplantation can possibly extend the lives of patients by 10–15 years compared to dialysis [6].
However, there are many more patients with end stage renal disease than there are renal transplantation donors. In 2014, there were 101,513 people in the United States on the waiting list for kidney transplantations. At the same time, there were only about 13,125 donor kidneys available [12]. Some patients with end stage renal disease receive a kidney from a living donor. In the donation process, there is a large number of volunteers that offer to donate their kidney. Most of the volunteered kidneys are excluded from becoming a kidney donor for medical reasons, including old age. In a recent study of kidney donors at Stanford University from 2007 to 2009, it was found that 92 % of potential donor kidneys were excluded from consideration, exacerbating the shortage of kidneys available for transplantation [13]. As a result of the shortage of donor kidneys, many patients with end stage renal disease do not have the opportunity to receive a donor kidney for renal transplantation, an operation that would extend their lives. In principle, improvements in the criteria for exclusion might allow one to rescue potential donor kidneys that might be suitable for renal transplantation even though they are currently excluded from renal transplantation.
2.1 Predictors of Renal Transplant Outcome
Currently, there are three main criteria affecting the success of renal transplantation: ABO blood type and HLA histocompatibility matching, kidney preservation time and donor age. Donor kidneys that do not match the recipient for ABO blood type and HLA histocompatibility are at risk for graft rejection. Many kidneys are stored cold while awaiting the transplantation procedure, especially kidneys from deceased donors.
The third factor, age, is important for renal transplantation success as greater age of the donor diminishes the chance of success of the renal transplant. On the average, kidneys from older donors have a shorter graft survival time than those from younger donors. The short-term difference is relatively minor, but is amplified with the passage of time: About 95 % of kidneys have a graft survival greater than 1 year when the donor was younger than age 50, and about 85 % of kidneys have a 1 year graft survival rate when the donor was over age 65 [14]. At 5 years after the transplantation, there is about a 25 % increase in renal survival in kidneys from younger donors compared to those from elder donors [15]. Thus, while on average there is a drop off in graft survival from elder donors, there is also a significant number of exceptions where a kidney from an elder donor has a long graft survival time [16–21].
If we could better understand why old kidney age affects graft survival, it might be possible to identify kidneys from the elderly population that are still fit for renal transplantation. One way to do this is to develop a set of biomarkers for physiological age that could predict renal transplant outcome better than chronological age, or at least that could be used to improve transplant outcome in combination with chronological age. That is, among elderly donors of the same age, the kidney aging biomarker should be able to identify donors with a higher chance for long term graft survival. This might be one way to expand the pool of donor kidneys available for renal transplantation. Several molecular assays are being developed as biomarkers for renal graft survival.
Recent studies have begun to identify biomarkers of aging that can be used to help predict how well a kidney will perform in renal transplantation. Aging is a complex process dependent on many different mechanisms and pathways. One of the cellular pathways that may contribute to aging of the kidney is cell senescence [22–26]. Cell senescence could impact renal function if it prevented cell division necessary to replace lost or damaged cells. However, cell turnover in the kidney is normally low compared to other tissues with high rates of cell turnover, such as the hematopoietic system or the lining of the gut [27]. Thus, it is unclear how strongly cell senescence would impact renal function under normal circumstances. However, disease or injury could result in loss of renal cells by apoptosis, and in this situation cell proliferation would be required in order to replace dead cells.
As kidneys grow old, there is an increased frequency of senescent cells. Senescence could contribute to renal aging in at least three ways. First, cell senescence may prevent new cells from replenishing cells that are lost from disease or damage. Second, cell senescence could lead to an increase of macromolecular damage. When a cell divides, there is a burst of new synthesis of all of the macromolecules needed to form the new cells (DNA, RNA, protein etc.). Conversely, in any post-mitotic cell such as a senescent cell, there is no net gain of RNA and protein, so new RNA and proteins are synthesized via transcription and translation at a much lower rate. Assuming that RNA and protein levels in senescent cells are at steady state, then the levels of transcription and translation are set to merely replace RNA as it is lost via RNA degradation and protein as it is lost by degradation via proteolysis machinery such as autophagosomes or the proteasome. As a result, macromolecules in a non-dividing cell have a longer molecular half-life than those in a dividing cell. The increased molecular half-life exposes all of the molecules to increased susceptibility for damage accumulation; for instance, macromolecules in a non-dividing cell would be expected to have higher levels of damage from reactive oxygen species. Third, senescent cells secrete a variety of signaling molecules and cytokines, a phenomenon referred to as the senescence-associated secretory phenotype [28]. The senescence-associated cytokines include factors such as interleukin-6 and interleukin-1β that can activate inflammatory signaling pathways. One possibility is increased abundance of senescent cells during aging contributes to chronic inflammation.
One marker of senescent cells is expression of the cell cycle regulator CDKN2A/p16. CDKN2A/p16 plays an important role in cell cycle regulation by decelerating progression from the G1 to the S phase [29]. In the normal cell cycle, CDKN2A/p16 acts to inhibit cell division by binding CDK4/6, which ultimately inhibits the activity of transcription factors such as E2F1 and arrests cell proliferation [30]. High levels of CDKN2A/p16 expression prevent cell division and are a hallmark of cell senescence [31]. Expression of CDKN2A/p16 increases with age in the kidney [32, 33]. Several studies have shown that expression levels of CDKN2A/p16 can be used as a biomarker of aging in order to predict renal transplant outcome. At the time of the renal transplant, a kidney biopsy was obtained and CDKN2A/p16 levels were measured [22, 34, 35]. Age, CDKN2A/p16 levels, and a combination of Age/ CDKN2A/p16 levels were evaluated as predictors for renal transplant success. CDKN2A expression (biological age) was found to be better than donor age (chronological age) in predicting organ function [35]. However, CDKN2A/p16 levels combined with chronological age was found to be the most powerful predictor for renal function following transplantation [22]. The key concept is that CDKN2A/p16 levels may be measuring biological age, and that biological age may be better than simple chronological age as a predictor of future renal function following transplantation.
