Abstract
Senescence is key component of plant development. It enables remobilisation of nutrients from organs that are no longer needed or have been compromised and ends with programmed cell death (PCD) of all the cells in the organ. Cytological features of senescence-associated PCD are distinct, though shared in part with other forms of plant PCD triggered by abiotic stress or pathogen attack. These cytological features define a form of PCD termed vacuolar cell death and share some characteristics of autophagy. Key features are enlargement of the vacuole through fusion of vesicles carrying cytoplasmic cargo and vacuolar rupture releasing lytic enzymes into the cytoplasm. Biochemical processes are common to many forms of plant PCD including the activation of proteases, lipases, nucleases and transporters. Vacuolar processing enzymes (VPEs) that have caspase activity have emerged as important players in several forms of PCD. However, less is known about upstream signals and regulatory mechanisms that control the initiation and progression of senescence and trigger the onset of PCD. Here, the signals and mechanisms triggering senescence-associated PCD are reviewed, and their conservation across different tissues is assessed. Key questions that remain to be answered in future research are discussed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
9.1 Introduction
Plant senescence is defined here as the process by which individual plant organs, or, in the case of annual plants, the whole individual, are programmed to die. Although death of the organ and its constituent cells is the final outcome of senescence, a key feature of senescence is the remobilisation of nutrients between organs that are no longer needed and other parts of the plant where nutrients can be used for growth or storage [1–3]. For example, the remobilisation of nutrients from leaves to the developing fruit in annual plants is important for optimal seed and fruit development. Leaf senescence is therefore an integral part of the ecological strategy of annual plants.
Programmed cell death (PCD) in the context of senescence is understood here to be distinct from developmental PCD, despite conservation of some cytological features. The latter generally relates to the removal of a small number of cells in an organ which is otherwise retained, such as the tapetum [4], nucellus [5], the suspensor [6], or the formation of holes in leaves of species such as Monstera or the lace plant Aponogeton madagascariensis [7]. These processes have, by some authors, been regarded as a form of programmed senescence [8]. This was based on the premise that there is no clear difference in the mechanisms of PCD occurring in isolated cells during early development and the larger-scale PCD occurring during whole organ senescence, and hence the terms are interchangeable. Conversely, it has been argued that the term PCD is appropriate both to the death of groups of cells during development and the PCD occurring at the end of senescence. In this chapter, I will consider PCD in the context of the regulation of whole organ senescence, focussing mainly on common mechanisms and signals but also assessing senescence and PCD in different organs to consider the commonality of mechanisms and signals. The regulation of senescence has been extensively reviewed (e.g. [1, 3, 9]) so only a brief overview will be presented here, trying to identify regulatory points relating to the later, PCD-related events and setting the context for senescence-associated PCD within the cell.
At the level of whole organ senescence, one of the key issues that have been widely discussed in this field is the distinction between the process of senescence and the process of cell death [3, 8, 10]. Are they a continuum, or are they distinct processes, regulated separately? Are they in fact opposing forces: senescence keeping the tissue alive until remobilisation is complete [11]? Clearly, remobilisation needs to be tightly coordinated to ensure that the structures required for nutrient mobilisation and transport are still functional while cell contents are broken down and cells enter PCD. A further difficulty in understanding senescence is that there obviously needs to be heterogeneity across organs at a cellular level to enable remobilisation to take place. For example, in apparently healthy petals in flowers at full bloom, electron microscopy reveals that many of the internal cells have already died [12]. This complicates studies, for example, of gene expression during senescence.
The timing of the remobilisation and hence PCD in the context of other developmental processes in the plant is also critical. Furthermore, species differ in the coordination of this timing. For example, in wheat where leaf senescence was disrupted by expression of the ipt gene driven by a senescence-induced promoter, leaves stayed green for longer, but seed yield was not increased. In contrast, transgenic tobacco, expressing the same transgene, produced a better yield of seed due to the extension of the photosynthetic period in the leaves [13]. Post-fertilisation floral senescence is also tightly regulated to remobilise nutrients towards developing seeds [14]. Hence, coordination of the initiation of senescence and ultimate PCD in different organs at the level of the whole plant is very important.
Another key issue is whether there is a senescence-associated PCD mechanism that is distinct from other forms of plant PCD and common to all senescent organs. The mechanisms of plant PCD have been studied in diverse systems such as xylogenesis, self-incompatible pollen responses, stress-induced PCD in cell cultures and the hypersensitive response [15] as well as developmental senescence. Are mechanisms of PCD conserved between senescence-associated PCD and PCD in these other systems or is PCD-associated senescence distinct? Furthermore, organ senescence is often classified as developmentally or stress induced. Numerous stresses cause premature leaf senescence thus removing leaves which are diseased or compromised [16]. So another question is whether the primary signal for senescence affects the PCD mechanism. Abscission is also a common feature in organ senescence [14, 17]. However, abscission of both floral organs and leaves can occur before the death of all the cells. So we can ask whether the PCD occurring in organs that are no longer attached to the plant is regulated in the same way as attached-organ senescence. An extreme example is that of postharvest biology. In a commercial setting, leaves and flowers are detached from the plant and kept in the cold and dark, often in a dehydrated state. Does this trigger the same type of PCD as developmental or attached-organ stress-induced senescence or even post-abscission senescence? In this review, I will try to address some of these key questions and identify areas for future study. Our ability to understand senescence and senescence-associated PCD and control them would benefit both agricultural production and limit postharvest waste. Many key seed crops are monocarpic; hence, seed production is linked to resource remobilisation from the leaves. Delaying senescence through functional stay-green varieties can result in increased yield [18]. Postharvest fruit and vegetables are perishable items and require careful handling throughout the supply chain to avoid damage leading to rapid senescence-like processes. It is estimated that in India, for example, up to 30 % of fresh fruit and vegetables are lost through lack of an efficient cold chain [19]. It is therefore important that we identify the gaps in our knowledge of both pre- and postharvest senescence-associated PCD and the tools that can help us to fill them.
9.2 Induction of Senescence and Senescence-Associated PCD
If we accept that senescence-associated PCD occurs as the final stage of senescence and as a direct result of the induction of senescence, then the first question we can ask is whether there are common signals that induce the initiation and progression of senescence within or between different organs.
9.2.1 Signals and Mechanisms: How Much Conservation Is There?
Organ senescence occurs as part of the natural developmental programme of a plant. In some organs such as floral organs and seed pods, the lifespan of the organ is species specific [20, 21]. A distinctive feature of pod senescence is the very close coordination between seed development and pod/silique senescence, and floral senescence is often closely linked to pollination. This means that senescence in these organs is not influenced to a great extent by environmental stresses. Other organs such as leaves are responsive to abiotic and biotic stress signals that can initiate senescence in a single leaf or the whole plant [17]. An interesting question is therefore whether different signals can independently activate the same mechanism of senescence and PCD. At a gene expression level, this has been recently investigated with an ambitious exercise in which microarrays from experiments using 28 different methods for inducing senescence in Arabidopsis organs were compared [22]. Array data were taken from experiments on leaves, young seedlings, siliques, petioles and cell suspensions and included induction of senescence by plant growth regulators (ethylene, abscisic acid (ABA), jasmonic acid (JA), salicylic acid (SA) and brassinolide), high glucose/low nitrogen, a range of abiotic stresses (dark, cold, genotoxic, heat, osmotic, oxidative, wounding, salt, UV-B, ozone and heavy metal stress), three pathogens (Phytophthora infestans, Pseudomonas syringae and Botrytis cinerea) as well as developmental senescence in leaves and siliques. An interesting finding was that although all the treatments induced symptoms of senescence, most of the treatments only shared less than 10 % of their up-regulated genes with developmentally induced senescence. Four treatments were identified as sharing most changes in gene expression (between one half and one third) with developmentally induced senescence: age-induced PCD in cell cultures, infection with Botrytis cinerea (48 h), high glucose/low nitrogen and dark stress (5 days). One feature these four induction methods share is that they were all imposed for longer periods of time. This suggests that the shared gene expression may relate to later events in senescence, perhaps associated to PCD rather than early induction of the senescence process. This was supported when time points within these treatments were analysed: more gene expression was shared with developmentally induced senescence at later time points. This suggests that though signals that initiate senescence differ between induction systems, the “executioner” genes are shared. This supports a model in which senescence-associated PCD mechanisms are common at least within similar organs.
9.2.2 Are Specific PGRs Associated with Senescence-Associated PCD?
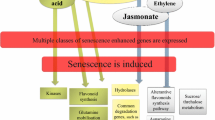
Aside from the method of inducing senescence, another aspect of senescence control and its associated PCD are the internal triggers that initiate or regulate it. Plant growth regulators are clearly key coordinators of some types of developmental plant PCD. For example, PCD in cereal aleurone cells is induced by gibberellins and delayed by ABA [23]. However, the regulation of senescence-associated PCD by PGRs is much less clear-cut. Senescence seems to be regulated negatively by cytokinins and positively by the main stress hormones: abscisic acid (ABA), salicylic acid, (SA), jasmonic acid (JA) and ethylene (Fig. 9.1). Gibberellins and auxin also contribute to retarding senescence, and brassinosteroids may accelerate it although mechanisms are less clear [24]. An important question is whether growth regulators regulate only senescence initiation or also onset of PCD.
