Abstract
Sickle cell disease (SCD) is a monogenic disorder caused by a single base mutation but despite its apparent genetic simplicity, the clinical phenotype is hugely variable. In addition to environmental factors, family and epidemiological studies indicate that genetic variants co-inherited with the sickle mutation have a key role in modifying the disease course. This article provides an overview of the genetic modifiers of SCD known to date. Co-inheritance of α-thalassemia and persistent fetal hemoglobin (HbF) production are established major genetic modifiers but they do not explain the full spectrum of the phenotypic variability of SCD. While characterization of some of the key variants and pathways involved in HbF regulation have provided new therapeutic targets for HbF reactivation, generation of a personalized genetic risk score to inform prognosis and guide management requires a larger panel of genetic modifiers yet to be discovered. Elucidation of new genetic modifiers may also provide an insight into other “druggable” targets for therapeutic intervention.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
15.1 Introduction
Sickle cell disease (SCD) has been heralded as the first “molecular disease” when Pauling ascribed its basis to the presence of an abnormal hemoglobin in 1949 (Pauling et al. 1949). In 1957, Ingram (1957) described the abnormal hemoglobin as being caused by a single amino acid substitution (glutamic acid changed to valine) at position 6 of the β-globin chain of hemoglobin and in 1963, Goldstein et al. (1963) showed that this arose from a single base change of T > A at codon 6. Figure 15.1 summarizes a timeline of the significant events that have contributed to the understanding of the genetic basis, and management of SCD.
A timeline of significant events that have contributed to the understanding of the genetic basis and management of SCD. SCA sickle cell anemia, mRNA messenger ribonucleic acid, PND prenatal diagnosis, RFLP restriction fragment length polymorphism, CSSCD co-operative study of sickle cell disease, PGD preimplantation genetic diagnosis, BMT bone marrow transplantation, PCR polymerase chain reaction, RE restriction enzyme, GWAS genome-wide association study
In addition to homozygosity for the βS allele (HbSS, also referred to as sickle cell anemia, SCA), the syndrome of SCD includes HbSC disease (compound heterozygosity of HbS with HbC, Glu6 to Lys6), and HbSβ thalassemia (HbSβ+ or HbSβ0 thalassemia, depending on the type of the β-thalassemia mutation). Generally, the compound heterozygotes have a milder disease than HbSS, but even within each genotypic and ethnic group, a spectrum of clinical variability is the recurring theme. For example, within the HbSS group, at the mild end, patients can be asymptomatic while early mortality, frequent hospital admissions with acute pain episodes, childhood strokes and other end organ damage, typify the severe end of the clinical spectrum.
Both environmental and genetic factors contribute to this clinical variability. The importance of weather changes such as cold and rain as triggers of acute pain have been recognized and reported for many years but the conclusions were not consistent due to logistical difficulties in conducting such studies. Environmental factors also include nutritional state, access to social support, and medical care, all of which influence risk factors such as infections. The impact of environmental factors is demonstrated most graphically on the differences in the natural history and outcomes of SCD between the high- and low-income countries.
Twin studies, based on the concordance and discordance of disease complications and severity, have traditionally been used to assess the relative contributions of genetic and environmental factors in complex disorders such as diabetes and schizophrenia. Since monozygotic twins have identical DNA sequences, variation in their disease course can be attributed largely to the effects of the environment. There are three reports of this kind in SCD; two were limited to single pairs of identical twins, one with HbSS and α-thalassemia, and the other with HbSβ thalassemia (Amin et al. 1991; Joishy et al. 1976). The third study investigated nine pairs of identical twins, six with HbSS and three HbSC, from Jamaica (Weatherall et al. 2005). These twins have been followed for 15 years or more, and as a comparison group for examining degrees of concordance between laboratory parameters, 350 gender- and age-matched sibling pairs were also studied. These studies reported that while the twins showed similarities and concordance in laboratory parameters, and attained height and weight, there was discordance in frequency of acute painful episodes and other clinically critical complications. The conclusion was that environmental factors are of great importance in defining the clinical course of SCD.
Family and epidemiological studies indicate that genes co-inherited with the sickle mutation have a key role in modifying the disease course, including higher incidence of stroke (Driscoll et al. 2003) and concordant response to hydroxycarbamide therapy in siblings (Steinberg et al. 1997). Co-inheritance of α-thalassemia and persistent fetal hemoglobin (HbF) production are established major genetic modifiers of SCD. Numerous candidate gene and genome-wide association studies (GWAS) have defined genetic differences in SCD patients and attempted to identify other genetic variants with particular disease complications. However, the disease complexity has presented immense challenges, and these studies have only provided a small amount of the variation in SCD severity observed in the clinic. Roles for other genetic modifiers of SCD severity have been proposed based on the pathophysiology downstream of the primary event (HbS polymerization), however, the majority of these putative markers have not been replicated.
The clinical diversity of SCD itself presents difficulties for genotype/phenotype correlation studies in terms of accurately defining clinical “sub-phenotypes” (Ballas et al. 2012; Smith-Whitley and Pace 2007; Rees et al. 2010). The clinical implications of a clearer understanding of the genetic variants and mechanisms responsible for the phenotypic variability of SCD are significant. First, the ability to predict disease severity based on a genetic SCD “panel” to facilitate risk stratification of patients: high risk patients might then be followed more intensively, and higher risk therapies (hematopoietic stem cell transplantation, hydroxycarbamide) could be targeted at these patients. Second, new modifying genetic variants might suggest new therapeutic targets for investigation.
We describe the current understanding in terms of determining this phenotypic diversity as well as provide an update of the genetic modifiers of severity in SCD.
15.2 Complications of SCD and Problems in Defining Severity of Phenotypes: Global vs Sub-phenotypes
While full genetic understanding of SCD remains incomplete, the emergence of genome-wide genotyping platforms, next generation sequencing, and rapid advances in genetic research has re-focused some attention back onto phenotyping. Clear and consistent definition of phenotypes is critical to the success of genetic association studies. This is particularly pertinent in SCD where there is profound variety in both the severity and nature of complications. Many of its complications are acute such as recurrent acute pain episodes, acute chest syndrome, priapism and stroke; some are intermittent, leading eventually to chronic complications and organ damage, such as chronic pain, pulmonary hypertension and sickle chronic lung disease, penile dysfunction and cognitive disability. The acute complications vary considerably not only between patients but also in the same patient with time.
Phenotypes can be clinical or laboratory parameters. While laboratory parameters are simple to measure, their values vary with the clinical state of the patient; for example, lactate dehydrogenase and white cell count which are normally elevated during steady-state, further increase during acute clinical events. Phenotypes are not always consistent or valid, e.g. pulmonary hypertension. Furthermore, some complications are uncommon (e.g. overt strokes) which makes related or “intermediate” traits a preferred endpoint (e.g. imaging results such as raised trans-cranial Doppler (TCD) velocities, or silent infarcts on magnetic resonance imaging).