Another strategy to develop biomarkers for renal transplantation is to identify gene expression signatures that can predict renal graft survival. In this approach, gene expression from the entire genome is measured from kidney biopsies at the time of transplantation using DNA chips. The renal transplants are then separated into two groups based on success of the graft, and the gene expression data are analyzed to identify differences in expression between kidneys that were or were not successful in the renal transplant. In one study, 31 renal allografts were separated into low and high GFR after 1 year following transplantation. Then, expression profiles taken at the time of transplantation were analyzed, resulting in the identification of 52 genes that showed significantly different expression profiles between the high- and low-functioning kidneys [36]. The gene expression profile of these 52 genes at the time of transplantation was able to predict the success of the renal transplantation over a medium term.
In a second study, 92 renal allografts were separated based on whether or not patients required dialysis during the first week (delayed graft function), and then gene expression data were analyzed to identify 206 genes whose expression showed a significant difference. This study suggests that preimplant gene expression profiles may be able to identify kidneys of poor quality that perform poorly in transplantation. This information may eventually improve organ allocation [37].
3 Hallmarks of Renal Aging
To better understand renal aging, it is useful to consider mechanisms that are involved in aging in other tissues, and even other species such as mice, flies, worms and yeast. The human aging process can be thought of as a clock that spans about 80 years. Aging mechanisms guide the rate at which this clock proceeds, and the most central pathways are part of the clock mechanism itself. As we grow old, aging affects many of the underlying networks in the kidney. There is accumulation of damage of diverse types to cellular components such as DNA, proteins and lipids. There are changes in gene expression and epigenetic networks with age. Cells lose their ability to divide and undergo senescence. There is a steady increase in the thickness of the extracellular matrix that is a major determinant of fibrosis. It is possible that changes in each of these networks serves as part of a molecular aging clock, that changes over time and dictates the rate of functional decline of the kidney.
A hallmark of aging not only changes as we grow old, but it also plays an important functional role in the physiological decline of the kidney with old age. Genetic and pharmacological experiments that reset the clock in old cells or organs to the young state should have a beneficial effect. By contrast, experiments in which the aging pathway has been reset in young cells to the old state should cause rapid aging to ensue.
These hallmarks of kidney aging might collectively form a type of aging clock that dictate the functional and physiological state of the kidney over a lifetime. Although these pathways progress during aging in everyone, the rate of progression can vary between people. For people of the same chronological age, the aging clock might be slightly more advanced in one person than the other. With additional studies, hallmarks of aging could one day become very important because they could be used as biomarkers to report the true physiological age of a person or tissue, rather than mere chronological age. Not only would the aging biomarkers associate with the current functional state of the kidney better than chronological age, but the aging biomarkers would be better than chronological age at predicting the future trajectory of renal decay.
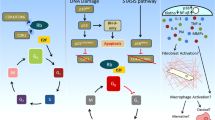
A recent review describes nine hallmarks of aging that form the conceptual pillars to understand changes as one grows old [38]. These hallmarks are: genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient-sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication. In addition to the general hallmarks, kidney aging is known to involve increased Klotho expression, chronic inflammation, and fibrosis (Fig. 3). Below, we consider each of these hallmarks and summarize what is known about how that hallmark may contribute to human renal aging.
One of the most important hallmarks of kidney aging is chronic inflammation [39]. Chronic inflammation occurs when there is an increased abundance of immune cells – B cells, T cells, neutrophils and macrophages. Low levels of activity of the immune cells lead to a low grade inflammatory response that contributes to fibrosis and tissue damage with age. One of the causes of chronic inflammation in the kidney may be increased systemic levels of inflammatory cytokines, such as IL1, IL6, and TNFα, in old age [40, 41]. The inflammatory response leads to increased production and accumulation of fibrinogen and C-reactive protein by the liver, leading to increased systemic levels of inflammatory biomarkers. The level of chronic inflammation is higher in patients with chronic kidney disease compared to healthy age-matched controls, suggesting that chronic inflammation may play a role in the etiology of this disease [39]. However, the relationship between cause and effect between chronic inflammation and renal aging is unclear; specifically, it is unclear whether chronic inflammation causes renal damage to accumulate with age, whether age-related renal damage causes chronic inflammation, or whether both act together in a complicated feedback loop with increasingly dire consequences.
The inflammatory cytokines that are responsible for chronic inflammation could arise from several sources. One source is from immune cells (B cells, T cells and macrophages) that become dysregulated in old age [42, 43]. A second source is from adipocytes, which are known to produce many inflammatory cytokines, including TNFα and IL-6 [44]. Cytokines secreted from adipose tissue would enter the blood system and increase chronic inflammation throughout the body, including the kidney. A third source is from senescent cells, which secrete many inflammatory cytokines as part of the senescence phenotype [45]. Senescent cells increase in number in old age, leading to increased production of the inflammatory cytokines, a phenomenon termed the senescent-associated secretory phenotype [28]. A fourth source is from cells within the kidney itself. DNA microarray analysis showed that aged human kidneys have increased expression of certain inflammatory cytokines and chemokines [46]. One of the results of chronic inflammation is the recruitment of immune cells that secrete inflammatory cytokines, which may further increase chronic inflammation in the kidney as part of a positive feedback loop.
The inflammatory cytokine TNFα activates initiates a signaling cascade leading to activation of the transcription factor NFκB. The TNFα signaling pathway is induced when TNFα binds the TNF receptor (TNFR1), which leads to the degradation of IKB, a protein that normally keeps the NFκB complex inactive in the cytoplasm. Once IKB is degraded, the NFκB complex is no longer tethered in the cytoplasm and enters the nucleus where it activates expression of its target genes, which include many inflammatory cytokines and chemokines. Activity of NFκB in the kidney has been found to increase with age in the rat [47]. One of the major functions of NFKB is to mediate inflammatory and innate immune responses. Besides TNFα, the inflammatory cytokine IL-1, lipopoysacharide, and reactive oxygen species can activate NFκB.
Another hallmark of renal aging is fibrosis of the interstitium or scarring of the glomeruli [48]. The glomeruli are the functional unit of the kidney responsible for filtering the blood. The tubules and interstitium constitute 90 % of the volume of the kidney. Interstitial fibrosis and glomerulosclerosis increase with age, characterized by an increased thickening of the extracellular matrix. Extracellular matrix is composed primarily of collagen. Matrix metalloproteases are zinc-dependent endopeptidases responsible for degrading collagen and proteoglycans, and may function to help remodel the extracellular matrix. Excess production of collagen or altered expression of matrix metalloproteases could play a role in thickening of the extracellular matrix in old age. Mesenchymal cells (i.e. fibroblasts and myofibroblasts) produce extracellular matrix. Increased numbers or altered functions of these cells in old age could be responsible for increased collagen deposition and fibrosis. Activated interstitial mesenchymal cells are thought to contribute directly to renal fibrosis by secreting fibrotic factors and extracellular matrix proteins that accumulate in the interstitial space and disrupt normal epithelial architecture [48].