Regulation of leaf senescence by developmental and stress signals, plant growth regulators and transcription factors. Adapted from Schippers et al. [24], Guiboileau et al. [1], and Fischer [9], integrating information on the levels of growth regulators and activation of genes from Breeze et al. [25] to attempt to infer timing of signals in relation to onset and spread of senescence-associated PCD
During senescence, cytokinin levels fall, and restoration of cytokinin levels by exogenous application delays senescence symptoms such as chlorophyll breakdown. An early study, over 40 years ago, showed that application of cytokinins to detached leaves delayed signs of senescence [26]. This is further supported by delayed senescence resulting from expression of the cytokinin biosynthesis ipt gene driven by the senescence-specific SAG12 promoter in tobacco [13]. In addition, mutants in ORE12-1 which encodes a cytokinin receptor AHK3 show delayed senescence. Phosphorylation of ARR2 by AHK3 is required for leaf longevity, and the mechanism of its action appears to be related to carbohydrate supply. ARR2 up-regulates extracellular invertase and hexose transporters resulting in increased sink activity [27]. This explains how the role of cytokinin in delaying senescence is common to both photosynthetic organs such as leaves and non-photosynthetic organs such as flowers. Chang et al. [28] showed delayed floral senescence in petunia in transgenic plants expressing the SAG12::ipt construct. These data suggest that cytokinins are involved in the early stages of senescence in regulating the source/sink balance which tips the organ into the senescence programme.
Conversely, all the stress-related PGRs, ABA, SA, JA and ethylene, have roles as positive regulators of senescence [1]. ABA was shown to mediate leaf senescence induced by low nitrogen/high sugar suggesting a link to metabolic signalling [29, 30]. In several species, ABA rises during floral senescence, often quite late in the senescence programme (reviewed in [21]), and it may play a particularly important role in the group of species in which ethylene is not a coordinator of senescence. Further analysis of whether ABA affects PCD in floral senescence would be useful.
SA mediates abiotic and biotic stress signals and affects gene expression during leaf senescence [29]. Reduction of SA levels through mutation of the SA biosynthetic gene sid2 or in transgenic plants through expression of the SA hydroxylase-encoding gene NahG resulted in delayed yellowing, reduced necrosis [31] and extended leaf lifespan when flowering was induced [32]. Furthermore, expression of important senescence-associated genes (SAGs) such as γVPE [33] and the WRKY6 [34] and WRKY53 [35] transcription factors (TFs) was delayed. The effect of SA on SAGs is conserved across leaves, petals and siliques in Arabidopsis [21]. However, effects of SA on gene expression during developmental leaf senescence are not shared with dark-induced senescence indicating a divergent role [36]. SA also appears to have a role in autophagy (discussed below) and the timing of its responses suggests perhaps a role in the progression of senescence as well as its initiation [25]. Less is known about the role of SA in floral senescence although it appears to rise in Lilium post-anthesis [37]. Its role in floral senescence and PCD merits further investigation.
Methyl jasmonate (derived from JA) was originally identified through its activity in promoting senescence [38]. JA and methyl jasmonate mainly promote leaf senescence through their inhibition of photosynthesis [39]. One of the two key enzymes that degrade chlorophyll, CHLOROPHYLLASE1, is strongly up-regulated by JA [40]. At the same time, JA down-regulates Rubisco activase which catalyses the light activation of Rubisco and is required for photosynthesis [41]. However, JA also induces jasmonate-induced proteins (JIPs), some of which inhibit protein synthesis. Effects of JA and SA on leaf senescence are linked by their modulation of WRKY53 action. Early in senescence, WRKY53 is induced by SA, while later as JA levels rise, WRKY53 expression is down-regulated (Hiderhofer and Zentgraf [42]). Hence, the effects of JA appear to influence both early and later events during senescence.
Ethylene also regulates leaf senescence, and mutants in ethylene perception such as etr-1 and ein2/ore3 show delayed senescence [43]. However, sensitivity to ethylene is age dependent in leaves and thus may regulate the stage of commitment to the senescence programme rather than downstream PCD (Fig. 9.1). Ethylene is also a major regulator of floral senescence in some species (reviewed in [14, 20, 44]), again initiating and coordinating the senescence programme.
To what extent any or all of these PGRs regulate senescence-associated PCD is not clear. Based on changes in sensitivity to ethylene, Schippers et al. [24] suggested a model for leaf senescence by which there is a window of sensitivity to all the PGRs. Cytokinin and ethylene act early at a stage when the organ is not fully committed to senescence, while JA, ABA and SA act later in a window that is no longer sensitive to ethylene and in which senescence is now initiated [45, 46]. A comparison of JA, SA and ABA levels during leaf senescence [25] indicates that although patterns differ, all three PGRs show peaks of expression late in senescence and thus might be associated with PCD events. However, JA- and ABA-responsive genes are also up-regulated at the earlier stages of senescence suggesting a role for these PGRs both in the initial stages and later execution of senescence/PCD. The approximate timing of PGR action and downstream TFs in relation to senescence progression and spread of PCD is summarised in Fig. 9.1, although clearly more data are needed to fully understand the regulation of these processes.
9.2.3 ROS as a Regulator of Senescence-Associated PCD
In Arabidopsis and other annual species, flowering is a trigger for leaf senescence under optimal conditions, although leaf age also independently regulates timing of senescence [24]. In both Arabidopsis and Brassica napus, bolting is associated with a transient rise in ROS which may be an important internal trigger in activating rosette senescence [47]. Furthermore, both leaf and petal senescence are associated with a rise in ROS (reviewed in [47, 48]). Developmentally induced PCD such as PCD in the abscission zone is also associated with a rise in ROS [49]. A fall in antioxidant capacity and rise in oxidative stress may thus be a component of the trigger in different forms of plant PCD. ROS levels are modulated through a complex network of ROS scavenging and ROS-producing enzymes, and changes in the balance of these enzymes occur during plant senescence. For example, both catalase and ascorbate peroxidase activities fall at an early stage of bolting in Arabidopsis, leading to a feedback amplification loop that results in an increase in H2O2 and activation of key TFs that initiate senescence in the leaves [47]. A very detailed analysis of transcriptional changes through leaf expansion and senescence [25] revealed that oxidative stress response genes are highly represented in the very early stages of leaf senescence. This supports a role for ROS both as an activator of senescence and senescence-associated PCD seen in other systems. Furthermore, recent evidence suggests that redox status may be important in activating PCD-related enzymes. For example, in developmentally induced PCD in spruce megagametophytes, production of H2O2 activates caspase-like proteins [50], and in ageing elm seeds, treatment with ascorbic acid delayed a rise in caspase-3-like activity [51]. A key parameter often missing though is the intracellular location of the ROS during PCD which has not been determined in most studies. However, a study of the effect of light and O2 levels in harpin-induced PCD in Nicotiana sylvestris leaves concluded that a rise in ROS in the apoplast or in chloroplasts was not required for PCD, while changes in mitochondrial ROS may be important in coordinating the PCD programme [52]. It will be very interesting to get a full picture of the sources of ROS in senescence-induced PCD in different organs which so far is not complete (reviewed in [48, 53]).
9.2.4 Can We Identify TFs Regulating Senescence-Associated PCD?
The initiation of leaf senescence involves a massive reprogramming of gene expression. This reprogramming is mediated by a complex network of TFs [25] involving over 100 TFs of different families. The main families involved are the NAC, WRKY, MYB, C2-H2 zinc finger, bZIP and AP2-EREBP families. NAC and WRKY TFs have been associated with senescence in several tissues including Arabidopsis leaves, petals and siliques [20, 21, 35, 54–57]. Direct evidence for TF control of senescence comes from mutants and over-expressing lines. For example, over-expression of ANAC029 [58] and ANAC092 [55], two important TFs involved in regulating senescence, results in premature senescence, while in their respective mutants, senescence is delayed, indicating that they are both positive regulators of senescence.
WRKY TFs are divided into three groups according to their structure, and the group III WRKY TFs are confirmed as senescence regulators. WRKY53 appears to act as a positive regulator of leaf senescence [35], whereas wrky70 and wrky54 mutants senesce early indicating that these TFs are negative regulators of senescence [56, 57]. All three of these group III WRKY TFs interact with another group III WRKY: WRKY30 by yeast 2-hybrid [56], suggesting that WRKY30 is an upstream regulator of the others. Furthermore, all four of these TFs are induced by SA, and both WRKY53 and WRKY30 are induced by ROS. This is probably a small part of a much more complex TF network, and these TFs may act quite early in the induction of leaf senescence. Numerous TFs are up-regulated before visible signs of leaf senescence including eight NAC domain and 4 WRKY TFs [25]. These TFs fell into two expression clusters: one group was up-regulated and then later down-regulated, whereas expression of the other group continued to rise into late senescence [25]. This latter group is likely to be more important in regulating the later stages of senescence including perhaps PCD events. WRKY TFs were overrepresented later in leaf senescence as were members of the large AP2-EREBP family of TFs. Members of both these TF families are induced by SA and JA [59], and AP2-EREBP TFs are also ethylene induced [60]. Thus, more focus on the roles of WRKY and AP2-EREBP TFs in regulating senescence-associated PCD may be fruitful. It will also be interesting to discover if the same networks of regulators operate in senescence of other organs or whether they differ.
The role of NAC TFs in leaf senescence has been recently reviewed [2] showing that this family of TFs plays an important role in integrating stress and developmental signals not only in Arabidopsis but also in crop species. Some of the NAC TFs may also be directly regulating expression of proteolytic enzymes. In soybean (Glycine max), two NAC TFs are associated with osmotic and ER stress-induced PCD [61]. Coordinately, they regulate common targets including a VPE gene [62], and expression of these TFs was associated with an increase in caspase-1 activity in soybean protoplasts. So in addition to regulating early stages of senescence, NAC TFs may also have a role in later events associated with PCD.