Many studies focus on specific complications of SCD—sometimes described as “sub-phenotypes”—i.e. particular end organ damage/failure (e.g. stroke, proteinuria, osteonecrosis, pulmonary hypertension) as separate, individual phenotypic endpoints. Global markers of severity—for example mortality—offer the potential for a more cohesive endpoint that may be more informative overall. However, global severity scores have proved difficult to define. Accurate end-point definitions are crucial to enable differentiation of “cases” and “controls”. Examples of proposed global severity scores include: (1) a “severity index” based on frequency of acute painful episodes, hospitalization, blood transfusion, infection and specific complications during the previous years starting from the birth of the child (el-Hazmi 1992); (2) the presence of dactylitis in infants, white cell count and hemoglobin (Hb) level to predict severe disease outcomes as defined by death, stroke, frequent pain and acute chest syndrome (Miller et al. 2000); and (3) a global severity score using a Bayesian network model (a “statistical” phenotype) (Sebastiani et al. 2007).
15.3 Genetic Methodologies
Generally, two approaches have been used to locate genetic variants in human disease: linkage analysis and association studies (Hirschhorn and Daly 2005). Linkage analysis studies aim to establish linkage between genes that co-segregate with a trait/disease within a family. This technique has been successful in highly penetrative single gene disorders, but has had limited success in many common diseases which comprise complex traits. Association studies look for differences in the frequencies of genetic variants between cases and controls to find genetic variants that are strongly associated with a trait/disease. If a variant is more common in cases than controls, an association is described. Such studies require large sample numbers and until recently have not been feasible due to genotyping cost. Crucially, SNPs identified in pilot studies (“discovery cohort”) should always be replicated in additional independent populations (“validation cohort”).
Prerequisites for any genetic association study include: (1) heritability (correlation of trait in sibling pairs, good r value); and (2) a clear distinction between cases and controls (or sufficient variability in a quantitative trait). These criteria present problems in many clinical manifestations of SCD. For example, hospital admissions and duration of stay have used for an objective definition of pain but these measures are influenced by cultural and social factors as well as intermittent illness, such as infections. For convenience, common or “pooled” controls have been used and this can compromize the analysis by contaminating cases in the controls. Adequate patient numbers are essential to allow robust statistical analysis and replication. Again, this presents problems in SCD genetic association studies; most institutions have small numbers of patients (in contrast to hypertensive or diabetic cohorts). Admixture of different ethnic groups is a confounder when different cohorts are pooled for analysis unless population stratification is accounted for prior to association analysis.
Two types of association studies have been utilized in SCD: candidate gene and genome wide association studies (GWAS). Candidate gene association studies look for differences in the frequencies of genetic variants in targeted genes between cases and controls, while GWAS involve an unbiased scan of the whole human genome (Manolio 2013). Many candidate gene association studies in SCD have been published, but often these associations have not been replicated/validated in independent cohorts. Furthermore, critics of candidate gene studies argue that our limited knowledge of SCD pathophysiology is inadequate to predict functional candidate genes (Manolio 2013). By design, GWAS are more likely to reveal unsuspected interactions as the GWAS approach delivers a “hypothesis free” method that could reveal new genes controlling SCD, and thereby exposing novel pathophysiological pathways.
GWAS will also confirm previous candidate genes if the association is robust (Menzel et al. 2007a; Milton et al. 2012). A case in point is the application of GWAS in the unexpected discovery of BCL11A (an oncogene that, hitherto, was not known to have a role in erythropoiesis) as a quantitative trait locus (QTL) controlling HbF (Menzel et al. 2007a; Uda et al. 2008). GWAS also confirmed association of the other two loci—Xmn1-HBG2 (rs782144) on chromosome 11p and HBS1L-MYB (HMIP) on chromosome 6q—with HbF production, that were previously discovered through candidate gene (Labie et al. 1985) and genetic linkage studies (Craig et al. 1996). Similarly, GWAS confirmed the association between bilirubin level and UGT1A1 polymorphism in SCD (Milton et al. 2012).
It has also become evident that simpler, “intermediate” phenotypes, such as HbF, that are reproducible and measurable, and disease-related, are much more successful in genetic association in SCD studies than clinical endpoints. Such intermediate endpoints or endo-phenotypes are often quantitative traits; they provide more power in genetic strategies. For example, blood flow velocity in the middle cerebral artery as detected by TCD screening is a biomarker of early cerebrovascular disease in SCD. Studies have shown that chronic blood transfusion therapy at this stage can prevent overt stroke (Adams et al. 1998). In this regard, TCD velocity would be an extremely attractive intermediate phenotype in studies for detecting genetic variants associated with sickle vasculopathy and stroke risk.
Whole genome or whole exome sequencing using next generation sequencing technology in combination with well-defined phenotypes offers the possibility of identifying new genetic variants (Bamshad et al. 2011). GWAS in combination with exome sequencing identified mutations in GOLGB1 and ENPP1 with stroke protection in sickle cell anemia (SCA) (Flanagan et al. 2013). In this study, overt stroke was the clinical marker but these variants have yet to be independently validated in a different population group.
15.4 Genetic Modifiers of SCD Severity
Global markers of SCD severity represent the “holy grail” of accurate clinical phenotyping. Multiple attempts at providing scoring systems, and using these for genetic associations have been made. Using the global severity index propounded by El-Hazmi (1992), Nishank identified three eNOS gene polymorphisms-eNOS 4a/b, eNOS 894G > T and eNOS -786 T > C associated with SCD severity (Nishank et al. 2013). A GWAS study (Sebastiani et al. 2010) utilized the global severity score devised by the same group (Sebastiani et al. 2007) in over 1200 SCD patients, and replicated in a validation set of samples. Validated SNPs included: KCNK6 (potassium channel gene) and TNKS (telomere length regulator gene).
15.4.1 Modifiers of Global SCD Severity at the Primary Level
The central mechanism underlying the pathophysiology of SCD is the polymerization of deoxygenated HbS and formation of irreversibly sickled erythrocytes that lead to the two hallmarks of the disease—recurrent episodes of vaso-occlusion and pain, and chronic hemolytic anemia. Factors that impact the primary event of the disease process will thus have a global effect on the disease phenotype. They include the causative genotype, co-existing α-thalassemia and the innate ability to produce HbF.
15.4.1.1 Causative Sickle Genotype
In African-descended populations, HbSS typically accounts for 65–70 %, and HbSC 30–35 % of the cases of SCD, with most of the remainder having HbSβ thalassemia. Other genotypes of SCD have been described, including compound heterozygotes of HbS with HbD, HbO-Arab, but these are rare. While presence of HbS is fundamental to the pathobiology, the likelihood of HbS polymerization and sickling is also highly dependent on the concentration of intra-erythrocytic HbS, as well as the presence of non-HbS hemoglobin (Noguchi et al. 1983). Thus, individuals with HbSS or HbSβ0 thalassemia, where the intra-cellular Hb is almost all HbS, tend to have the most severe disease, followed by HbSC and HbSβ+ thalassemia. Most studies discussed below consider the homozygous SCD state of HbSS disease only.
HbA (α2β2) or HbA2 (α2δ2) do not participate in HbS polymerization. Since the β+ thalassemia alleles in Africans are of the milder type with minimal deficit in β globin production, Africans with HbSβ+ thalassemia have substantial proportions of intra-erythrocytic HbA and the SCD tends to be very mild. In contrast, individuals with HbSβ+ thalassemia in the Mediterranean, have SCD almost as severe as that of HbSS (Serjeant and Serjeant 2001). Subjects with sickle cell trait (HbAS) with HbS of 35–40 %, rarely suffer from symptoms of SCD. Under exceptional circumstances, however, such as intense physical activity and dehydration, the consequent increased intracellular HbS concentration can induce vaso-occlusive pain (Bonham et al. 2010).