Changes in DNA methylation and DNA repair play a role in fibrosis. Bechtel et al. found that increased DNA methylation plays a key role in renal fibrosis. They performed a genome-wide screen to look for DNA methylation differences between fibroblasts from fibrotic and non-fibrotic kidneys, and found 12 genes that showed increased levels of DNA methylation in the fibrotic kidneys [49]. They then inhibited DNA methylation using 5-azacytidine and found that fibrosis was lessened [49]. These results indicate that increased DNA methylation plays a causative role in causing renal fibrosis.
Another clue about the mechanisms responsible for renal fibrosis was obtained by studying a heriditary form of chronic kidney disease. Karyomegalic interstitial nephritis is a rare hereditary form of chronic kidney disease with only 12 known families worldwide [50]. Kidneys in patients with karyomegalic interstitial nephritis have enlarged hyperchromatic nuclei and develop chronic kidney disease in their third decade. The specific diagnostic traits associated with chronic kidney disease in these patients include interstitial fibrosis, tubular atrophy and microcyst formation. These symptoms are seen in normal elder subjects as part of kidney aging, suggesting that karyomegalic interstitial nephritis involves an accelerated rate of fibrosis in the kidney.
The genetic cause for karyomegalic interstitial nephritis has recently been found to be due to mutations in the FAN1 gene. FAN1 plays a role in DNA repair. Specifically, FAN1 is required to repair interstrand DNA crosslinks, so that covalently cross-linked DNA strands cannot separate during S-phase in patients with a non-functional form of FAN1 [51, 52]. In renal tissue from patients with karyomegalic interstitial nephritis, there was an increased level of double strand breaks. These results show that DNA damage caused by reduced activity of FAN1 can potentiate renal fibrosis.
Nutrient sensing is a general hallmark of aging in all tissues and most species. Caloric restriction can extend lifespan of nearly every species tested, including yeast, worms, flies, mice and primates [53]. Recent work has begun to unravel the molecular mechanisms underlying lifespan extension due to caloric restriction [54–56]. These molecular mechanisms involve key modulators of aging such as the sirtuins and mTOR.
Sirtuins are a family of protein deacetylases that are involved in a diverse array of cellular processes such as life span regulation, fat mobilization in human cells, insulin secretion and caloric restriction [57]. In mammals, there are seven sirtuin genes (SIRT1 to SIRT7). Activation of sirtuins has been shown to extend lifespan in diverse organisms including yeast, worms, flies and mice [58]. In the kidney, SIRT1 is abundantly expressed in renal medullary interstitial cells, where it is cytoprotective and participates in the regulation of blood pressure and sodium balance [59, 60]. Sirtuins require nicotinamide adenine dinucleotide (NAD+) as a cofactor, and hence could be responsive to changes in metabolic state of the cell. Levels of nicotinamide adenine dinucleotide show a marked decline in the kidney in old age, which may decrease activity of sirtuins thereby contributing to cellular dysfunction in old age [59, 61]. Regulation of sodium balance by SIRT1 involves repression of expression of the α-subunit of the epithelial sodium channel, ENaC [59].
Genetic experiments by He et al., 2010 have shown that Sirt1 activity plays an important role in kidney function [60]. On the one hand, lowering Sirt1 activity impairs renal function. For example, reduced Sirt1 expression in mouse renal cells in vitro leads to reduced resistance to oxidative stress. Genetically mutant mice with reduced Sirt1 activity (Sirt1+/−) show lower levels of cyclooxygenase-2 (COX2) expression and impaired kidney function under several conditions. Sirt1+/− mice show more renal apoptosis and fibrosis in mice that have suffered kidney injury by unilateral ureteral obstruction. On the other hand, increasing Sirt1 activity improves renal function. Sirt1 activity can be pharmacologically increased either by resveratrol or by the drug SRT2183. Pharmacologic activation of Sirt1 improves cell survival in response to oxidative stress, attenuates renal apoptosis and fibrosis, and increases expression of COX2 in the renal medulla.
mTOR is a conserved kinase that acts in a nutrient sensing pathway to regulate cell growth and longevity. In yeast, worms and flies, mutants with reduced TOR activity have longer lifespans [56]. In mice, inhibition of mTOR activity with the drug rapamycin results in extended lifespan [56]. During normal kidney aging, mTOR expression shows a marked increase in glomerular mesangial cells in old age [62]. This result suggests that increased levels of mTOR in the kidney may contribute to poor cell regulation and cell senescence in old age.
Klotho plays an important role in aging of the kidney [63]. Klotho is a transmembrane protein primarily expressed in the distal tubule cells of the kidney and the brain choroid plexus. Loss-of-function mutations in the Klotho gene in mice are associated with symptoms of premature aging, including hyperphosphatemia and a shorter lifespan [64]. Conversely, overexpression of Klotho extends lifespan [64]. In humans, serum levels of Klotho decrease with age after age 40 years and there are low levels of Klotho in patients with Chronic Kidney Disease [65–67]. One of the main functions of Klotho is to act as a co-receptor for Fibroblast Growth Factor 23 [68]. However, there is also evidence that Klotho can also regulate the Insulin-like Growth Factor signaling pathway, can participate in Ca2+ homeostasis, phosphate homeostasis and can relieve oxidative stress [69].
Changes in gene expression occur with age, and can be used to predict the physiological age of kidneys. DNA microarrays have been used to define the changes in gene expression that accompany the renal aging process. One study analyzed RNA from 74 patients ranging in age from 27 to 92 years, and found 985 genes to change expression with age [46]. Among these 74 individuals, there was a good correlation between the gene expression signature and the biological age of the kidney. For example, for some kidney samples, the gene expression signatures did not resemble kidneys of their chronological age but rather kidneys from individuals that were younger. Subsequent histological examination of these renal samples showed that they had lower levels of interstitial fibrosis, arterial hyalinosis and glomerulosclerosis than typically found in kidneys of that chronological age. That is, the gene expression signatures were able to accurately predict that these renal samples had a biological age that was younger than their chronological age. Similarly, the gene expression signatures were also able to predict renal samples with a biological age that is older than their chronological age. A second study analyzed RNA isolated from 20 kidney samples and identified about 500 genes that changed expression with age [70]. The sets of kidney aging genes showed a large degree of overlap in the two DNA microarray studies.