9.2.5 Regulation of Leaf Senescence-Associated PCD by MicroRNAs
Another level of control for senescence and senescence-associated PCD has been recently discovered in microRNAs. MicroRNAs (miRNAs) are small genome-encoded RNAs, approximately 21 nucleotides in length that regulate levels of target transcripts by promoting their degradation or inhibiting their translation [63]. More than a thousand miRNAs have been discovered in plants, and they are involved in the regulation of many developmental processes including hormonal biosynthesis and signalling, pattern formation and biogenesis as well as responses to stress [64, 65]. It is thought that most TFs are under miRNA control [66]. Recently, at least two miRNAs regulating Arabidopsis leaf senescence have been identified. It will be interesting to see if these miRNAs also regulate senescence in other organs.
miR164 family members regulate the expression of ANAC092 (ORE1), a positive regulator of senescence and cell death [67]. In triple mutants in which all three miR164 family members are mutated, senescence-associated PCD was accelerated, including more rapid chlorophyll degradation and an accelerated increase in ion leakage, indicative of increased PCD. In miR164 over-expressing lines, senescence-associated PCD is delayed. The regulation is complicated by the role of another TF: EIN2. ANAC092 is up-regulated with leaf senescence by EIN2 and negatively regulated by miR164. As leaves age, levels of miR164 decline, inhibited by EIN2 thus resulting in ANAC092 up-regulation. This has been described as a trifurcate feed-forward regulatory pathway [67].
Another miRNA controlling senescence is miR319 [68]. This microRNA controls (TEOSINTE BRANCHED1, CYCLOIDEA and PCF) TCP TF genes which in turn regulate JA biosynthesis. In jawD plants, in which miR319 is over-expressed, senescence is delayed, but this effect can be reversed by application of JA. However, a defect in JA alone such as that found in the coi1 mutant does not affect senescence, suggesting that other factors are involved.
9.3 Mechanisms and Regulators of Senescence-Associated PCD
9.3.1 Cytology and Overall Mechanisms of Plant PCD
A central question relating to senescence-associated PCD is the cellular mechanism by which cell death is executed. PCD is now accepted as a rather broad term indicating active cell destruction and encompassing several distinct mechanisms of action. Four morphologically distinct forms of PCD are identifiable in eukaryotic cells: apoptosis, autophagy, oncosis and pyroptosis [69]. Additionally, PCD with necrosis-like morphology has been added to the list [70]. Of these, van Doorn et al. [15] suggest that only two can be identified in plants: vacuolar cell death and necrosis. Vacuolar cell death is defined as essentially autophagic PCD with a contribution to cellular destruction from collapsed lytic vacuoles. Necrosis is typified by early collapse of the plasma membrane and protoplast shrinkage. Furthermore, van Doorn et al. [15] suggest that PCD associated with development and senescence is by the vacuolar cell death route, whereas necrotic PCD occurs during abiotic stress-induced PCD. However, they also accept that some types of PCD such as those that occur during the hypersensitive response, during the formation of starchy endosperm and during self-incompatible pollen–pistil interactions show features of both types of PCD mechanism. A key point of this classification is that other forms of PCD identified in animal cells such as apoptosis are not found in plant cells.
Vacuolar cell death as defined by van Doorn et al. [15] is generally associated with an increase of the vacuolar volume. Fusion of vesicles with the vacuole and invaginations of the vacuolar membrane are seen in electron micrographs, reminiscent of micro- or macroautophagy. Both are likely occurring in the same cells. For example, Toyooka et al. [71] suggested that in fact there may be two different autophagy-like mechanisms involved in PCD occurring during cotyledon senescence. Firstly, there is a process that resembles micropexophagy (microautophagy involving peroxisomes) in yeast. Starch granules are wrapped in small acidic vesicles and then transferred to a lytic vacuole. The second process resembles macroautophagy with the formation of autophagosomes. The final step in this form of PCD is rupture of the tonoplast delivering vacuolar hydrolases into the cytoplasm which degrade the contents. Other morphological features include nuclear fragmentation and the formation of actin cables while mitochondria and other organelles remain relatively intact until tonoplast rupture. Vacuolar cell death is further associated with expression of autophagic markers such as Atg8, acidification of the vacuole, cytoskeletal reorganisation and activation of VPEs. However, at least two mechanisms for vacuole-mediated PCD have been proposed [72]: one in which the vacuolar membrane collapses and the other in which it does not. The latter is associated with the hypersensitive response and involves fusion of the vacuolar membrane with the plasma membrane and delivery of lytic enzymes into the apoplastic space. Destructive vacuolar cell death is associated with developmental PCD and is similar to that described by van Doorn et al. [15].
9.3.2 Autophagy and Senescence-Induced PCD
Vacuolar PCD shows strong similarities to macroautophagy, a process that is well studied in yeast and animal cells and has recently been reviewed in plant cells [73]. The defining feature of macroautophagy is the formation of autophagosomes. These are double-membraned vesicles that enclose portions of the cytoplasm and deliver their content to the vacuole where it is degraded by lytic enzymes. Two pathways are involved which resemble ubiquitin conjugation systems. In yeast, Atg8 is cleaved by Atg4 and thus made available to bind Atg7. Atg8 is then transferred to Atg3 and finally to phosphatidylethanolamine. Atg7 also activates Atg12 and transfers it to Atg10 and finally to Atg5. These two proteins form a complex with Atg16 which is also required for autophagosome assembly. In Arabidopsis, homologous autophagy-related ATG genes have been identified to all of those found in yeast, with the added complexity that in yeast some components such as Atg8 are encoded by single genes while in Arabidopsis they comprise gene families of several members with differential expression patterns.
However, although autophagosome-like structures are visible in cells undergoing senescence-associated PCD, mutation of many of the autophagy-related genes (ATG2, ATG4s, ATG5, ATG6, ATG7, ATG8s, ATG9, ATG10, ATG12s and ATG18a) results in accelerated leaf senescence when plants are grown at normal nutrient levels [73]. Furthermore, Japanese morning glory (Ipomoea nil) mutants, in which autophagy genes were down-regulated, showed accelerated PCD symptoms in senescent petals [74]. This calls into question whether the autophagic machinery is responsible for senescence-associated PCD or whether it is associated with the remobilisation phase of senescence. Of course, it may act in both.
In leaves, autophagy-related genes are up-regulated surprisingly early before leaves are fully expanded and continue to increase in expression throughout senescence [25]. However, different ATG genes were up-regulated at different times: 14 were already up-regulated during leaf expansion, whereas ATG7 was not up-regulated until levels of total protein and chlorophyll were declining in the leaves [25]. ATG7, ATG8A ATG8B and ATG8H, were coordinately up-regulated in the early stages of senescence. Since in yeast Atg8 and Atg7 are required in the final steps in the assembly of autophagosomes [73], the up-regulation of these genes in Arabidopsis leaves may activate senescence-associated autophagy. In pollinated Petunia petals, four ATG8 homologues were up-regulated quite late, once wilting was already visible, and paralleled increases in ethylene, suggesting that ethylene may be regulating their expression [75]. Autophagy in Arabidopsis leaves also seems to be under hormonal control. ROS, SA, JA and ABA were all elevated in mutants of ATG5 [76]. However, the accelerated leaf senescence associated with the atg5 mutant was reversed in sid2 and NahG plants but not in mutants that affect JA or ethylene signalling, indicating a regulatory role for SA. Furthermore, the SA analogue BTH induced the accumulation of autophagosomes. ATG5, ATG8 and ATG12 were up-regulated in all three Arabidopsis senescent tissues (petal, leaf and silique) studied by Wagstaff et al. [21], suggesting a common function across senescent organs. More sophisticated experiments using inducible RNAi constructs to down-regulate ATG genes at later stages in senescence may help to resolve whether these increases in expression later in senescence are required for PCD.
9.3.3 Other Vesicles in Senescence-Associated PCD
In addition to autophagosome-like vesicles, a number of other vesicles have been seen during plant PCD, many also in developmental and senescence-associated PCD (Table 9.1) which either transport cargo from the ER or cytoplasm to the vacuole or deliver their cargo directly to the cytoplasm following tonoplast rupture.
Ricinosomes were originally described from observations of seed endosperm in the castor oil plant Ricinus communis [77–79] and more recently in tomato anthers [80]. They have also been seen in endosperm and suspensor cells of quinoa (Chenopodium quinoa) acting as early markers for PCD [81]. Ricinosomes derive from the ER where they pick up their cargo of KDEL-cysteine proteases, and unlike autophagosomes, the cargo is delivered directly into the cytoplasm of cells undergoing PCD. Of relevance to senescence-associated PCD is that ricinosomes were also seen in senescent day lily (Hemerocallis) petals [77]. However, there have been no further reports of them in senescent tissues. Furthermore, Battelli et al. [88] did not see the KDEL protein expressed in Lilium longiflorum senescent petals associated with ricinosome-like vesicles, but did see the KDEL protease in the vacuole. In seeds of Vigna mungo, KDEL proteins were also seen inside KDEL vesicles (KV) [71]. These appear to be very similar to ricinosomes in size (200–500 nm) but deliver their cargo to storage vacuoles instead of the cytoplasm. So clearly further work is needed to establish whether ricinosome-mediated delivery of KDEL proteases to the cytoplasm post-tonoplast rupture is a conserved feature of senescence-associated PCD or whether delivery to vacuoles in senescence-associated PCD also occurs.