The HbS gene is found on a genetic background of four common β-globin haplotypes: Senegal, Benin, Central African Republic (or Bantu), and Arab-Indian. Clinical studies demonstrate variation in SCD severity between the βS haplotypes, with decreasing severity from the Bantu > Benin > Senegal > Arab-Indian haplotypes. Disease severity correlates inversely with the HbF levels seen in these groups; lowest HbF seen in individuals with Bantu haplotype, and highest HbF in individuals with Arab-Indian haplotype (Nagel et al. 1985, 1987, 1991; Powars 1991; Figueiredo et al. 1996). The differences in clinical severity were ascribed to the difference in HbF levels implicating the Xmn1-HBG2 site which is linked to the Senegal and Arab-Indian βS haplotype but not to the Bantu haplotype (Labie et al. 1985) (see below for further discussion on modifying effects of HbF on SCD).
Other major genetic factors that influence the primary event of HbS polymerization include the co-inheritance of α-thalassemia and HbF (α2γ2) levels.
15.4.1.2 Alpha Genotype
About one-third of African-descended patients with SCD have co-existing α-thalassemia (Steinberg and Embury 1986). Most commonly, this is due to the deletion variant (−α3.7/); the majority of patients are heterozygous (αα/–α3.7) with 3–5 % homozygous for the deletion (−α3.7/−α3.7) (Steinberg and Embury 1986; Vasavda et al. 2007). Co-inheritance of α-thalassemia affects SCD red cell phenotype; it reduces intracellular HbS concentration and the propensity of HbS polymerization, reducing the number of irreversibly sickled cells and decreasing hemolysis (Embury et al. 1982; Ballas 2001).
Clinically, co-inherited α-thalassemia protects against complications related to severe hemolysis including pulmonary hypertension, leg ulceration, priapism and albuminuria (Steinberg 2009; Buchanan et al. 2004). Conversely, the increased hematocrit and associated blood viscosity in α-thalassemia predispose to an increased likelihood of developing osteonecrosis, acute chest syndrome (ACS), retinopathy and acute painful vaso-occlusive episodes (Embury et al. 1994). Several studies have also demonstrated association of α-thalassemia with lower TCD measurements and, hence, reduced risk for stroke (Bernaudin et al. 2008; Rees et al. 2009; Flanagan et al. 2011; Cox et al. 2014) while another study could not demonstrate association between α-thalassemia and magnetic resonance angiography (MRA)-defined vasculopathy in paediatric patients with HbSS disease (Thangarajh et al. 2012). The lack of association in the latter study could be related to patient selection. Co-existing α-thalassemia also reduces bilirubin with a quantitative effect that is independent to that of the UGT1A1 promoter polymorphism (Vasavda et al. 2007). Co-inheritance of α-thalassemia blunts the response to hydroxycarbamide therapy in SCD; this may be explained by its effect on HbF levels and MCV, two key parameters associated with hydroxycarbamide response (Vasavda et al. 2008).
In Jamaicans, the absence of α-thalassemia and higher HbF levels appeared to predict a more benign disease (Thomas et al. 1997), while a subsequent study reported that α-thalassemia did not promote survival in older Jamaicans with HbSS SCD (Serjeant et al. 2007).
15.4.1.3 Fetal Hemoglobin
Fetal hemoglobin (HbF, α2γ2) is a major ameliorating factor in SCD. Understanding fetal hemoglobin control and its therapeutic reactivation (pharmacological and genetic approaches) remains a top research priority. HbF reduces the propensity for HbS polymerization and its sequelae in two major ways: (1) the hybrid tetramers (α2γβS) do not partake in HbS polymerization, and (2) the presence of intra-erythocytic HbF dilutes the concentration of HbS (Noguchi et al. 1988). The clinical phenotype of SCD becomes evident within 6 months to 2 years of age as HbF levels decline.
HbF levels impact the “primary” level of disease pathology—HbS polymerization—thus HbF levels have a global beneficial effect. Indeed, in SCD, high HbF levels are a major predictor of survival (Platt et al. 1994), and reduced pain (Platt et al. 1991; Dampier et al. 2004); conversely, low levels of HbF have been associated with increased risk of brain infarcts in young children (Wang et al. 2008). At the sub-phenotype level, there appear to be disparities in its effects on complications such as renal impairment, retinopathy and priapism (Thein 2011; Steinberg and Sebastiani 2012). The failure of HbF to modulate all complications of SCD uniformly in the different reports may be related to the small sample sizes in genetic studies and even smaller numbers of end complications, and to ascertainment of phenotypes.
15.4.1.3.1 Update on the Genetic Control of Fetal Hemoglobin (HbF)
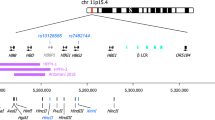
Developmental stage-specific expression of the β-like globin genes appears to be governed by two principles: competition for the upstream β-locus control region (LCR), and autonomous silencing of the embryonic and fetal globin genes involving various ubiquitous and erythroid-specific transcription factors (Wilber et al. 2011; Stamatoyannopoulos 2005). Although the fetal globin genes are autonomously silenced in adults, genetic variants lying both within and outside the HBB locus lead to natural variation in the level of expression of the fetal globin genes and HbF, of over 20-fold (Thein and Craig 1998). Some of these variants significantly ameliorate the clinical symptoms of the β-hemoglobinopathies. These variants account for 89 % of the quantitative variation but the genetic etiology is complex with no clear Mendelian inheritance patterns (Garner et al. 2000). Three known quantitative trait loci (QTLs) for the common HbF variation in adults include: Xmn1-HBG2 (rs782144) within the β-globin gene cluster on chromosome 11p, HBS1L-MYB intergenic region (HMIP) on chromosome 6q23, and BCL11A on chromosome 2p16 (Thein and Menzel 2009; Thein et al. 2009; Sankaran et al. 2010).
Variants in the HBB, HMIP and BCL11A loci account for 10–50 % of the variation in HbF levels in adults, healthy or with SCD or β-thalassemia, depending on the population studied (Menzel et al. 2007a; Lettre et al. 2008; Galanello et al. 2009; Bhatnagar et al. 2011; Makani et al. 2011; Badens et al. 2011; Bae et al. 2012; Mtatiro et al. 2014). The remaining variation (‘missing heritability’) is likely to be accounted for by many loci with relatively small effects, and/or rare variants with significant quantitative effects on γ-globin gene expression that are typically missed.
15.4.1.3.1.1 HBB Cluster on Chromosome 11p
Xmn1-HBG2 (rs782144) in the HBB cluster was the first known QTL for HbF and long-implicated by clinical genetic studies (Labie et al. 1985) (see section “Causative Sickle Genotype” above). The differences in clinical severity of SCD were ascribed to the difference in HbF levels implicating the Xmn1-HBG2 site which is linked to the Senegal and Arab-Indian βS haplotype but not to the Bantu haplotype (Labie et al. 1985). Recent high resolution genotyping, however, suggests that rs782144 is not likely to be the variant itself, but in tight linkage disequilibrium with causal element(s) that remain to be discovered in the β-globin cluster. In vitro reporter gene assays suggest that Gγ globin promoters isolated from Asian and Senegal chromosomes exert higher transcriptional activity than their counterparts from Benin and Bantu chromosomes (Ofori-Acquah et al. 2001). In particular, the Bantu Gγ promoter is 10 times weaker than the Asian promoter (Ofori-Acquah et al. 2001). However, the association between haplotypes, HbF levels and disease severity in SCD remains somewhat contentious due to the wide variation in HbF levels among individuals of the same haplotype.