What are the upstream transcription factors responsible for causing the changes in expression of the age-regulated kidney genes? In order to identify the upstream regulators, ChIP-seq data from the ENCODE consortium were examined. The ENCODE consortium has defined the in vivo binding sites for a large number of transcription factors (about 160) [71]. For each transcription factor, they used ChIP seq to first immunoprecipitate the transcription factor from tissue culture cells, and then sequenced the bound DNA in the immunoprecipitate. Each experiment resulted in a list of target genes bound by that transcription factor in specific cell lines. To find transcription factors that bind to the age-related kidney genes, a bioinformatics screen was performed to search for transcription factors that had ChIP seq datasets showing a large degree of overlap with the kidney age-related genes [72]. The top three transcription factors that showed enrichment to binding the aging-regulated genes were STAT1, STAT3 and NFκB. These three transcription factors are all known to be involved in mediating the inflammatory response.
STAT1, STAT3 and NFκB appear to mediate transcriptional changes during the kidney aging process in vivo [72]. All three transcription factors show higher levels of activation in old age. When the transcription factors are activated by inflammatory cytokines in human renal epithelial cells, the resulting changes in gene expression recapitulate the gene expression changes that occur during kidney aging to a large extent. These data indicate that activation of these three transcription factors during kidney aging may contribute to a large fraction of the aging transcriptional program. The finding that NFκB binds to and regulates genes during aging confirms and extends previous work showing that increased NFκB activity contributes to aging phenotypes in many tissues [73–75].
4 Genetic Differences May Explain Some of the Individual Variation in Kidney Aging
There is a great deal of variation in the rate of loss of kidney function with age between different individuals. In old age, many people have lost a considerable fraction of kidney function, leading to chronic kidney disease or end stage renal disease. Other people have relatively mild loss of kidney function. Genetic variation accounts for some of the individual variation in kidney aging. Genome-wide association studies (GWAS) have begun to define DNA polymorphisms that are associated with either reduced glomerular filtration rate or chronic kidney disease. In principle, genetic algorithms may one day be used in the young to help predict which individuals are at risk for a rapid loss of renal function as they grow old, and which individuals are more likely to have a slower rate of loss of kidney function with age. These genetic risk scores would be able to look-up the information from each of the individual polymorphisms and create one combined score to be used to predict future renal rates of aging.
Genome wide association studies have recently been successful at identifying single nucleotide polymorphisms (SNPs) associated with a variety of renal phenotypes associated with age. An example is glomerular filtration rate, which is an estimate of renal function based on levels of creatinine in the urine. The studies are controlled for age, so DNA variants associated with GFR in these studies would identify loci that predispose one to having higher or lower GFR for a given age. There are at least two ways in which a SNP could be associated with variation in GFR. First, the SNP could be associated with a different baseline of renal function (higher or lower GFR) but not with a difference in the rate of renal aging. In this case, GFR would decline similarly in individuals that vary at the SNP, but the GFR would have a different baseline so that those with a higher baseline would be less likely to have renal disease as they grow old. Second, the SNP could be associated with a different rate of renal aging. In this case, allelic variation at this SNP may have little effect on young adults as all might have a similar starting point for renal function. However, individuals with one allele could show more rapid rates of renal decline leading to larger and larger differences in GFR as one ages.
Another powerful phenotype used in kidney GWA studies is chronic kidney disease. Chronic kidney disease is diagnosed when an individual has glomerular filtration rate below 60 ml/min/1.73 m2 for 3 or more months, or if the individual has other signs of kidney damage. The gene association studies using chronic kidney disease as a phenotype are similar to the GWAS using glomerular filtration rate as a trait because one of the main criteria for diagnosing chronic kidney disease is glomerular filtration rate. Chronic kidney disease splits up individuals based on GFR levels above or below the 60 ml/min/1.73 m2 cutoff and GFR assigns a continuous variable to each individual.
Kidney GWA studies have also been performed for albuminuria and diabetic nephropathy. Albuminuria refers to albumin in the urine, which is another indication of poor kidney function. Diabetic nephropathy is characterized by three related conditions: albuminuria, low glomerular filtration rate, and high blood pressure.
A total of 67,073 individuals have been examined to search for SNPs associated with GFR, chronic kidney disease, albuminuria and diabetic nephropathy [76, 77]. These studies have found 27 SNPs associated with these parameters of kidney function. These SNPs may be useful in helping to identify individuals with high and low renal function. This information could be useful to individuals when they are alive by providing information useful for precision medicine. In addition, the genetic information could be useful to help identify which kidneys from elderly donors may still be viable for use in renal transplantations.
In order to predict renal function, one would not evaluate each of the kidney SNPs one at a time, but rather one would develop an algorithm that could evaluate all of the SNPs together and provide a score that summarizes the information from all of the SNPs combined. A preliminary study was performed combining information from 16 kidney SNPs to create a genetic risk score for chronic kidney disease [76]. The genotype score was evaluated in 2129 test subject and was partially successful in predicting chronic kidney disease risk. Carrying a high number of risk alleles was partially able to predict those at increased risk for chronic kidney disease. However, the effect of the genotype was small, and not necessarily an improvement over clinical factors such as lifestyle, blood pressure and the presence of Type 2 Diabetes. Beyond renal disease, genetic algorithms have been developed for Type 2 Diabetes mellitus, incident myocardial infarction, coronary heart disease, myocardial infarction and stroke in women [78–81]. In each case, the genetic algorithms had only a mild effect in predicting disease risk beyond current clinical tests, indicating that improved methods or more complete data will be required for these algorithms to become widely used. Future studies may be able to expand the number of SNPs known to be associated with renal function, leading to improved genetic algorithms with greater predictive power.