Senescence-associated vesicles (SAVs) are seen in senescent tobacco, Arabidopsis and soybean leaves [82–84] and form independently from the autophagosome-required gene ATG7. These are highly acidic vacuoles containing high cysteine peptidase activity, of similar size to ricinosomes (700 nm) but are located in the peripheral cytoplasm around plastids. They are only visible in senescent leaves and appear to contain the majority of cellular cysteine protease activity in senescent leaf cells [84]. They contain the senescence marker protease SAG12, and in fact all cells expressing SAG12 contain SAVs; however, SAG12 is not required for their formation. They are involved in breakdown of chloroplasts and are only found in senescent leaf cells that contain chloroplasts. They therefore may be specifically required for chloroplast disassembly. However, SAG12 is also expressed in non-photosynthetic senescent tissues such as petals [21, 89], so it will be interesting to discover whether SAVs are also important in non-photosynthetic senescent tissues.
Two forms of ER bodies containing different sets of β-glucosidases have been identified in Arabidopsis. They are surrounded by ribosomes, and one form was found constitutively in Arabidopsis seedling epidermal cells. On wounding or salt stress, another form of ER body is induced. The ER bodies fuse with each other and then with the vacuole [85, 86]. Of particular interest is that ER bodies carry precursors to γVPE and a stress-inducible protease RD21. It would be interesting to verify whether these vesicles are ever detected during senescence-associated PCD.
ATI1 bodies are formed in response to carbon starvation from the ER network [87]. These compartments are marked by ATI1 and ATI2, plant-specific proteins which associate with the autophagosome protein ATG8. ATI1 bodies are clearly distinct from ER bodies and also from Golgi, mitochondria and peroxisomes and do not contain ER-lumenal markers. They are also distinct from autophagosomes as they did not co-localise with ATG8f-marked vesicles [86]. Their destination is the central vacuole, and it is proposed that they may be carrying mis-folded proteins for destruction. It will be important to identify their cargo and also to discover whether they play a role in stress-induced or senescence-associated PCD.
9.3.4 Role of the Mitochondrion and Cytochrome c Leakage
The mitochondrion plays a central role in the regulation of animal cell apoptosis [90]. Pro-apoptotic members of the Bcl-2 family cause permeabilisation of the outer mitochondrial membrane and release of cytochrome c into the cytosol. Cytochrome c binds with Apaf1 promoting a caspase cascade which results in death of the cell [91]. Hence, cytochrome c leakage was searched for and found in many plant PCD systems [92]. In plant cells, cytochrome c leakage into the cytoplasm is categorised by van Doorn et al. [15] as a feature of necrosis, associated with abiotic stress-induced PCD. In support of this, it has been reported, for example, in heat-induced PCD in tobacco BY2 cultures [93] and cucumber cotyledons [94], whereas in pollination-induced senescence of petunia flowers, the release of cytochrome c did not occur [95]. It was reported however during induced ageing in elm seeds [51]. This induction method aims to simulate seed ageing or senescence, by the induction of accelerated ageing through elevated temperatures (controlled deterioration treatment). It therefore calls into question whether this is PCD by senescence or abiotic stress induction. On the other hand, leakage of cytochrome c to the cytoplasm was also noted during cotyledon senescence in mung bean (Vigna radiata [96]) and during tulip petal senescence [97] processes which are clearly developmental and not induced by abiotic stress. Hence, whether cytochrome c release is a key component of senescence-associated PCD mechanisms remains an open question. Analysis of cytosolic cytochrome c levels in many more classical organ senescence systems at different time points is needed to resolve this question. Ideally, this should be done using techniques such as immunocytochemistry to visualise the cytochrome c levels spatially and correlate them with other PCD markers.
In animal cells, cytochrome c release can be triggered by increases in cytosolic calcium which activate the permeability transition pore (PTP), composed of VDAC and ANT. In plants, homologues to both VDAC and ANT are present, indicating that the PTP may be functional (recently reviewed by Diamond and McCabe [92]). VDAC expression is up-regulated in abiotic-induced PCD systems such as induction of PCD in pearl millet by salinity, drought, cold and SA [98]. However, in leaf senescence (eFP browser, [99, 100]) and ageing seeds [101], VDAC genes were either down-regulated or did not change in expression. This further supports a role for cytochrome c release, at least via the PTP, in abiotic stress induced but not senescence-induced plant PCD.
9.3.5 What Is the Role of the Endoplasmic Reticulum ?
The endoplasmic reticulum (ER) performs an important role in the cell in monitoring protein folding, both in plants and other eukaryotes. In plants, ER stress can be induced by a range of abiotic and biotic stresses that result in an imbalance between the rate at which proteins are synthesised and the capacity of the ER to process them. If unfolded proteins accumulate in the ER, this triggers the unfolded protein response (UPR) [102]. Initially, the cell mitigates the stress by up-regulating ER chaperones, reducing the rate of translation and increasing the rate of degradation of mis-folded proteins. In plants, these processes are mediated by two pathways. One involves ER-membrane associated TFs (bZIP17 and bZIP28). The other requires a protein kinase: RNA-splicing factor IRE1. However, under prolonged ER stress, autophagy or PCD can be triggered. Autophagosomes were formed in response to ER-stress in Arabidopsis seedlings [103], and this process required ATG18a. Autophagosome production also required IRE1 indicating a dual role for IRE1 in protecting against ER-stress and also in activating autophagy. However how PCD is triggered by ER-stress is less clear [102]. One possibility is that this is also via IRE1. In animal cells, the ER-located Bax inhibitor-I (BI-1) binds to IRE1 and decreases its activity [104]. BI-1 is conserved across eukaryotes, including plants, and seems to inhibit plant PCD [105–107]. Furthermore, it seems to play a role in mitigating ER stress [108]. BI-1 is highly up-regulated in senescent leaves (eFP browser, [100]) and is also up-regulated during silique senescence [21] although less so in petals. BI-1 is also linked with JA signalling: atbi1-2 knockout mutants show accelerated MeJA-induced leaf senescence, induced by an increase in calcium levels [109] probably released from the ER into the cytoplasm. So this provides a significant link between the ER and senescence-associated PCD.
Recently, a key role for the ER in plant senescence-associated PCD has been proposed [110]. DMP1 is a membrane protein localised both to the ER and tonoplast. When expressed under its native promoter, which drives senescence-specific expression, over-expression of DMP1 results in ER fragmentation, during both natural and dark-induced leaf senescence. The sequence of these membrane rearrangements indicates that ER disintegration precedes vacuolar collapse during senescence-associated PCD. So whether DMP1 and the ER are playing a regulatory role in senescence-associated PCD merits further investigation.
9.3.6 Proteolysis as a Regulator of Senescence-Associated PCD
Caspases are key regulators of animal apoptosis, and therefore extensive research was carried out to determine whether they had a role in the regulation of plant PCD [111]. It has been known for some time that caspase activity is associated with plant PCD; however, direct homologues of caspase genes have not been discovered in sequenced plant genomes. Attention was drawn to VPEs due to their caspase-1-like activity; however, unlike animal caspases that are located in the cytosol, VPEs are vacuolar. The Arabidopsis genome contains four VPE genes: α, β, γ and δ [72]. Mutation of all four VPEs in Arabidopsis inhibited PCD induced by the mycotoxin fumonisin B1 [112] and by infection with the mutualistic fungus Piriformospora indica [113]. Tonoplast rupture was also inhibited by mutation of all four VPE genes [112], and YVAD-ase (caspase-1-like) activity was abolished [112]. Only γVPE (AT4G32940) is up-regulated during leaf senescence (eFP browser, [100]), suggesting that it is this class that is most likely involved in senescence-associated PCD, although in Arabidopsis petals both γ- and βVPE genes were up-regulated [21]. Up-regulation of VPE genes has also been reported in petals of other species (carnation: [114]; daffodil: [115]; Ipomea: [116]). It will be interesting to see whether only γ- and βVPE genes are up-regulated in species other than Arabidopsis and critically identify their substrates.
According to van Doorn et al. [15], activation of VPEs is diagnostic of the vacuolar mechanisms of plant PCD. However, caspase activities are associated with a wide range of plant PCD systems including abiotic stress induced by heat (e.g. [93]), pathogens (e.g. [117]) and developmental processes during xylogenesis-associated PCD [118] as well as in petal senescence (e.g. [119]) and caspase-3-like activity in cotyledon senescence [96]. This suggests that if VPEs are indeed diagnostic of vacuolar cell death, then this mechanism is also contributing to PCD in these different PCD systems. However, caution must be exercised in the interpretation of these studies to infer that caspase activity is indeed involved in plant PCD. This is because in virtually all studies, substrates used to demonstrate caspase activity were synthetic tetra peptides containing the target sequence for mammalian caspases (such as YVAD as a target for caspase-1) and not whole proteins. These substrates are not fully specific for individual caspases [111] making the identification of specific caspase activities difficult.
Although the search for caspase genes in plant genomes was in vain, a related family of genes, metacaspases, were found [120]. Metacaspases are cysteine-dependent proteases falling into two structural classes (I and II). Although they do not have caspase activity, metacaspases have been associated with different forms of plant PCD including both abiotic (AtMC8, [121]; AtMC4, [122]) and biotic (AtMC1 and AtMC2, [123]) stress-induced PCD in Arabidopsis, as well as embryogenesis-related developmental PCD [124] in Picea. Of particular interest in Picea is the finding that autophagy is required for death of the suspensor, and furthermore it lies downstream of a class II metacaspase gene which is also required for suspensor PCD [125]. In addition, if this metacaspase–autophagy PCD pathway is suppressed by silencing ATG gene expression or the class II metacaspase, there is a switch from vacuolar PCD to necrotic cell death.