15.4.1.3.1.2 BCL11A on Chromosome 2p16
Functional studies in primary human erythroid progenitor cells and transgenic mice demonstrated that BCL11A acts as a repressor of γ-globin gene expression that is effected by SNPs in intron 2 of this gene (Sankaran et al. 2008). Fine-mapping demonstrated that these HbF-associated variants, in particular rs1427407, localized to an enhancer that is erythroid-specific and not functional in lymphoid cells (Bauer et al. 2013). BCL11A does not interact with the γ-globin promoter but occupies discrete regions in the HBB complex (Jawaid et al. 2010). The silencing effect of BCL11A involves re-configuration of the HBB locus through interaction with GATA-1 and SOX6 that binds the proximal γ globin promoters (Xu et al. 2010, 2013). In a proof-of-principle, bcl11a knock-out in sickle mice increased HbF up to 30 %, reversing end-organ damage caused by the SCD (Xu et al. 2011).
15.4.1.3.1.3 HMIP on Chromosome 6q23
High resolution genetic mapping and resequencing refined the 6q QTL to a group of variants in tight linkage disequilibrium (LD) in a 24-kb block between the HBS1L and MYB gene, referred to as HMIP-2 (Thein et al. 2007). The causal SNPs are likely to reside in two clusters within the block, at −84 and −71 kb respectively, upstream of MYB (Stadhouders et al. 2014; Menzel et al. 2014). Functional studies in transgenic mice and primary human erythroid cells provide overwhelming evidence that the SNPs at these two regions disrupt binding of key erythroid enhancers affecting long-range interactions with MYB and MYB expression, providing a functional explanation for the genetic association of the 6q HBS1L-MYB intergenic region with HbF and F cell levels (Stadhouders et al. 2012, 2014; Suzuki et al. 2013). A three-base pair (3-bp) deletion in HMIP-2 -84 region is one functional element in the MYB enhancers accounting for increased HbF expression in individuals who have the sentinel SNP rs9399137 that was found to be common in European and Asian populations, although less frequently in African-derived populations (Farrell et al. 2011).
The HBS1L-MYB intergenic enhancers do not appear to affect expression of HBS1L, the other flanking gene (Stadhouders et al. 2014). HBS1L was excluded as having a role in the regulation of HbF and erythropoiesis in a recent report of rare uncharacterized disorders, where whole-exome sequencing revealed mutations in the HBS1L gene leading to a loss-of-function in the gene (Sankaran et al. 2013). The individual had normal blood counts and normal HbF levels. Thus, HMIP-2 is likely to affect HbF and hematopoietic traits via regulation of MYB. MYB was also causally implicated by fine-mapping which identified rare missense MYB variants associated with HbF production (Galarneau et al. 2010).
MYB expression is also reduced by GATA-1 (Welch et al. 2004) and micro (mi)RNA-15a and -16-1 (Sankaran et al. 2011). Elevated levels of the latter have been proposed as the mechanism for the persistently elevated HbF levels, one of the unique features in infants with trisomy 13 (Huehns et al. 1964). These infants have increased expression of miRNAs 15a and 16-1 produced from an extra copy of the genes encoding miRNAs 15a and 16-1 on the triplicated chromosome 13. A recent study provided evidence that the increased HbF effect is mediated, at least in part, through down-modulation of MYB via targeting of its 3′ UTR by the miRNAs 15a and 16-1 (Sankaran et al. 2011).
The MYB transcription factor is a key regulator of erythropoiesis, and modulates HbF expression via two mechanisms: (1) indirectly through alteration of the kinetics of erythroid differentiation: low MYB levels accelerate erythroid differentiation leading to release of early erythroid progenitor cells that are still synthesizing predominantly HbF (Jiang et al. 2006), and (2) directly via activation of KLF1 and other γ-globin repressors (e.g., nuclear receptors TR2/TR4) (Bianchi et al. 2010; Suzuki et al. 2013; Tallack and Perkins 2013).
Modulation of MYB expression also provides a functional explanation for the pleiotropic effect of the HMIP-2 SNPs with other erythroid traits such as red cell count, MCV, MCH, HbA2 levels, and also with platelet and monocyte counts (Menzel et al. 2007b, 2013; Soranzo et al. 2009; van der Harst et al. 2012).
15.4.1.3.1.4 KLF1 on Chromosome 19p13
KLF1 (previously termed EKLF), discovered by Jim Bieker in 1993 (Miller and Bieker 1993), re-emerged as a key transcription factor controlling HbF through genetic studies in a Maltese family with β-thalassemia and hereditary persistence of HbF (HPFH). Linkage studies identified a locus for the HPFH that segregated independently of the HBB locus on chromosome 19p13 which encompassed KLF1 (Borg et al. 2010). Subsequent studies, which included expression profiling of erythroid progenitor cells, confirmed KLF1 as the γ-globin gene modifier in this family. Family members with HPFH were heterozygous for the nonsense K288X mutation in KLF1 that disrupted the DNA-binding domain of KLF1, a key erythroid gene regulator. Collective studies have now confirmed that KLF1 is key in the switch from HBG to HBB expression; it not only activates HBB directly, providing a competitive edge, but also silences the γ-globin genes indirectly via activation of BCL11A (Siatecka and Bieker 2011; Zhou et al. 2010; Esteghamat et al. 2013). KLF1 may also play a role in the silencing of embryonic globin gene expression (Viprakasit et al. 2014; Magor et al. 2015).
Although there have been numerous reports of association of KLF1 variants with increased HbF either as a primary phenotype, or in association with other red cell disorders (Borg et al. 2011), several GWASs of HbF (including ones in SCD patients of African descent) failed to identify common variants (Bhatnagar et al. 2011; Mtatiro et al. 2014).
The emerging network of HbF regulation also includes SOX6, chromatin-modeling factor FOP and the NURD complex, the orphan nuclear receptors TR2/TR4 (part of DRED) and the protein arginine methyltransferase PRMT5, involving DNA methylation and histone deacetylases 1 and 2 epigenetic modifiers. Regulators of the key transcription factors, such as miRNA-15a and 16-1 in controlling MYB, could also have a potential role in regulating HbF levels (Suzuki et al. 2014).
15.4.2 Glucose-6-Phosphate Dehydrogenase Deficiency
Glucose-6-phosphate dehydrogenase deficiency (G6PD) is common in patients with SCD of African ancestry (Bouanga et al. 1998). There is controversy about the effects of G6PD on TCD velocities, a biomarker for stroke risk in SCD; some studies report that G6PD increases the risk for high cerebral blood flow velocities (Bernaudin et al. 2008; Thangarajh et al. 2012) but others observed no effects (Rees et al. 2009; Cox et al. 2014; Flanagan et al. 2011). These conflicting reports could be related to the methodology used in the assay of the enzyme, or the panel of G6PD variants genotyped (Thangarajh et al. 2012; Flanagan et al. 2011). An earlier study showed that G6PD deficiency did not influence SCD clinical endpoints including survival, Hb levels, hemolysis, rate of acute pain or acute anemic episodes (Steinberg et al. 1988).