One study has specifically searched for SNPs that are associated with different levels of kidney function, independent of age [82]. To do this study, longitudinal data was used from the Baltimore Longitudinal Study of Aging and from the InCHIANTI cohort. Both of these cohorts have followed individuals over a number of years, so it was possible to follow the loss of renal function with age for each individual. However, the total number of people in each cohort was relatively small (1–3 thousand), precluding a straightforward GWA study for association with different rates of loss of GFR.
Instead of searching through every SNP in the genome, the study used a genomic convergence approach to systematically select SNPs that are most likely to be involved in kidney aging. The first assumption was that genes whose expression changes with age would be enriched for those with functional effect on the rate of renal aging or renal function (630 aging-related genes). The next step was to search for SNPs that are associated with differential expression of these 630 genes (expression quantitative trait loci; eQTLs) because higher or lower expression levels may affect the age at which the expression of a gene dips below a functional threshold (110 SNPs associated with differential expression in kidney aging related genes). The last step was to determine whether any of the 110 SNPs was associated with different levels of renal function independent of age in the Baltimore Longitudinal Study of Aging or the InCHIANTI cohort. Two linked SNPs (rs1711437 and rs1784418) located within the matrix metalloproteinase 20 gene (MMP20) were found to associate with loss of GFR independent of age at a significant level following correction for multiple hypothesis testing. For an individual who carries the A allele at rs1711437, his or her creatinine clearance is approximately that of someone 4–5 years younger who does not carry the A allele. In the BLSA population, the genotype of rs1711437 explains 2.1 % of the variation in creatinine clearance and in the InCHIANTI population, the genotype explains 0.9 % of the variation.
Future work will help find the missing heritability in DNA variants associated with kidney function, disease and aging. These studies may use full genome sequence rather than DNA chip analysis, enabling one to analyze rare variants. Larger cohorts may become available that will increase statistical power. Genetic algorithms may become available that are more powerful and able to better predict renal function. These algorithms may gain effectiveness by capitalizing on the complex genetic interactions among the SNPs associated with renal aging. These interactions are termed epistatic interactions, and refer to all of the possible ways in which two SNPs may be associated with renal aging that are not merely the additive sum of each individual SNP. For instance, the two SNPs may act redundantly, such that a protective allele in one SNP is as effective as a protective allele in both SNPs. Alternatively, the two SNPs may act synergistically, such that protective alleles in both SNPs are far more effective than would be predicted by adding up the effects of each allele individually. Future work may lead to ways to use DNA information to predict current and future renal function. For the purposes of renal transplantation, this could be key as a criterion to be used to help select which potential donor kidneys are most likely to retain adequate function following renal transplantation.
5 Conclusion
As we enter the era of precision medicine, individual genetic and biomarker data can be used to provide information about the physiological age of a person, rather than simple chronological age. Recent work has begun to unravel some of the mechanisms underlying kidney aging (Fig. 3). These mechanisms include increasing levels of cell senescence, chronic inflammation, fibrosis, and transcriptional regulation of the aging gene network. A better and more complete understanding of the molecular underpinnings of aging may one day enable the development of biomarkers that are able to report true biological age, as opposed to chronological age. These aging biomarkers could be used to more accurately ascertain which kidneys are most suitable for renal transplantation. This precision medicine approach of using personalized aging biomarkers may enable one to expand the pool of available kidneys for transplantation without diminishing the length of graft survival or the quality of the transplant.
References
Lindeman RD (1990) Overview: renal physiology and pathophysiology of aging. Am J Kidney Dis 16(4):275–282. doi:S0272638690001184 [pii]
Davies DF, Shock NW (1950) Age changes in glomerular filtration rate, effective renal plasma flow, and tubular excretory capacity in adult males. J Clin Invest 29(5):496–507. doi:10.1172/JCI102286
Hoang K, Tan JC, Derby G, Blouch KL, Masek M, Ma I, Lemley KV, Myers BD (2003) Determinants of glomerular hypofiltration in aging humans. Kidney Int 64(4):1417–1424. doi:kid207 [pii] 10.1046/j.1523-1755.2003.00207.x
Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J (2009) A new equation to estimate glomerular filtration rate. Ann Intern Med 150(9):604–612
K/DOQI (2002) K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis 39(2 Suppl 1):S1–S266
Wolfe RA, Ashby VB, Milford EL, Ojo AO, Ettenger RE, Agodoa LY, Held PJ, Port FK (1999) Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N Engl J Med 341(23):1725–1730. doi:10.1056/NEJM199912023412303
Nyengaard JR, Bendtsen TF (1992) Glomerular number and size in relation to age, kidney weight, and body surface in normal man. Anat Rec 232(2):194–201. doi:10.1002/ar.1092320205