In Arabidopsis, there are nine metacaspase genes, and a lack of phenotypes from single mutants suggests that there may be functional redundancy between the family members, making it difficult to verify their role in different forms of PCD. Of the nine Arabidopsis metacaspase genes, only AtMC9 is highly expressed in senescence and PCD [126], and this gene is strongly up-regulated in older Arabidopsis petals (eFP browser, [100]). In their transcriptomic analysis of leaf senescence, Breeze et al. [25] found only two class II genes (AtMC6 and AtMC9) were up-regulated but were different to those up-regulated during other forms of plant PCD in Arabidopsis (see above). Their up-regulation coincided with the up-regulation of the ATG7 and ATG8 genes suggesting that this may be an important time-point in the activation of senescence-associated PCD. AtMC9 is mainly localised in the nucleus [126] though GFP fusion protein was also detected in the cytoplasm, so it will be very interesting to understand the timing of its enzymatic activity during senescence-associated PCD.
Recently, caspase-3 activity has been associated in very different systems with the proteasome. Caspase-3 activity associated with xylem differentiation in Populus was derived from the 20S proteasome subunit [118]. Furthermore, a caspase-3 activity linked to the release of antibacterial proteins into the intercellular space during infection was associated more specifically with the PBA1 (β1) subunit of the 20S proteasome core [117]. The caspase-3 activity in both systems seems likely to be involved in tonoplast breakdown [117, 118]. It will therefore be interesting to map more precisely the timing and location of the caspase-3 activity during senescence-associated PCD to establish whether it plays the same role here. Of course, the crucial point is the detection of the substrates, which presumably are ubiquitinated prior to direction to the proteasome.
The proteasome also appears to have an important role as a means of removing negative and positive regulators of senescence. ORE9 which is an F-box protein [127] and ATE [128] which is part of the N-rule ubiquitination pathway are both positive regulators of Arabidopsis leaf senescence as mutants in these genes show delayed senescence. In contrast, a RING-type ubiquitin ligase NLA and the PUB-ARM E3 ubiquitin ligase SAUL1 are negative leaf senescence regulators [129, 130]. The proteasome machinery has been implicated in PCD associated with the hypersensitive response, particularly the PUB-ARM proteins [131]. Hence, the finding that SAUL1 regulates senescence suggests a role for this protein also in senescence-associated PCD. Vogelmann et al. [132] showed up-regulation of leaf senescence-associated TFs such as WRKY53, WRKY6 ANAC092 and ANAC029 as well as premature PCD in saul1-1 mutants. Detailed analysis of the timing of changes in gene expression revealed up-regulation of SA-associated genes. A role for SA in SAUL1-induced senescence and senescence-associated PCD was confirmed by the finding that the effects of SAUL1 on senescence require PAD4, an enzyme required for SA signalling.
9.3.7 Downstream Effectors and Markers of Senescence-Associated PCD
It has long been known (e.g. [133]) that genes encoding degradative enzymes are up-regulated during senescence, and it is assumed that these are the effectors of senescence-associated PCD. The cysteine protease gene SAG12 has emerged as a key marker for senescence, expressed as the first signs of leaf yellowing become visible [25] and somewhat earlier in petal senescence [89]. Later in leaf senescence, as yellowing becomes more widespread, a high proportion (44 %) of expressed genes are involved in metabolism, presumably involved in the degradation of cell contents and nutrient remobilisation [25]. Metabolic genes were also highly expressed in petal senescence in all species studied (reviewed in [20]). Another feature of gene expression in late leaf senescence is the reactivation of cytoskeleton-related genes. In yeast and mammalian cells, microtubules are required for efficient autophagy [134], so this late up-regulation of cytoskeleton-associated genes may further support for an autophagy-like model of senescence-associated PCD.
9.3.8 Chloroplasts During Senescence-Associated PCD
One important division in senescing organs is between photosynthetic organs where chloroplasts play a key role in ROS generation and are a source of photosynthate, and non-photosynthetic organs which lack functional chloroplasts. In photosynthetic organs, senescence can be considered as a form of starvation which is imposed by a withdrawal of nutrients that are redirected to other parts of the plant. For example, many seed pods such as those of legumes and siliques in Brassica species are photosynthetic organs that undergo a senescence programme which has parallels with that in leaves. Chloroplasts are dismantled resulting in yellowing, and nutrients are transferred to the developing seeds [135]. Alternatively, a reduction in photosynthetic capacity may be a result of reduced light levels, reduced water availability or disease and stresses which tip the balance towards senescence.
Chlorophyll degradation is a key marker of the senescence of photosynthetically active tissues. Genes associated with chlorophyll breakdown are switched on relatively early, and the morphology of plastids changes to a form termed “gerontoplasts”. This process is reversible, and Thomas et al. [10] proposed that this is the stage that should be called senescence, whereas PCD starts after this, once a point of no return has been reached (Fig. 9.1). Although reversibility has been demonstrated in some systems, including cells in which downstream processes associated with PCD-like expression of SAG12 have been initiated [136], it is in fact relatively rare [8]. Thomas et al. [10] suggested that the presence of functioning chloroplasts is a key feature maintaining life of the cell and that a reversal of senescence and early PCD processes requires chloroplast function. A second peak in expression of chlorophyll breakdown genes late in senescence [25] may regulate the final breakdown of chlorophyll–protein complexes and the cessation of photosynthesis during the process that leads to irreversible cell death.
More recently, it has been shown that the reduction in size and number of chloroplasts occurring as a result of dark-induced senescence requires functional autophagy genes [137]. Furthermore, both chlorophyll and Rubisco were seen in the vacuole indicating that chloroplasts were being transported to the vacuole for degradation via autophagy. However, stromal proteins have also been seen in senescence-associated vesicles (SAVs) [83] which do not require a functional autophagic machinery [82]; hence, it would seem that at least two independent mechanisms are involved in chloroplast breakdown during senescence-associated PCD.
9.3.9 Progression of Senescence-Associated PCD Across Organs
One of the difficulties in studying the gene regulation of senescence-associated PCD is the asynchrony of PCD within an organ. In leaves, PCD often starts at the tip of the leaves (Fig 9.2a) seen as the areas that first yellow or turn brown in both dicots and monocots [3]. Staining with trypan blue that specifically stains dead cells shows the increase in dead cells with time during developmental leaf senescence [2]. In many autumnal leaves, the loss of chlorophyll is evidently delayed around the major veins. In common with leaves, senescence and PCD in cotyledons are not uniform across the organ. Thus, the first signs of PCD, using TUNEL staining as a marker, were seen in the margins of cotton cotyledons [138] followed later by the centre. The asynchrony of PCD is also evident in other organs such as siliques and petals. In petals, the loss of internal cells is remarkable in flowers that still appear in full bloom or early senescence (Fig 9.2b).
Heterogeneity of PCD across tissues in Arabidopsis leaves (a) and Lilium longiflorum senescent tepals (b–d). (a) Leaves from 10-week-old Arabidopsis plants: yellowing and browning typically starts at the tip of the leaf and spreads downwards (scale bar = 5 mm); (b) at loose closed bud stage, L. longiflorum tepal mesophyll cells are round in shape and rich in amyloplasts, without signs of autolysis. Magnification 10 K, scale bar, 1 μm; (c) at open flower stage, epidermal cells show a diffuse cytoplasm with vesiculation. Magnification 3.2 K, scale bar, 3 μm; (d) in early senescence (as tepals become translucent and show signs of browning). Intact vascular tissues and a completely collapsed mesophyll layer. Magnification 2 K, scale bar, 3 μm. Nuclei (n), vacuole (v), cell wall (cw), plastids (p), mitochondria (m), amyloplasts (a), vesicles (vs) and oleosomes (o). Electron microscopy images of L. longiflorum tepals are reproduced from R. Battelli PhD thesis. See Battelli et al. [119] for further details and images of tepal senescence in L. longiflorum
In organs other than leaves, a key point is the coordination of senescence and PCD between different cell types and related organs. In pods, for example, specialised sets of cells perform specific functions such as dehiscence to release the seeds [139]. The walls of Arabidopsis siliques comprise inner and outer epidermal cells enclosing three mesophyll layers and adjacent to the inner epidermis a layer of cells that become the encocarp. Cells of the inner epidermis start to undergo PCD first, while the endocarp cell walls thicken, and the cytoplasm of inner epidermal cells disappears completely by the time the siliques are fully yellow but well before dehiscence [21].
In many flowers, pollination triggers senescence and ultimate PCD (reviewed in [20]); however, PCD is regulated within the individual organs and cell types. In pistils, senescence starts in the transmitting tract shown by early cellular deterioration and activation of expression of the senescence marker gene BNF1 [140]. BNF1 is then activated in the stigma followed by the ovules. In the absence of pollination, pistils do senesce, although the timing of this event is species specific. In some species such as pea [141], this occurs after just 3 days, while flowers can last for months in orchids [14]. A key indicator of pistil senescence is a loss of responsiveness to GA3 which is able to induce parthenocarpic fruit development [140]; this in turn is regulated by ethylene which seems to be produced by the ovules [142]. The coordination of pistil senescence with petal senescence also seems to vary. In some species, such as almond, the stigma remains receptive even when petals are already senescing [143], while in others such as the economically important grass Leymus chinensis, the stigma remains receptive to pollen for only 3 h [144]. So another area in need of further research is the coordination of PCD within and between related senescing organs.