15.4.2.1 Genetic Modifiers of Organ-Specific Complications
The striking phenomenon in SCD is its clinical diversity. Multiple complications are common in SCD, both acute (frequent pain episodes, acute chest syndrome, strokes) and chronic (pulmonary hypertension, sickle nephropathy, gallstones, osteonecrosis). The variation in global severity of the disease, as well as the incidence of specific end-organ complications (“sub-phenotypes”) in SCD, cannot be explained by these three major genetic modifiers—causative sickle genotype, HbF level and α-globin genotype—alone. While the primary etiology in SCD is HbS polymerization, multiple different (but inter-related) downstream pathological mechanisms contribute to SCD phenotype: hemolysis/heme damage, inflammation, oxidant injury, nitric oxide biology, vaso-regulation, cell adhesion and blood coagulation. These factors have modifying effects independent of HbS polymerization and are likely be multi-genic traits. All of these downstream pathways suggest candidate genes that could plausibly affect the different sickle-related complications. Based on this pathophysiology, researchers have identified candidate genes for gene association studies related to specific sickle complications or “sub-phenotypes”.
Genetic association studies (both candidate gene studies and GWAS) have identified multiple possible genetic associations with SCD complications (Table 15.1).
15.4.2.1.1 Acute Pain Episodes
Acute pain episodes (APE) are the hallmark clinical feature in SCD. They are a measure of disease severity and a predictor of early mortality (Platt et al. 1991). Frequency of APE varies widely in SCD patients, with highest pain rates seen in those with high hematocrit and low HbF (Platt et al. 1991). Outwith these associations, there is no concrete further understanding of the genetic basis of APE frequency in SCD. It is probably the complication most affected by environmental factors. A compounding problem with pain studies is the clinical definitions of phenotypes. Nearly all patients with SCD have pain, and it is often difficult to quantitate objectively both frequency and severity of individual APE. Furthermore, the standard treatment for pain in APE is parenteral opioids, and individual response to opioid analgesia is itself related to genetic variability of their metabolism (Ballas 2007), making it harder still to dissect and measure APE accurately. As a result of these complicating features, many genetic studies on pain in SCD are poor, in particular because of lack of clear-cut definitions of cases versus controls required to make objective associations. Furthermore, some of the studies described are poorly conducted and not corrected for other key modifying factors including genotype and HbF levels. In African American patients and patients from Cameroon, association of HbF with the 3 loci (BCL11A, HBS1L-MYB, and XmnI-HBG2) was accompanied by a corresponding reduction in APEs and hospitalization (Lettre et al. 2008; Wonkam et al. 2014).
Published studies have chosen candidate genes based on APE pathology, itself a complex event involving: red cell deformation, enhancement of white cell adhesion, inflammation, endothelial injury and activation of the coagulation and complement pathways. Examples of studies relating to APE in SCD include genes related to:
-
Oxidative stress. SCD complications, and notably APE, are associated with oxidative stress. Glutathione S-transferases (GSTs) are a group of enzymes that protect against oxidative stress. Shiba found the GSTM1 null genotype to be associated with increased risk of severe APE in Egyptian SCD patients (Shiba et al. 2014)
-
Vasculopathy. Vascular endothelial growth factors (VEGF) are known to contribute to the pathogenesis of APE in SCD. A study in Bahrain associated multiple VEGF gene polymorphisms with the risk of APE (Al-Habboubi et al. 2012). Unfortunately, the differences between cases and controls was not clear cut (compared patients with SCD having had a recent APE or not).
-
Thrombosis. Cystathionine beta-synthase (CBS) enzyme gene mutations are a risk factor for thromboembolic disorders. CBS 844ins68 was three times more frequent among SCD patients with APE (Alves Jacob et al. 2011). Again, there was poor clarification of the difference between “severe” and “mild” individuals with APE.
-
Infections. MBL2 codes for mannose-binding lectin (MBL), and is associated with modifications in the progression of infectious and inflammatory vascular diseases. Using better definitions of APE severity (using APE frequency), MBL2 polymorphisms have been associated with APE in children with SCD (Oliveira et al. 2009; Mendonça et al. 2010). Unexpectedly, studies have observed no association of MBL2 variants with susceptibility to infections (Oliveira et al. 2009) (Dossou-Yovo et al. 2009).
15.4.2.1.2 Gallstones
Jaundice and a predisposition to gallstones is associated with a variant in the promoter (TA repeats) of uridine diphosphate (UDP)-glucuronosyl-transferase 1A (UGT1A1), also referred to as Gilbert’s syndrome. Co-inheritance of Gilbert’s syndrome with SCD has been shown in multiple populations to increase the risk for developing gallstones (Passon et al. 2001; Fertrin et al. 2003; Vasavda et al. 2007). The influence of UGT1A1 polymorphism became more evident in patients while on hydroxycarbamide therapy; children with 6/6 UGT1A1 genotype achieved normal bilirubin levels while children with 6/7 or 7/7 UGT1A1 genotypes did not (Heeney et al. 2003).
The association of Gilbert’s syndrome with gallstones has also been validated in other populations with different hemolytic anemias e.g. hereditary spherocytosis (del Giudice et al. 1999), HbE/β-thalassemia (Premawardhena et al. 2001) and β-thalassemia (Galanello et al. 2001). Thus, the association of UGT1A1 polymorphisms and gallstones in SCD is a well-replicated phenomenon. GWAS also confirmed the association between bilirubin level and UGT1A1 polymorphism in SCD (Milton et al. 2012).
The triad of Gilbert’s syndrome, SCD and gallstones presents a possible clinical context where genetic information may aid clinical decision-making. More widely in SCD, the role of elective cholecystectomy in asymptomatic gallstones remains controversial. While one study of SCD patients with asymptomatic gallstones showed significant increased morbidity in patients who were not electively cholecystectomized and subsequently had a symptomatic cholecystectomy (Curro et al. 2007), another study of SCD patients with gallstones demonstrated that the large majority remained asymptomatic over a 13-year follow up period (Attalla et al. 2013).
Thus, the addition of the (UGT1A1) genotype to the clinical phenotype of gallstones in SCD presents the question of whether these patients should have elective cholecystectomy.
15.4.2.1.3 Sickle Nephropathy
Renal impairment as measured by either proteinuria or glomerular filtration rate (GFR) are common complications of SCD (Sharpe and Thein 2014; Nath and Hebbel 2015), and in some cases sickle renal disease progresses to end-stage renal failure. Renal dysfunction is associated with severity of hemolysis (Becton et al. 2010; Maier-Redelsperger et al. 2010; Day et al. 2012). As a result of this, co-inheritance of α-thalassemia is protective against albuminuria (Nebor et al. 2010a).
The MYH9-APOL1 locus, an important genetic risk factor for end-stage renal failure in non-SCD populations of African ancestry (Genovese et al. 2010), has also been shown to be associated with sickle cell nephropathy (Ashley-Koch et al. 2011). It is broadly considered that the true association is with APOL1, due both to the stronger statistical association with that gene and the lack of identification of causal functional variants in MYH9. The original association with MYH9 has been attributed to the strong linkage disequilibrium between MYH9 and APOL1.