Tan JC, Busque S, Workeneh B, Ho B, Derby G, Blouch KL, Sommer FG, Edwards B, Myers BD (2010) Effects of aging on glomerular function and number in living kidney donors. Kidney Int 78(7):686–692. doi:10.1038/ki.2010.128ki2010128 [pii]
Anderson S, Halter JB, Hazzard WR, Himmelfarb J, Horne FM, Kaysen GA, Kusek JW, Nayfield SG, Schmader K, Tian Y, Ashworth JR, Clayton CP, Parker RP, Tarver ED, Woolard NF, High KP (2009) Prediction, progression, and outcomes of chronic kidney disease in older adults. J Am Soc Nephrol 20(6):1199–1209. doi:10.1681/ASN.2008080860 ASN.2008080860 [pii]
Hariharan S, Johnson CP, Bresnahan BA, Taranto SE, McIntosh MJ, Stablein D (2000) Improved graft survival after renal transplantation in the United States, 1988 to 1996. N Engl J Med 342(9):605–612. doi:MJBA-420901 [pii] 10.1056/NEJM200003023420901
Gjertson DW (1996) A multi-factor analysis of kidney graft outcomes at one and five years posttransplantation: 1996 UNOS update. Clin Transpl 10:343–360
United Network for Organ Sharing (UNOS) (2015) http://www.unos.org/donation/index.php?topic=data. Accessed 22 Feb 2015
Lapasia JB, Kong SY, Busque S, Scandling JD, Chertow GM, Tan JC (2011) Living donor evaluation and exclusion: the Stanford experience. Clin Transplant 25(5):697–704. doi:10.1111/j.1399-0012.2010.01336.x
de Fijter JW (2005) The impact of age on rejection in kidney transplantation. Drugs Aging 22(5):433–449, doi:2257 [pii]
Terasaki PI, Gjertson DW, Cecka JM, Takemoto S, Cho YW (1997) Significance of the donor age effect on kidney transplants. Clin Transplant 11(5 Pt 1):366–372
Lee CM, Scandling JD, Shen GK, Salvatierra O, Dafoe DC, Alfrey EJ (1996) The kidneys that nobody wanted: support for the utilization of expanded criteria donors. Transplantation 62(12):1832–1841
Prommool S, Jhangri GS, Cockfield SM, Halloran PF (2000) Time dependency of factors affecting renal allograft survival. J Am Soc Nephrol 11(3):565–573
Smits JM, Persijn GG, van Houwelingen HC, Claas FH, Frei U (2002) Evaluation of the Eurotransplant Senior Program. The results of the first year. Am J Transplant 2(7):664–670
Oppenheimer F, Aljama P, Asensio Peinado C, Bustamante Bustamante J, Crespo Albiach JF, Guirado Perich L (2004) The impact of donor age on the results of renal transplantation. Nephrol Dial Transplant (19 Suppl 3):iii11–iii15. doi:10.1093/ndt/gfh1008 19/suppl_3/iii11 [pii]
Cohen B, Smits JM, Haase B, Persijn G, Vanrenterghem Y, Frei U (2005) Expanding the donor pool to increase renal transplantation. Nephrol Dial Transplant 20(1):34–41. doi:gfh506 [pii] 10.1093/ndt/gfh506
Frei U, Noeldeke J, Machold-Fabrizii V, Arbogast H, Margreiter R, Fricke L, Voiculescu A, Kliem V, Ebel H, Albert U, Lopau K, Schnuelle P, Nonnast-Daniel B, Pietruck F, Offermann R, Persijn G, Bernasconi C (2008) Prospective age-matching in elderly kidney transplant recipients–a 5-year analysis of the Eurotransplant Senior Program. Am J Transplant 8(1):50–57. doi:10.1111/j.1600-6143.2007.02014.x AJT2014 [pii]
McGlynn LM, Stevenson K, Lamb K, Zino S, Brown M, Prina A, Kingsmore D, Shiels PG (2009) Cellular senescence in pretransplant renal biopsies predicts postoperative organ function. Aging Cell 8(1):45–51. doi:ACE447 [pii] 10.1111/j.1474-9726.2008.00447.x
Yang H, Fogo AB (2010) Cell senescence in the aging kidney. J Am Soc Nephrol 21(9):1436–1439. doi:10.1681/ASN.2010020205 ASN.2010020205 [pii]
Melk A, Halloran PF (2001) Cell senescence and its implications for nephrology. J Am Soc Nephrol 12(2):385–393
Wills LP, Schnellmann RG (2011) Telomeres and telomerase in renal health. J Am Soc Nephrol 22(1):39–41. doi:10.1681/ASN.2010060662 22/1/39 [pii]
Naesens M (2011) Replicative senescence in kidney aging, renal disease, and renal transplantation. Discov Med 11(56):65–75
Fontana L, Partridge L (2015) Promoting health and longevity through diet: from model organisms to humans. Cell 161(1):106–118. doi:10.1016/j.cell.2015.02.020
Campisi J, Robert L (2014) Cell senescence: role in aging and age-related diseases. Interdiscip Top Gerontol 39:45–61. doi:000358899 [pii] 10.1159/000358899 [doi]
Kamb A (1995) Cell-cycle regulators and cancer. Trends Genet 11(4):136–140. doi:S0168-9525(00)89027-7 [pii]
Vidal A, Koff A, Kamb A (2000) Cell-cycle inhibitors: three families united by a common cause Cell-cycle regulators and cancer. Gene 247(1–2):1–15. doi:S0378-1119(00)00092-5 [pii] S0168-9525(00)89027-7 [pii]
Chandler H, Peters G (2013) Stressing the cell cycle in senescence and aging. Curr Opin Cell Biol 25(6):765–771. doi:10.1016/j.ceb.2013.07.005
Melk A, Schmidt BM, Takeuchi O, Sawitzki B, Rayner DC, Halloran PF (2004) Expression of p16INK4a and other cell cycle regulator and senescence associated genes in aging human kidney. Kidney Int 65(2):510–520. doi:10.1111/j.1523-1755.2004.00438.x
Melk A, Schmidt BM, Vongwiwatana A, Rayner DC, Halloran PF (2005) Increased expression of senescence-associated cell cycle inhibitor p16INK4a in deteriorating renal transplants and diseased native kidney. Am J Transplant 5(6):1375–1382. doi:AJT [pii] 10.1111/j.1600-6143.2005.00846.x
Koppelstaetter C, Schratzberger G, Perco P, Hofer J, Mark W, Ollinger R, Oberbauer R, Schwarz C, Mitterbauer C, Kainz A, Karkoszka H, Wiecek A, Mayer B, Mayer G (2008) Markers of cellular senescence in zero hour biopsies predict outcome in renal transplantation. Aging Cell 7(4):491–497. doi:ACE398 [pii] 10.1111/j.1474-9726.2008.00398.x
Gingell-Littlejohn M, McGuinness D, McGlynn LM, Kingsmore D, Stevenson KS, Koppelstaetter C, Clancy MJ, Shiels PG (2013) Pre-transplant CDKN2A expression in kidney biopsies predicts renal function and is a future component of donor scoring criteria. PLoS One 8(7):e68133. doi:10.1371/journal.pone.0068133, PONE-D-12-38457 [pii]