9.3.10 Senescence-Associated PCD On and Off the Plant
If remobilisation to other plant organs is the defining feature of senescence, then postharvest senescence or off-the-plant senescence must be categorised separately. However, both terminate in PCD; thus, it is of interest to review the progression and final stages of the process. Postharvest storage of plant material such as leafy vegetables and flowers results in deterioration which closely resembles physiological features of on-the-plant senescence including protein degradation, leaf yellowing or petal wilting (e.g. [20, 145]).
Perhaps surprisingly, processes very similar to those evoked during developmental or stress-induced senescence on the plant are also activated in organs that are detached from the plant. Even a leaf that has been ingested by a ruminant activates similar biochemical and cellular processes as those activated during senescence [146] including chlorophyll and protein breakdown, increase of protease and nuclease activity and increasing membrane leakage indicating progressive PCD. Both SAG12 and γVPE were up-regulated postharvest in broccoli [145], suggesting similar PCD processes.
The majority of studies on petal senescence are performed with detached flowers, and remarkably few studies have compared on- and off-the-plant senescence. Arrom and Munné-Bosch [37] did show differences in endogenous growth regulator levels; however, these were difficult to interpret. An interesting study on PCD in detached petals [147] indicates that nuclear morphology in abscised petals resembles more closely the effects of dehydration-induced PCD than developmental PCD in petals. Differing morphologies of petal PCD were found in different species [148] in which either DNA fragmented within the nucleus or membrane-bound chromatin-containing bodies indicated nuclear fragmentation. Further detailed comparisons of the cytology of petal PCD on and off the plant in abscising and non-abscising species would be interesting, to determine whether the vacuolar/autophagic mechanism is operating in all cases.
Other forms of senescence such as fruit and seed senescence necessarily occur off the plant. The duration of seed viability is a species-specific character which is also affected by environmental effects such as temperature and humidity [149, 150], and viability declines with age. Several studies have shown that seed ageing is associated with nucleic acid degradation (e.g. [151, 152]) and increased oxidative stress resulting in lipid peroxidation (e.g. [153]). Recently, a transcriptomic survey of artificially accelerated pea seed ageing showed that an early change, prior to loss of viability, is up-regulation of genes related to PCD including BI-1, oxidative stress and protein ubiquitination [101]. Markers of ER-stress such as BiP were also up-regulated. A reduction in the expression of antioxidant-related genes was also reported in other systems (e.g. sunflower seeds [154]). In a controlled deterioration treatment experiment with elm seeds [51], changes in ROS were also associated with PCD markers. Furthermore, a rise in caspase activity and cytochrome c release could be blocked by treatment with ascorbic acid, suggesting a link between oxidative stress and PCD-linked caspase activity.
9.4 Future Prospects and Tools Needed
One of the difficulties in deriving mechanisms applicable across senescence systems is a lack of comparable analyses in different organs within the same species. At a molecular level, these have been attempted comparing different organs within model species such as Arabidopsis [21] and ornamentals such as wallflowers [89]. Given new perspectives on the cytological mechanisms of senescence-associated PCD [15], it would be very useful to revisit well-studied senescence systems such as leaves and petals as well as the less studied senescence systems such as other floral organs within the same species to establish common mechanisms.
Another issue that remains to be resolved is the spatial and temporal development of PCD through individual organs. There is a need to study gene expression and signal perception and transduction at a cellular rather than an organ level to understand how PCD is being regulated within the cell and across the organ. The use of transgenic lines expressing fluorescent proteins fused to markers for cell types and cell status may help to resolve this.
The input signals that start the senescence process seem to operate through overlapping growth regulators and downstream TFs. More careful comparisons of different induction systems such as that carried out by the PRESTA project (http://www2.warwick.ac.uk/fac/sci/lifesci/research/presta/) examining the transcriptomic patterns induced by different stress treatments provide a valuable resource. This needs to be expanded since comparisons of existing transcriptomic data carried out in different labs with different parameters run the risk of being misleading. Unravelling networks of TFs is telling us a lot about upstream regulation of senescence and PCD; however, even downstream genes may have multiple input signals from several TFs, so the network may be very complex. TF networks will also need to be integrated with an understanding of epigenetic mechanisms and posttranslational protein–protein interactions to get a full picture of the network.
However, next-generation sequencing opens the way to study these mechanisms beyond the well-studied model organisms into crop species. This is especially needed in horticultural crops that are represented by so many different species and varieties. These provide perishable fresh produce, and potential benefits include improving nutritional quality while reducing waste in support of a healthy diet and food security.
References
Guiboileau A, Sormani R, Meyer C, Masclaux-Daubresse C (2010) Senescence and death of plant organs: nutrient recycling and developmental regulation. C R Biol 333:382–391
Woo HR, Kim HJ, Nam HG, Lim PO (2013) Plant leaf senescence and death – regulation by multiple layers of control and implications for aging in general. J Cell Sci 126:4823–4833
Thomas H (2013) Senescence, ageing and death of the whole plant. New Phytol 197:696–711
Bedinger P (1992) The remarkable biology of pollen. Plant Cell 4:879–887
Lombardi L, Casani S, Ceccarelli N, Galleschi L, Picciarelli P, Lorenzi R (2007) Programmed cell death of the nucellus during Sechium edule Sw. seed development is associated with activation of caspase-like proteases. J Exp Bot 58:2949–2958
Lombardi L, Ceccarelli N, Picciarelli P, Lorenzi R (2007) Caspase-like proteases involvement in programmed cell death of Phaseolus coccineus suspensor. Plant Sci 172:573–578
Gunawardena AHLAN (2008) Programmed cell death and tissue remodelling in plants. J Exp Bot 59:445–451
van Doorn WG, Woltering EJ (2004) Senescence and programmed cell death: substance or semantics? J Exp Bot 55:2147–2153
Fischer AM (2012) The complex regulation of senescence. Crit Rev Plant Sci 31:124–147
Thomas H, Ougham HJ, Wagstaff C, Stead AD (2003) Defining senescence and death. J Exp Bot 54:1127–1132
Reape TJ, McCabe PF (2008) Tansley review: Apoptotic-like programmed cell death in plants. New Phytol 180:13–26
Wagstaff C, Malcolm P, Rafiq A, Leverentz M, Griffiths G, Thomas B, Stead A, Rogers H (2003) Programmed cell death (PCD) processes begin extremely early in Alstroemeria petal senescence. New Phytol 160:49–59
Gan S, Amasino RM (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270:1986–1988
Rogers HJ (2006) Programmed cell death in floral organs: how and why do flowers die? Ann Bot (Lond) 97:309–315
van Doorn WG, Beers EP, Dangl JL, Franklin-Tong VE, Gallois P, Hara-Nishimura I, Jones AM, Kawai-Yamada M, Lam E, Mundy J, Mur LAJ, Petersen M, Smertenko A, Taliansky M, Van Breusegem F, Wolpert T, Woltering E, Zhivotovsky B, Bozhkov PV (2011) Morphological classification of plant cell deaths. Cell Death Differ 18:1241–1246
Gepstein S, Glick BR (2013) Strategies to ameliorate abiotic-stress induced plant senescence. Plant Mol Biol 82:623–633
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136
Gregersen PL, Culetic A, Boschian L, Krupinska K (2013) Plant senescence and crop productivity. Plant Mol Biol 82:603–622
Mittal S (2007) Strengthening backward and forward linkages in horticulture: some successful initiatives. Agric Econ Res Rev 20:457–469
Rogers HJ (2013) From models to ornamentals: how is flower senescence regulated? Plant Mol Biol 82:563–574
Wagstaff C, Yang TJW, Stead AD, Buchanan-Wollaston V, Roberts JA (2009) A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J 57:690–705
Guo Y, Gan SS (2012) Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ 35:644–655
Sabelli PA (2012) Replicate and die for your own good: endoreduplication and cell death in the cereal endosperm. J Cereal Sci 56:9–20
Schippers JHM, Jing HC, Hille J, Dijkwel PP (2007) Developmental and hormonal control of leaf-senescence. In: Plants SG (ed) In senescence processes. Blackwell, Oxford, pp 145–170
Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim Y, Penfold CA, Jenkins D, Zhang C, Morris K, Jenner C, Jackson S, Thomas B, Tabrett A, Legaie R, Moore JD, Wild DL, Ott S, Rand D, Beynon J, Denby K, Mead A, Buchanan-Wollaston V (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23:873–894
Richmond AE, Lang A (1957) Effect of kinetin on protein content and survival of detached Xanthium leaves. Science 125:650–651
Hwang I, Sheen J, Müller B (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63:353–380
Chang H, Jones ML, Banowetz GM, Clark DG (2003) Overproduction of cytokinins in petunia flowers transformed with PSAG12:IPT delays corolla senescence. Plant Physiol 132:2174–2183
Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJ (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42:567–585
Pourtau N, Mares M, Purdy S, Quentin N, Ruel A, Wingler A (2004) Interactions of abscisic acid and sugar signalling in the regulation of leaf senescence. Planta 219:765–772
Morris K, Mackerness SAH, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23:677–685
Abreu M, Munné-Bosch S (2009) Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana. J Exp Bot 60:1261–1271
Kinoshita T, Yamada K, Hiraiwa N, Kondo M, Nishimura M, Hara-Nishimura I (1999) Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J 19:43–53
Robatzek S, Somssich E (2001) A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J 28:123–133
Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55:853–867
van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge U, Kunze R (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141:776–792
Arrom L, Munné-Bosch S (2012) Hormonal changes during flower development in floral tissues of Lilium. Planta 236:343–354
Ueda J, Kato J (1980) Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol 66:246–249
Reinbothe C, Springer A, Samol I, Reinbothe S (2009) Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J 276:4666–4681
Tsuchiya T, Ohta H, Okawa K et al (1999) Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA 96:15262–15367
Shan X, Wang J, Chua L, Jiang D, Peng W, Xie D (2011) The role of Arabidopsis rubisco activase in jasmonate-induced leaf senescence. Plant Physiol 155:751–764
Hiderhofer K, Zentgraf U (2001) Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213:469–473
Oh SA, Park JH, Lee GI, Paek KH, Park S, Nam HG (1997) Identification of three genetic loci controlling leaf-senescence in Arabidopsis thaliana. Plant J 12:527–535
van Doorn WG, Woltering EJ (2008) Physiology and molecular biology of petal senescence. J Exp Bot 59:453–480
Jing HC, Sturre MJ, Hille J, Dijkwel PP (2002) Arabidopsis onset of leaf death mutants identify a regulatory pathway controlling leaf-senescence. Plant J 32:51–63
Jing H, Schippers J, Hille J, Dijkwel P (2005) Ethylene-induced leaf senescence depends on age-related changes and OLD genes in Arabidopsis. J Exp Bot 56:2915–2923
Bieker S, Riester L, Stahl M, Franzaring J, Zentgraf U (2012) Senescence-specific alteration of hydrogen peroxide levels in Arabidopsis thaliana and oilseed rape spring variety Brassica napus L. cv. Mozart. J Integr Plant Biol 54:540–554
Rogers HJ (2012) Is there an important role for reactive oxygen species and redox regulation during floral senescence? Plant Cell Environ 35:217–233
Bar-Dror T, Dermastia M, Kladnik A, Tušek Žnidarič M, Pompe Novak M, Meir S, Burd S, Philosoph-Hadas S, Ori N, Sonego L, Dickman MB, Lers A (2011) Programmed cell death occurs asymmetrically during abscission in tomato. Plant Cell 23:4146–4163
He X, Kermode AR (2010) Programmed cell death of the megagametophyte during post-germinative growth of white spruce (Picea glauca) seeds is regulated by reactive oxygen species and the ubiquitin-mediated proteolysis system. Plant Cell Physiol 51:1707–1720
Hu D, Ma G, Wang Q, Yao J, Wang Y, Pritchard HW, Wang X (2012) Spatial and temporal nature of reactive oxygen species production and programmed cell death in elm (Ulmus pumila L) seeds during controlled deterioration. Plant Cell Environ 35:2045–2059
Garmier M, Priault P, Vidal G, Driscoll S, Djebbar R, Boccara M, Mathieu C, Foyer CH, Paepe RD (2007) Light and oxygen are not required for harpin-induced cell death. J Biol Chem 282:37556–37566
Zentgraf U, Hemleben V (2008) Molecular cell biology: are reactive oxygen species regulators of leaf senescence? Prog Bot 69:117–138
Hickman R, Hill C, Penfold CA, Breeze E, Bowden L, Moore JD, Zhang P, Jackson A, Cooke E, Bewicke-Copley F, Mead A, Beynon J, Wild DL, Denby KJ, Ott S, Buchanan-Wollaston V (2013) A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J 75:26–39
Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue G-P, Mueller-Roebera B (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4:346–360
Besseau S, Li J, Tapio Palva E (2012) WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot 63:2667–2679
Ulker B, Shahid Mukhtar M, Somssich IE (2007) The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226:125–137
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371
Gutterson N, Reuber TL (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7:465–471
Faria JAQA, Reis PAB, Reis MTB, Rosado GL, Pinheiro GL, Mendes GC, Fontes EPB (2011) The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress- and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol 11:129
Mendes GC, Reis PAB, Calil IP, Carvalho HH, Aragão FJL, Fontes EPB (2013) GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc Natl Acad Sci USA 110:19627–19632
Naqvi AR, Sarwat M, Hasan S, Choudhury NR (2012) Biogenesis, functions and fate of plant microRNAs. J Cell Physiol 227:3163–3168
Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 35:21–44
Sunkar R, Chinnusamy V, Zhu J, Zhu JK (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12:301–309
Rajagopal R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20:3407–3425
Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323:1053–1057
Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6:230
Fink SL, Cookson BT (2005) Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun 73:1907–1916
Kitanaka C, Kuchino Y (1999) Caspase-independent programmed cell death with necrotic morphology. Cell Death Differ 6:508–515
Toyooka K, Okamoto T, Minamikawa T (2001) Cotyledon cells of Vigna mungo seedlings use at least two distinct autophagic machineries for degradation of starch granules and cellular components. J Cell Biol 154:973–982
Hara-Nishimura I, Hatsugai N (2011) The role of vacuole in plant cell death. Cell Death Differ 18:1298–1304
Liu Y, Bassham DC (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63:215–237
Shibuya K, Yamada T, Suzuki T, Shimizu K, Ichimura K (2009) InPSR26, a putative membrane protein, regulates programmed cell death during petal senescence in Japanese morning glory. Plant Physiol 149:816–824
Shibuya K, Niki T, Ichimura K (2013) Pollination induces autophagy in petunia petals via ethylene. J Exp Bot 64:1111–1120
Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21:2914–2927
Schmid M, Simpson D, Gietl C (1999) Programmed cell death in castor bean endosperm is associated with the accumulation and release of a cysteine endopeptidase from ricinosomes. Proc Natl Acad Sci USA 96:14159–14164
Schmid M, Simpson DJ, Sarioglu H, Lottspeich F, Gietl C (2001) The ricinosomes of senescing plant tissue bud from the endoplasmic reticulum. Proc Natl Acad Sci USA 98:5353–5358
Greenwood JS, Helm M, Gietl C (2005) Ricinosomes and endosperm transfer cell structure in programmed cell death of the nucellus during Ricinus seed development. Proc Natl Acad Sci USA 102:2238–2243
Senatore A, Trobacher CP, Greenwood JS (2009) Ricinosomes predict programmed cell death leading to anther dehiscence in tomato. Plant Physiol 149:775–790
López-Fernández MP, Maldonado S (2013) Ricinosomes provide an early indicator of suspensor and endosperm cells destined to die during late seed development in quinoa (Chenopodium quinoa). Ann Bot (Lond) 112:1253–1262
Otegui MS, Noh YS, Martínez DE, Vila Petroff MG, Staehelin LA, Amasino RM, Guiamet JJ (2005) Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J 41:831–844
Martínez DE, Costa ML, Gomez FM, Otegui MS, Guiamet JJ (2008) ‘Senescence-associated vacuoles’ are involved in the degradation of chloroplast proteins in tobacco leaves. Plant J 56:196–206
Carrión CA, Lorenza Costa M, Martínez DE, Mohr C, Humbeck K, Guiamet JJ (2013) In vivo inhibition of cysteine proteases provides evidence for the involvement of ‘senescence-associated vacuoles’ in chloroplast protein degradation during dark-induced senescence of tobacco leaves. J Exp Bot 64:4967–4980
Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa N, Nishimura M, Hara-Nishimura I (2001) A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol 42:894–899
Ogasawara K, Yamada K, Christeller JT, Kondo M, Hatsugai N, Hara-Nishimura I, Nishimura M (2009) Constitutive and inducible ER bodies of Arabidopsis thaliana accumulate distinct β-glucosidases. Plant Cell Physiol 50:480–488
Honig A, Avin-Wittenberg T, Ufaz S, Galili G (2012) A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 24:288–303
Battelli R, Lombardi L, Picciarelli P, Lorenzi R, Frigerio L, Rogers HJ (2014) Expression and localisation of a senescence-associated KDEL-cysteine protease from Lilium longiflorum tepals. Plant Sci 214:38–46
Price A, Orellana D, Salleh F, Stevens R, Acock R, Buchanan-Wollaston V, Stead A, Rogers H (2008) A comparison of leaf and petal senescence in wallflower reveals common and distinct patterns of gene expression and physiology. Plant Physiol 147:1898–1912
Martinou J-C, Youle RJ (2011) Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell 21:92–101
Tait SWG, Green DR (2010) Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 11:621–632
Diamond M, McCabe PF (2011) Mitochondrial regulation of plant programmed cell death. Adv Plant Biol 1:439–465
Vacca RA, Valenti D, Bobba A, Merafina RS, Passarella S, Marra E (2006) Cytochrome c is released in a reactive oxygen species-dependent manner and is degraded via caspase-like proteases in tobacco bright-yellow 2 cells en route to heat shock-induced cell death. Plant Physiol 141:208–219
Balk J, Leaver CJ, McCabe PF (1999) Translocation of cytochrome c from the mitochondria to the cytosol occurs during heat-induced programmed cell death in cucumber plants. FEBS Lett 463:151–154
Xu Y, Hanson MR (2000) Programmed cell death during pollination-induced petal senescence in Petunia. Plant Physiol 122:1323–1333