15.4.2.1.4 Stroke
A familial predisposition to stroke in HbSS SCD was first identified by Driscoll et al. (2003). This has prompted numerous gene association studies where a variety of associations have been established between multiple genes and stroke—VCAM1/G1238C, VCAM1/T1594C, IL4R/S503P, TNFA/G-308S, TNF-α/-308G > A allele, LDLR/Ncol +/, ADRB2/Q/27E, AGT/AG repeats, HLA genes (Hoppe et al. 2004; Taylor et al. 2002; Belisario et al. 2015; Tang et al. 2001; Styles et al. 2000). In some studies, stroke was subdivided into large and small vessel disease based on imaging studies (Hoppe et al. 2004). Of the 38 published SNPs associated with stroke, the effects of α-thalassemia and SNPs in four genes (ADYC9, ANXA2, TEK and TGFBR3) could be replicated, although only nominally significant association results were obtained (Flanagan et al. 2011). More recently, GWAS in combination with whole exome sequencing have identified mutations in two genes—GOLGB1 and ENPP1—associated with reduced stroke risk in pediatric patients but, again, this needs validation in independent studies (Flanagan et al. 2013).
15.4.2.1.5 Priapism
In males, priapism remains a common manifestation of SCD, found in about 35 % of men (Adeyoju et al. 2002). Independent association studies have identified KLOTHO (KL) with priapism in different populations (Nolan et al. 2005; Elliott et al. 2007). Separately, the TGFβ/SMAD pathway has also been implicated in priapism risk (Elliott et al. 2007).
15.4.2.1.6 Osteonecrosis
Osteonecrosis (avascular necrosis of the bone) occurs in about half of all adults in HbSS. Higher hematocrits are a predisposing factor, hence an increased incidence in HbSS patients with co-existing α-thalassemia, and patients with HbSC and HbSβ+ thalassemia genotypes. Association with bone morphogenic protein 6 (BMP6) have been replicated across populations (Baldwin et al. 2005; Ulug et al. 2009). This relates to TGF-β/SMAD/BMP pathway in bone metabolism. As for BMP6, regulating the activity of the TGF-β pathway to modulate its effects on bone may be possible (Callahan et al. 2002). Studies suggesting that factors in the coagulation pathway may be involved, such as MTHFR and platelet adhesion (HPA-5B allele), have been inconclusive (Castro et al. 2004; Zimmerman and Ware 1998; Galanello et al. 2001; Kutlar et al. 2001; Andrade et al. 1998).
15.4.2.1.7 Leg Ulcers
Leg ulceration varies widely in SCD with much higher prevalence in Jamaican patients than other cohorts (Alexander et al. 2004). This complication is closely associated with hemolysis severity, and therefore co-existing α-thalassemia is protective. Genetic association studies have implicated several genes in the TGF-β/SMAD/BMP pathway (Nolan et al. 2006). Duffy antigen receptor for chemokines (DARC) has also been shown to be associated with persistence of leg ulcers. It was suggested that the relatively higher white cell and neutrophil counts potentiate inflammation in the Duffy positive patients (Drasar et al. 2013).
15.4.2.1.8 Pulmonary Hypertension
Pulmonary hypertension has been defined in SCD studies using echocardiography, with tricuspid regurgitant jet velocity >2.5 m/s when right heart catheterization is unavailable (when it is defined as mean pulmonary artery pressure ≥25 mmHg and pulmonary capillary wedge pressure ≤15 mmHg). A tricuspid regurgitant jet (TRJ) velocity of >2.5 m/s occurs in about 30 % adults with SCD and is a risk factor for premature death (Gladwin et al. 2004). Pulmonary hypertension is associated with “hemolytic” sickle-complications—renal dysfunction, leg ulceration and priapism—which suggests a vasculopathy driven by chronic hemolysis underlies pulmonary hypertension, too (Taylor et al. 2008). Association studies have suggested multiple gene associations including: the TGF-β/BMP signalling pathway (ACVRL1, BMPR2 and BMP6) (Ashley-Koch et al. 2008) and polymorphisms previously implicated in primary idiopathic pulmonary hypertension (Machado et al. 2001).
A more recent multi-center study (Zhang et al. 2014) considered the hypoxic response as contributory to pulmonary hypertension. To identify genes regulated by the hypoxic response and not other effects of chronic anemia, individuals with SCD were compared with patients with Chuvash polycythemia (constitutive upregulation of hypoxia-inducible factors in the absence of anemia or hypoxia). A SNP associated with reduced MAPK8 expression (encoding a mitogen-activated protein kinase important for apoptosis, T-cell differentiation, and inflammatory responses), correlated with pulmonary hypertension. The association was further confirmed in an independent cohort (Walk-Treatment of Pulmonary Hypertension and Sickle Cell Disease With Sildenafil Therapy (walk-PHaSST) population). The homozygous AA genotype of rs10857560 was present in all 14 patients with pulmonary hypertension.
15.4.2.1.9 Acute Chest Syndrome
Acute chest syndrome (ACS) represents a severe acute manifestation of SCD that is potentially life-threatening. One study showed increased susceptibility to ACS associated with a SNP in endothelial NO synthase gene (eNOS or NOS3) (Sharan et al. 2004), albeit in female patients only. Separately, low exhaled nitric oxide and a polymorphism in the NOS1 gene has been implicated in ACS (Sullivan et al. 2001).
Galarneau et al. (2013) performed a gene-centric association study for ACS with individuals from the Cooperative Study of Sickle Cell Disease (CSSCD), with replication in independent cohorts. In the combined analysis, an association was found between ACS and rs6141803. This SNP is located 8.2 kb upstream of COMMD7, a gene highly expressed in the lung that interacts with nuclear factor-κB signalling.
Another candidate gene is Heme oxygenase-1 (HMOX1) which produces the protein HO-1, the rate-limiting enzyme in the catabolism of heme; HMOX1 might attenuate the severity of APE and hemolysis. Bean et al. (2012) investigated a highly polymorphic (GT)n dinucleotide repeat in the promoter of HMOX1 and showed that children with two shorter alleles had lower rates of ACS.
15.4.2.1.10 Splenic Sequestration
Cajado et al. (2011) identified an association between inflammatory markers TNF-α and IL-8 and splenic sequestration in children with SCD. Specifically, the A allele of the TNF-α -308G > A gene polymorphism was associated with an increased risk of splenic sequestration; and the T allele of the IL-8 -251A > T gene polymorphism was considered to be a protective factor for splenomegaly.
15.4.2.1.11 Infection
Infections are common events in SCD, especially in children. Studies have suggested that the incidence may be modulated by polymorphisms in the HLA locus, MBL2 gene which encodes the mannose binding protein, MPO (gene encoding myeloperoxidase), Duffy antigen receptor for chemokines (DARC) and TGF-β/BMP pathway (BMP6, TGFBR3, BMPR1A, SMAD6 and SMAD3) (Costa et al. 2005; Nebor et al. 2010b; Tamouza et al. 2002, 2007; Neonato et al. 1999; Cordero et al. 2009; Al-Ola et al. 2008; Adewoye et al. 2006).
15.4.2.1.12 Variable Response to Hydroxycarbamide Therapy
Hydroxycarbamide remains a major treatment option for SCD (Ware 2010; Yawn et al. 2014; National Institutes of Health: National Heart Lung and Blood Institute 2014). Clinical and laboratory response to hydroxycarbamide therapy however, is variable, a main determinant of response appears to be the baseline HbF level. Numerous association studies on HbF response to hydroxycarbamide have been reported, of which the association with baseline HbF levels and Xmn1-HBG2 seems to be the most robust (Ware et al. 2002; Green et al. 2013).