Kainz A, Perco P, Mayer B, Soleiman A, Steininger R, Mayer G, Mitterbauer C, Schwarz C, Meyer TW, Oberbauer R (2007) Gene-expression profiles and age of donor kidney biopsies obtained before transplantation distinguish medium term graft function. Transplantation 83(8):1048–1054. doi:10.1097/01.tp.0000259960.56786.ec 00007890-200704270-00011 [pii]
Mas VR, Scian MJ, Archer KJ, Suh JL, David KG, Ren Q, Gehr TWB, King AL, Posner MP, Mueller TF, Maluf DG (2011) Pretransplant transcriptome profiles identify among kidneys with delayed graft function those with poorer quality and outcome. Mol Med 17(11–12):1311–1322. doi:10.2119/molmed.2011.00159
Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G (2013) The hallmarks of aging. Cell 153(6):1194–1217. doi:10.1016/j.cell.2013.05.039
Mei C, Zheng F (2009) Chronic inflammation potentiates kidney aging. Semin Nephrol 29(6):555–568. doi:10.1016/j.semnephrol.2009.07.002 S0270-9295(09)00144-2 [pii]
Cesari M, Penninx BW, Pahor M, Lauretani F, Corsi AM, Rhys Williams G, Guralnik JM, Ferrucci L (2004) Inflammatory markers and physical performance in older persons: the InCHIANTI study. J Gerontol A Biol Sci Med Sci 59(3):242–248
Krabbe KS, Pedersen M, Bruunsgaard H (2004) Inflammatory mediators in the elderly. Exp Gerontol 39(5):687–699. doi:10.1016/j.exger.2004.01.009 S0531556504000531 [pii]
Dorshkind K, Montecino-Rodriguez E, Signer RA (2009) The ageing immune system: is it ever too old to become young again? Nat Rev Immunol 9(1):57–62. doi:10.1038/nri2471, nri2471 [pii]
Wu D, Meydani SN (2008) Age-associated changes in immune and inflammatory responses: impact of vitamin E intervention. J Leukoc Biol 84(4):900–914. doi:10.1189/jlb.0108023 jlb.0108023 [pii]
Dixit VD (2008) Adipose-immune interactions during obesity and caloric restriction: reciprocal mechanisms regulating immunity and health span. J Leukoc Biol 84(4):882–892. doi:10.1189/jlb.0108028 jlb.0108028 [pii]
Coppe JP, Desprez PY, Krtolica A, Campisi J (2010) The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol 5:99–118. doi:10.1146/annurev-pathol-121808-102144
Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, Xiao W, Mindrinos M, Crane E, Segal E, Myers BD, Brooks JD, Davis RW, Higgins J, Owen AB, Kim SK (2004) A transcriptional profile of aging in the human kidney. PLoS Biol 2(12):e427. doi:10.1371/journal.pbio.0020427
Jung KJ, Go EK, Kim JY, Yu BP, Chung HY (2009) Suppression of age-related renal changes in NF-kappaB and its target gene expression by dietary ferulate. J Nutr Biochem 20(5):378–388. doi:10.1016/j.jnutbio.2008.04.008 S0955-2863(08)00099-5 [pii]
Hewitson TD (2009) Renal tubulointerstitial fibrosis: common but never simple. Am J Physiol Renal Physiol 296(6):F1239–F1244. doi:10.1152/ajprenal.90521.2008 90521.2008 [pii]
Bechtel W, McGoohan S, Zeisberg EM, Muller GA, Kalbacher H, Salant DJ, Muller CA, Kalluri R, Zeisberg M (2010) Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med 16(5):544–550. doi:10.1038/nm.2135 nm.2135 [pii]
Zhou W, Otto EA, Cluckey A, Airik R, Hurd TW, Chaki M, Diaz K, Lach FP, Bennett GR, Gee HY, Ghosh AK, Natarajan S, Thongthip S, Veturi U, Allen SJ, Janssen S, Ramaswami G, Dixon J, Burkhalter F, Spoendlin M, Moch H, Mihatsch MJ, Verine J, Reade R, Soliman H, Godin M, Kiss D, Monga G, Mazzucco G, Amann K, Artunc F, Newland RC, Wiech T, Zschiedrich S, Huber TB, Friedl A, Slaats GG, Joles JA, Goldschmeding R, Washburn J, Giles RH, Levy S, Smogorzewska A, Hildebrandt F (2012) FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet 44(8):910–915. doi:ng.2347 [pii] 10.1038/ng.2347
Deans AJ, West SC (2011) DNA interstrand crosslink repair and cancer. Nat Rev Cancer 11(7):467–480. doi:nrc3088 [pii] 10.1038/nrc3088
Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, Elledge SJ (2010) A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell 39(1):36–47. doi:S1097-2765(10)00464-8 [pii] 10.1016/j.molcel.2010.06.023
Fontana L, Partridge L, Longo VD (2010) Extending healthy life span–from yeast to humans. Science 328(5976):321–326. doi:328/5976/321 [pii] 10.1126/science.1172539 [doi]
Guarente L (2007) Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol 72:483–488. doi:10.1101/sqb.2007.72.024
Imai S, Guarente L (2014) NAD+ and sirtuins in aging and disease. Trends Cell Biol 24(8):464–471. doi:S0962-8924(14)00063-4 [pii] 10.1016/j.tcb.2014.04.002 [doi]
Kaeberlein M (2013) mTOR inhibition: from aging to autism and beyond. Scientifica (Cairo) 2013:849186. doi:10.1155/2013/849186
Sauve AA, Wolberger C, Schramm VL, Boeke JD (2006) The biochemistry of sirtuins. Annu Rev Biochem 75:435–465. doi:10.1146/annurev.biochem.74.082803.133500
Guarente L (2013) Calorie restriction and sirtuins revisited. Genes Dev 27(19):2072–2085. doi:27/19/2072 [pii] 10.1101/gad.227439.113
Hao CM, Haase VH (2010) Sirtuins and their relevance to the kidney. J Am Soc Nephrol 21(10):1620–1627. doi:ASN.2010010046 [pii] 10.1681/ASN.2010010046 [doi]
He W, Wang Y, Zhang M-Z, You L, Davis LS, Fan H, Yang H-C, Fogo AB, Zent R, Harris RC, Breyer MD, Hao C-M (2010) Sirt1 activation protects the mouse renal medulla from oxidative injury. J Clin Invest 120(4):1056–1068. doi:10.1172/jci41563
Braidy N, Guillemin GJ, Mansour H, Chan-Ling T, Poljak A, Grant R (2011) Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS One 6(4):e19194. doi:10.1371/journal.pone.0019194
Zhuo L, Cai G, Liu F, Fu B, Liu W, Hong Q, Ma Q, Peng Y, Wang J, Chen X (2009) Expression and mechanism of mammalian target of rapamycin in age-related renal cell senescence and organ aging. Mech Ageing Dev 130(10):700–708. doi:S0047-6374(09)00112-2 [pii] 10.1016/j.mad.2009.08.005 [doi]