Egorova VP, Zhao Q, Lo Y-S, Jane W-N, Cheng N, Hou S-Y, Dai H (2010) Programmed cell death of the mung bean cotyledon during seed germination. Bot Stud 51:439–449
Azad AK, Ishikawa T, Ishikawa T, Sawa Y, Shibata H (2008) Intracellular energy depletion triggers programmed cell death during petal senescence in tulip. J Exp Bot 59:2085–2095
Desai MK, Mishra RN, Verma D, Nair S, Sopory SK, Reddy MK (2006) Structural and functional analysis of a salt stress inducible gene encoding voltage dependent anion channel (VDAC) from pearl millet (Pennisetum glaucum). Plant Physiol Biochem 44:483–493
Swidzinski JA, Sweetlove LJ, Leaver CJ (2002) A custom microarray analysis of gene expression during programmed cell death in Arabidopsis thaliana. Plant J 30:431–446
Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. Plos One 8:e718
Chen H, Osuna D, Colville L, Lorenzo O, Graeber K, Küster H, Leubner-Metzger G, Kranner I (2013) Transcriptome-wide mapping of pea seed ageing reveals a pivotal role for genes related to oxidative stress and programmed cell death. PLoS One 8:e78471
Howell SH (2013) Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol 64:477–499
Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC (2013) Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24:4635–4651
Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, Walter P, Reed JC, Glimcher LH, Hetz C (2009) BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1α. Mol Cell 33:679–691
Kawai-Yamada M, Ohori Y, Uchimiya H (2004) Dissection of Arabidopsis Bax Inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell 16:21–32
Matsumura H, Nirasawa S, Kiba A, Urasaki N, Saitoh H, Ito M, Kawai-Yamada M, Uchimiya H, Terauchi R (2003) Overexpression of Bax inhibitor suppresses the fungal elicitor-induced cell death in rice (Oryza sativa L.) cells. Plant J 33:425–434
Watanabe N, Lam E (2006) Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J 45:884–894
Watanabe N, Lam E (2008) BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J Biol Chem 283:3200–3210
Yue H, Nie S, Xing D (2012) Over-expression of Arabidopsis Bax inhibitor-1 delays methyl jasmonate-induced leaf senescence by suppressing the activation of MAP kinase 6. J Exp Bot 63:4463–4474
Kasaras A, Melzer M, Kunze R (2012) Arabidopsis senescence-associated protein DMP1 is involved in membrane remodeling of the ER and tonoplast. BMC Plant Biol 12:54
Bonneau L, Ge Y, Drury GE, Gallois P (2008) What happened to plant caspases? J Exp Bot 59:491–499
Kuroyanagi M, Yamada K, Hatsugai N, Kondo M, Nishimura M, Hara-Nishimura I (2005) Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J Biol Chem 280:32914–32920
Qiang X, Zechmann B, Reitz MU, Kogel KH, Schafer P (2012) The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an endoplasmic reticulum stress-triggered caspase-dependent cell death. Plant Cell 24:794–809
Hoeberichts FA, van Doorn WG, Vorst O, Hall RD, van Wordragen MF (2007) Sucrose prevents up-regulation of senescence-associated genes in carnation petals. J Exp Bot 58:2873–2885
Hunter DA, Steele BC, Reid MS (2002) Identification of genes associated with perianth senescence in daffodil (Narcissus pseudonarcissus L. ‘Dutch Master’). Plant Sci 163:13–21
Yamada T, Ichimura K, Kanekatsu M, van Doorn WG (2009) Homologues of genes associated with programmed cell death in animal cells are differentially expressed during senescence of Ipomoea nil petals. Plant Cell Physiol 50:610–625
Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, Hara-Nishimura I (2009) A novel membrane fusion-mediated plant immunity against bacterial pathogens. Gene Dev 23:2496–2506
Han J-J, Lin W, Oda Y, Cui K-M, Fukuda H, He X-Q (2012) The proteasome is responsible for caspase-3-like activity during xylem development. Plant J 72:129–141
Battelli R, Lombardi L, Rogers HJ, Picciarelli P, Lorenzi R, Ceccarelli N (2011) Changes in ultrastructure, protease and caspase-like activities during flower senescence in Lilium longiflorum. Plant Sci 180:716–725
Tsiatsiani L, Van Breusegem F, Gallois P, Zavialov A, Lam E, Bozhkov PV (2011) Metacaspases. Cell Death Differ 18:1279–1288
He R, Drury GE, Rotari VI, Gordon A, Willer M, Farzaneh T, Woltering EJ, Gallois P (2008) Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J Biol Chem 283:774–783
Watanabe N, Lam E (2011) Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J 66:969–982
Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P (2010) Arabidopsis type I metacaspases control cell death. Science 330:1393–1397
Suarez MF, Filonova LH, Smertenko A, Savenkov EI, Clapham DH, von Arnold S, Zhivotovsky B, Bozhkov PV (2004) Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr Biol 14:R339–R340
Minina EA, Filonova LH, Fukada K, Savenkov EI, Gogvadze V, Clapham D, Sanchez-Vera V, Suarez MF, Zhivotovsky B, Daniel G, Smertenko A, Bozhkov PV (2013) Autophagy and metacaspase determine the mode of cell death in plants. J Cell Biol 203:917–927
Kwon SI, Hwang DJ (2013) Expression analysis of the metacaspase gene family in Arabidopsis. J Plant Biol 56:391–398
Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13:1779–1790
Yoshida S, Ito M, Callis J, Nishida I, Watanabe A (2002) A delayed leaf senescence mutant is defective in arginyl-tRNA:protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J 32:129–137
Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ (2007) A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J 50:320–337
Raab S, Drechsel G, Zarepour M, Hartung W, Koshiba T, Bittner F, Hoth S (2009) Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J 59:39–51
Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16:2795–2808
Vogelmann K, Drechsel G, Bergler J, Subert C, Philippar K, Soll J, Engelmann JC, Engelsdorf T, Voll LM, Hoth S (2012) Early senescence and cell death in Arabidopsis saul1 mutants involves the PAD4-dependent salicylic acid pathway. Plant Physiol 159:1477–1487
Smart CM (1994) Gene expression during senescence. New Phytol 126:419–448
Monastyrska I, Rieter E, Klionsky DJ, Reggiori F (2009) Multiple roles of the cytoskeleton in autophagy. Biol Rev Camb Philos Soc 84:431–448
Hocking PJ, Mason L (1993) Accumulation, distribution and redistribution of dry matter and mineral nutrients in fruits of canola (oilseed rape), and the effects of nitrogen fertilizer and windrowing. Aust J Agric Res 44:1377–1388
Belenghi B, Salomon M, Levine A (2004) Caspase-like activity in the seedlings of Pisum sativum eliminates weaker shoots during early vegetative development by induction of cell death. J Exp Bot 55:889–897
Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A (2009) Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 149:885–893
Xie QE, Liu ID, Yu SX, Wang RF, Fan ZX, Wang YG, Shen FF (2008) Detection of DNA ladder during cotyledon senescence in cotton. Biol Plant 52:654–659
Spence J, Vercher Y, Gates P, Harris N (1996) ‘Pod shatter’ in Arabidopsis thaliana, Brassica napus and B. juncea. J Microsci 181:195–203
Carbonell-Bejerano P, Urbez C, Carbonell J, Granell A, Perez-Amador MA (2010) A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol 154:163–172
Carbonell J, Garcia-Martínez JL (1985) Ribulose-1,5-bisphosphate carboxylase and fruit set or degeneration of unpollinated ovaries of Pisum sativum L. Planta 164:534–539
Carbonell-Bejerano P, Urbez C, Granell A, Carbonell J, Perez-Amador MA (2011) Ethylene is involved in pistil fate by modulating the onset of ovule senescence and the GA-mediated fruit set in Arabidopsis. BMC Plant Biol 11:84
Yi W, Law SE, Mccoy D, Wetzstein HY (2006) Stigma development and receptivity in Almond (Prunus dulcis). Ann Bot (Lond) 97:57–63
Huang Z, Zhu J, Mu X, Lin J (2004) Pollen dispersion, pollen viability and pistil receptivity in Leymus chinensis. Ann Bot (Lond) 93:295–301
Page T, Griffiths G, Buchanan-Wollaston V (2001) Molecular and biochemical characterization of postharvest senescence in Broccoli. Plant Physiol 125:718–727
Kingston-Smith AH, Davies TE, Edwards JE, Theodorou MK (2008) From plants to animals; the role of plant cell death in ruminant herbivores. J Exp Bot 59:521–532
Yamada T, van Ichimura K, Doorn WG (2007) Relationship between petal abscission and programmed cell death in Prunus yedoensis and Delphinium. Planta 226:1195–1205
Yamada T, Ichimura K, van Doorn WG (2006) DNA degradation and nuclear degeneration during programmed cell death in petals of Antirrhinum, Argyranthemum, and Petunia. J Exp Bot 57:3543–3552
Walters C (1998) Understanding the mechanisms and kinetics of seed aging. Seed Sci Res 8:223–244
Kranner I, Minibayeva FV, Beckett RP, Seal CE (2010) What is stress? Concepts, definitions and applications in seed science. New Phytol 188:655–673
Osborne DJ, Sharon R, Ben-Ishai R (1981) Studies on DNA integrity and DNA repair in germinating embryos of rye (Secale cereale). Israel J Bot 29:259–272
Kranner I, Chen HY, Pritchard HW, Pearce SR, Birtic S (2011) Inter-nucleosomal DNA fragmentation and loss of RNA integrity during seed aging. Plant Growth Regul 63:63–72
McDonald MB (1999) Seed deterioration: physiology, repair and assessment. Seed Sci Technol 27:177–237
Kibinza S, Bazin J, Bailly C, Farrant JM, Corbineau F, El-Maarouf-Bouteau H (2011) Catalase is a key enzyme in seed recovery from ageing during priming. Plant Sci 181:309–315
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Rogers, H.J. (2015). Senescence-Associated Programmed Cell Death. In: Gunawardena, A.N., McCabe, P.F. (eds) Plant Programmed Cell Death. Springer, Cham. https://doi.org/10.1007/978-3-319-21033-9_9
Download citation
DOI: https://doi.org/10.1007/978-3-319-21033-9_9
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-21032-2
Online ISBN: 978-3-319-21033-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)