15.5 Conclusion
Although environmental factors are important in determining the clinical outcome of SCD, it is evident that the genetic background of the affected individual imparts a substantial contribution to the clinical severity and response to medication. The attraction of being able to generate a personalized genetic risk score as prognostic marker, and to guide therapeutics, plus the relative ease of genotyping and reducing costs, has been a major driver underlying the recent output of genetic association studies in SCD. But the results are questionable in the majority of these genetic association studies because of lack of replication. Nonetheless, genetic studies have been successful in characterizing some of the key variants and pathways involved in HbF regulation, providing new therapeutic targets for HbF reactivation.
We must continue the quest to discover key modifier genes of SCD as a major research priority. This requires taking advantage of whole genome sequencing and the new genomic platforms, but much larger sample sizes (and therefore multi-center collaborations) are required to tease out small statistical differences. Care must be taken to consider, and appropriately classify, different ethnicities.
Additionally, we must focus on developing rigorous clinical phenotypes and the importance of identification of “cases” and “controls”. Clinical researchers need to address the issue of defining and quantifying global sickle severity, as well as precise sub-phenotype definitions. As well as using clinical end points (stroke), it may be useful to use intermediate end points (trans-cranial Doppler velocities) with association studies. Many of the described association studies have highlighted the importance of identification of “cases” and “controls”.
Finally, for those variants already identified, we must endeavour to: validate the variants in independent, large populations; identify the causal variants; support the genetic evidence by functional assays or relevant models to uncover the underlying pathogenesis. Understanding of the underlying mechanisms may guide translation of these genetic discoveries into clinical benefit as targeted, novel therapies.
References
Adams RJ, McKie VC, Hsu L et al (1998) Prevention of a first stroke by transfusions in children with sickle cell anemia and abnormal results on transcranial doppler ultrasonography. N Engl J Med 339(1):5–11
Adewoye AH, Nolan VG, Ma Q et al (2006) Association of polymorphisms of IGF1R and genes in the transforming growth factor- beta /bone morphogenetic protein pathway with bacteremia in sickle cell anemia. Clin Infect Dis 43(5):593–598
Adeyoju AB, Olujohungbe AB, Morris J et al (2002) Priapism in sickle-cell disease; incidence, risk factors and complications – an international multicentre study. BJU Int 90(9):898–902
Alexander N, Higgs D, Dover G, Serjeant GR (2004) Are there clinical phenotypes of homozygous sickle cell disease? Br J Haematol 126(4):606–611
Al-Habboubi HH, Mahdi N, Abu-Hijleh TM et al (2012) The relation of vascular endothelial growth factor (VEGF) gene polymorphisms on VEGF levels and the risk of vasoocclusive crisis in sickle cell disease. Eur J Haematol 89(5):403–409
Al-Ola K, Mahdi N, Al-Subaie AM et al (2008) Evidence for HLA class II susceptible and protective haplotypes for osteomyelitis in pediatric patients with sickle cell anemia. Tissue Antigens 71(5):453–457
Alves Jacob M, da Cunha Bastos C, Regina Bonini-Domingos C (2011) The 844ins68 cystathionine beta-synthase and C677T MTHFR gene polymorphism and the vaso-occlusive event risk in sickle cell disease. Arch Med Sci 7(1):97–101
Amin BR, Bauersachs RM, Meiselman HJ et al (1991) Monozygotic twins with sickle cell anemia and discordant clinical courses: clinical and laboratory studies. Hemoglobin 15(4): 247–256
Andrade FL, Annichino-Bizzacchi JM, Saad ST, Costa FF, Arruda VR (1998) Prothrombin mutant, factor V Leiden, and thermolabile variant of methylenetetrahydrofolate reductase among patients with sickle cell disease in Brazil. Am J Hematol 59(1):46–50
Ashley-Koch AE, Elliott L, Kail ME et al (2008) Identification of genetic polymorphisms associated with risk for pulmonary hypertension in sickle cell disease. Blood 111(12):5721–5726
Ashley-Koch AE, Okocha EC, Garrett ME et al (2011) MYH9 and APOL1 are both associated with sickle cell disease nephropathy. Br J Haematol 155(3):386–394
Attalla BA, Karrar ZA, Ibnouf G et al (2013) Outcome of cholelithiasis in Sudanese children with Sickle Cell Anaemia (SCA) after 13 years follow-up. Afr Health Sci 13(1):154–159
Badens C, Joly P, Agouti I et al (2011) Variants in genetic modifiers of beta-thalassemia can help to predict the major or intermedia type of the disease. Haematologica 96(11):1712–1714
Bae HT, Baldwin CT, Sebastiani P et al (2012) Meta-analysis of 2040 sickle cell anemia patients: BCL11A and HBS1L-MYB are the major modifiers of HbF in African Americans. Blood 120(9):1961–1962
Baldwin C, Nolan VG, Wyszynski DF et al (2005) Association of klotho, bone morphogenic protein 6, and annexin A2 polymorphisms with sickle cell osteonecrosis. Blood 106(1):372–375
Ballas SK (2001) Effect of alpha-globin genotype on the pathophysiology of sickle cell disease. Pediatr Pathol Mol Med 20(2):107–121
Ballas SK (2007) Current issues in sickle cell pain and its management. Hematology Am Soc Hematol Educ Program 97–105
Ballas SK, Kesen MR, Goldberg MF et al (2012) Beyond the definitions of the phenotypic complications of sickle cell disease: an update on management. ScientificWorldJournal 2012:949535
Bamshad MJ, Ng SB, Bigham AW et al (2011) Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 12(11):745–755
Bauer DE, Kamran SC, Lessard S et al (2013) An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342(6155):253–257
Bean CJ, Boulet SL, Ellingsen D et al (2012) Heme oxygenase-1 gene promoter polymorphism is associated with reduced incidence of acute chest syndrome among children with sickle cell disease. Blood 120(18):3822–3828
Becton LJ, Kalpatthi RV, Rackoff E et al (2010) Prevalence and clinical correlates of microalbuminuria in children with sickle cell disease. Pediatr Nephrol 25(8):1505–1511
Belisario AR, Nogueira FL, Rodrigues RS et al (2015) Association of alpha-thalassemia, TNF-alpha (−308G>A) and VCAM-1 (c.1238G>C) gene polymorphisms with cerebrovascular disease in a newborn cohort of 411 children with sickle cell anemia. Blood Cells Mol Dis 54(1):44–50
Bernaudin F, Verlhac S, Chevret S et al (2008) G6PD deficiency, absence of alpha-thalassemia, and hemolytic rate at baseline are significant independent risk factors for abnormally high cerebral velocities in patients with sickle cell anemia. Blood 112(10):4314–4317
Bhatnagar P, Purvis S, Barron-Casella E et al (2011) Genome-wide association study identifies genetic variants influencing F-cell levels in sickle-cell patients. J Hum Genet 56(4):316–323
Bianchi E, Zini R, Salati S et al (2010) c-Myb supports erythropoiesis through the transactivation of KLF1 and LMO2 expression. Blood 116(22):e99–e110