Kuro-o M (2011) Klotho and the aging process. Korean J Intern Med 26(2):113–122. doi:10.3904/kjim.2011.26.2.113
Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, Iwasaki H, Iida A, Shiraki-Iida T, Nishikawa S, Nagai R, Nabeshima YI (1997) Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390(6655):45–51. doi:10.1038/36285
Koh N, Fujimori T, Nishiguchi S, Tamori A, Shiomi S, Nakatani T, Sugimura K, Kishimoto T, Kinoshita S, Kuroki T, Nabeshima Y (2001) Severely reduced production of klotho in human chronic renal failure kidney. Biochem Biophys Res Commun 280(4):1015–1020. doi:10.1006/bbrc.2000.4226 [doi] S0006291X00942268 [pii]
Pedersen L, Pedersen SM, Brasen CL, Rasmussen LM (2013) Soluble serum Klotho levels in healthy subjects. Comparison of two different immunoassays. Clin Biochem 46(12):1079–1083. doi:S0009-9120(13)00242-7 [pii] 10.1016/j.clinbiochem.2013.05.046 [doi]
Yamazaki Y, Imura A, Urakawa I, Shimada T, Murakami J, Aono Y, Hasegawa H, Yamashita T, Nakatani K, Saito Y, Okamoto N, Kurumatani N, Namba N, Kitaoka T, Ozono K, Sakai T, Hataya H, Ichikawa S, Imel EA, Econs MJ, Nabeshima Y (2010) Establishment of sandwich ELISA for soluble alpha-Klotho measurement: age-dependent change of soluble alpha-Klotho levels in healthy subjects. Biochem Biophys Res Commun 398(3):513–518. doi:S0006-291X(10)01250-7 [pii] 10.1016/j.bbrc.2010.06.110 [doi]
Olauson H, Vervloet MG, Cozzolino M, Massy ZA, Urena Torres P, Larsson TE (2014) New insights into the FGF23-Klotho axis. Semin Nephrol 34(6):586–597. doi:S0270-9295(14)00145-4 [pii] 10.1016/j.semnephrol.2014.09.005 [doi]
Xu Y, Sun Z (2015) Molecular basis of klotho: from gene to function in aging. Endocr Rev 36(2):174–193. doi:10.1210/er.2013-1079
Melk A, Mansfield ES, Hsieh SC, Hernandez-Boussard T, Grimm P, Rayner DC, Halloran PF, Sarwal MM (2005) Transcriptional analysis of the molecular basis of human kidney aging using cDNA microarray profiling. Kidney Int 68(6):2667–2679. doi:KID738 [pii] 10.1111/j.1523-1755.2005.00738.x
Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O’Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, Myers RM, Weissman SM, Snyder M (2012) Architecture of the human regulatory network derived from ENCODE data. Nature 489(7414):91–100. doi:nature11245 [pii] 10.1038/nature11245 [doi]
O’Brown ZK, Van Nostrand EL, Higgins JP, Kim SK (2015) The inflammatory transcription factors NFκB, STAT1 and STAT3 drive aging of the human kidney (submitted)
Adler AS, Kawahara TL, Segal E, Chang HY (2008) Reversal of aging by NFkappaB blockade. Cell Cycle 7(5):556–559, doi:5490 [pii]
Adler AS, Sinha S, Kawahara TL, Zhang JY, Segal E, Chang HY (2007) Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev 21(24):3244–3257. doi:gad.1588507 [pii] 10.1101/gad.1588507 [doi]
Southworth LK, Owen AB, Kim SK (2009) Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet 5(12):e1000776. doi:10.1371/journal.pgen.1000776
Kottgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O’Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Pare G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tonjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstatter A, Kollerits B, Kedenko L, Magi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Volzke H, Kroemer HK, Nauck M, Volker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Kramer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS (2010) New loci associated with kidney function and chronic kidney disease. Nat Genet 42(5):376–384. doi:ng.568 [pii] 10.1038/ng.568
O’Seaghdha CM, Fox CS (2012) Genome-wide association studies of chronic kidney disease: what have we learned? Nat Rev Nephrol 8(2):89–99. doi:nrneph.2011.189 [pii] 10.1038/nrneph.2011.189 [doi]
Meigs JB, Shrader P, Sullivan LM, McAteer JB, Fox CS, Dupuis J, Manning AK, Florez JC, Wilson PW, D’Agostino RB Sr, Cupples LA (2008) Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med 359(21):2208–2219. doi:359/21/2208 [pii] 10.1056/NEJMoa0804742
Talmud PJ, Hingorani AD, Cooper JA, Marmot MG, Brunner EJ, Kumari M, Kivimaki M, Humphries SE (2010) Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ 340:b4838
Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, Guiducci C, Perola M, Jula A, Sinisalo J, Lokki ML, Nieminen MS, Melander O, Salomaa V, Peltonen L, Kathiresan S (2010) A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet 376(9750):1393–1400. doi:S0140-6736(10)61267-6 [pii] 10.1016/S0140-6736(10)61267-6
Paynter NP, Chasman DI, Pare G, Buring JE, Cook NR, Miletich JP, Ridker PM (2010) Association between a literature-based genetic risk score and cardiovascular events in women. JAMA 303(7):631–637. doi:303/7/631 [pii] 10.1001/jama.2010.119
Wheeler HE, Metter EJ, Tanaka T, Absher D, Higgins J, Zahn JM, Wilhelmy J, Davis RW, Singleton A, Myers RM, Ferrucci L, Kim SK (2009) Sequential use of transcriptional profiling, expression quantitative trait mapping, and gene association implicates MMP20 in human kidney aging. PLoS Genet 5(10):e1000685. doi:10.1371/journal.pgen.1000685
Acknowledgments
We would like to thank Zachary O’Brown for helpful comments. This work was funded by NIA RO1 AG025941.
Editor: John Williams, National Institute on Aging (NIA), NIH.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing
About this chapter
Cite this chapter
Higgins, J.P., Kim, S.K. (2016). Renal Aging and Transplantation. In: Sierra, F., Kohanski, R. (eds) Advances in Geroscience. Springer, Cham. https://doi.org/10.1007/978-3-319-23246-1_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-23246-1_13
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-23245-4
Online ISBN: 978-3-319-23246-1
eBook Packages: MedicineMedicine (R0)