Bonham VL, Dover GJ, Brody LC (2010) Screening student athletes for sickle cell trait--a social and clinical experiment. N Engl J Med 363(11):997–999
Borg J, Papadopoulos P, Georgitsi M et al (2010) Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat Genet 42(9):801–805
Borg J, Patrinos GP, Felice AE, Philipsen S (2011) Erythroid phenotypes associated with KLF1 mutations. Haematologica 96(5):635–638
Bouanga JC, Mouele R, Prehu C et al (1998) Glucose-6-phosphate dehydrogenase deficiency and homozygous sickle cell disease in Congo. Hum Hered 48(4):192–197
Buchanan GR, DeBaun MR, Quinn CT, Steinberg MH (2004) Sickle cell disease. Hematology Am Soc Hematol Educ Program 35–47
Cajado C, Cerqueira BA, Couto FD et al (2011) TNF-alpha and IL-8: serum levels and gene polymorphisms (−308G>A and -251A>T) are associated with classical biomarkers and medical history in children with sickle cell anemia. Cytokine 56(2):312–317
Callahan JF, Burgess JL, Fornwald JA et al (2002) Identification of novel inhibitors of the transforming growth factor beta1 (TGF-beta1) type 1 receptor (ALK5). J Med Chem 45(5): 999–1001
Castro V, Alberto FL, Costa RN et al (2004) Polymorphism of the human platelet antigen-5 system is a risk factor for occlusive vascular complications in patients with sickle cell anemia. Vox Sang 87(2):118–123
Cordero EA, Veit TD, da Silva MA et al (2009) HLA-G polymorphism influences the susceptibility to HCV infection in sickle cell disease patients. Tissue Antigens 74(4):308–313
Costa RN, Conran N, Albuquerque DM et al (2005) Association of the G-463A myeloperoxidase polymorphism with infection in sickle cell anemia. Haematologica 90(7):977–979
Cox SE, Makani J, Soka D et al (2014) Haptoglobin, alpha-thalassaemia and glucose-6-phosphate dehydrogenase polymorphisms and risk of abnormal transcranial Doppler among patients with sickle cell anaemia in Tanzania. Br J Haematol 165(5):699–706
Craig JE, Rochette J, Fisher CA et al (1996) Dissecting the loci controlling fetal haemoglobin production on chromosomes 11p and 6q by the regressive approach. Nat Genet 12(1):58–64
Curro G, Meo A, Ippolito D, Pusiol A, Cucinotta E (2007) Asymptomatic cholelithiasis in children with sickle cell disease: early or delayed cholecystectomy? Ann Surg 245(1):126–129
Dampier C, Ely E, Eggleston B, Brodecki D, O’Neal P (2004) Physical and cognitive-behavioral activities used in the home management of sickle pain: a daily diary study in children and adolescents. Pediatr Blood Cancer 43(6):674–678
Day TG, Drašar ER, Fulford T, Sharpe CC, Thein SL (2012) Association between hemolysis and albuminuria in adults with sickle cell anemia. Haematologica 97(2):201–205
del Giudice EM, Perrotta S, Nobili B et al (1999) Coinheritance of Gilbert syndrome increases the risk for developing gallstones in patients with hereditary spherocytosis. Blood 94(7): 2259–2262
Dossou-Yovo OP, Zaccaria I, Benkerrou M et al (2009) Effects of RANTES and MBL2 gene polymorphisms in sickle cell disease clinical outcomes: association of the g.In1.1T>C RANTES variant with protection against infections. Am J Hematol 84(6):378–380
Drasar ER, Menzel S, Fulford T, Thein SL (2013) The effect of Duffy antigen receptor for chemokines on severity in sickle cell disease. Haematologica 98(8):e87–e89
Driscoll MC, Hurlet A, Styles L et al (2003) Stroke risk in siblings with sickle cell anemia. Blood 101(6):2401–2404
El-Hazmi MA (1992) Clinical and haematological diversity of sickle cell disease in Saudi children. J Trop Pediatr 38(3):106–112
Elliott L, Ashley-Koch AE, De Castro L et al (2007) Genetic polymorphisms associated with priapism in sickle cell disease. Br J Haematol 137(3):262–267
Embury SH, Dozy AM, Miller J et al (1982) Concurrent sickle-cell anemia and alpha-thalassemia: effect on severity of anemia. N Engl J Med 306(5):270–274
Embury SH, Hebbel RP, Mohandas N, Steinberg MH (eds) (1994) Sickle cell disease: basic principles and clinical practice. Raven, New York
Esteghamat F, Gillemans N, Bilic I et al (2013) Erythropoiesis and globin switching in compound Klf1::Bcl11a mutant mice. Blood 121(13):2553–2562
Farrell JJ, Sherva RM, Chen ZY et al (2011) A 3-bp deletion in the HBS1L-MYB intergenic region on chromosome 6q23 is associated with HbF expression. Blood 117(18):4935–4945
Fertrin KY, Melo MB, Assis AM, Saad ST, Costa FF (2003) UDP-glucuronosyltransferase 1 gene promoter polymorphism is associated with increased serum bilirubin levels and cholecystectomy in patients with sickle cell anemia. Clin Genet 64(2):160–162
Figueiredo MS, Kerbauy J, Goncalves MS et al (1996) Effect of alpha-thalassemia and beta-globin gene cluster haplotypes on the hematological and clinical features of sickle-cell anemia in Brazil. Am J Hematol 53(2):72–76
Flanagan JM, Frohlich DM, Howard TA et al (2011) Genetic predictors for stroke in children with sickle cell anemia. Blood 117(24):6681–6684
Flanagan JM, Sheehan V, Linder H et al (2013) Genetic mapping and exome sequencing identify 2 mutations associated with stroke protection in pediatric patients with sickle cell anemia. Blood 121(16):3237–3245
Galanello R, Piras S, Barella S et al (2001) Cholelithiasis and Gilbert’s syndrome in homozygous beta-thalassaemia. Br J Haematol 115(4):926–928
Galanello R, Sanna S, Perseu L et al (2009) Amelioration of Sardinian beta-zero thalassemia by genetic modifiers. Blood 114(18):3935–3937
Galarneau G, Palmer CD, Sankaran VG et al (2010) Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat Genet 40(12):1049–1051
Galarneau G, Coady S, Garrett ME et al (2013) Gene-centric association study of acute chest syndrome and painful crisis in sickle cell disease patients. Blood 122(3):434–442
Garner C, Tatu T, Reittie JE et al (2000) Genetic influences on F cells and other hematologic variables: a twin heritability study. Blood 95(1):342–346
Genovese G, Friedman DJ, Ross MD et al (2010) Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329(5993):841–845
Gladwin MT, Sachdev V, Jison ML et al (2004) Pulmonary hypertension as a risk factor for death in patients with sickle cell disease. N Engl J Med 350(9):886–895
Goldstein J, Konigsberg W, Hill RJ (1963) The structure of human hemoglobin. VI. The sequence of amino acids in the tryptic peptides of the beta chain. J Biol Chem 238:2016–2027
Green NS, Ender KL, Pashankar F et al (2013) Candidate sequence variants and fetal hemoglobin in children with sickle cell disease treated with hydroxyurea. PLoS One 8(2):e55709
Heeney MM, Howard TA, Zimmerman SA, Ware RE (2003) UGT1A promoter polymorphisms influence bilirubin response to hydroxyurea therapy in sickle cell anemia. J Lab Clin Med 141(4):279–282
Hirschhorn JN, Daly MJ (2005) Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 6(2):95–108
Hoppe C, Klitz W, Noble J et al (2003) Distinct HLA associations by stroke subtype in children with sickle cell anemia. Blood 101(7):2865–2869
Hoppe C, Klitz W, Cheng S et al (2004) Gene interactions and stroke risk in children with sickle cell anemia. Blood 103(6):2391–2396
Huehns ER, Hecht F, Keil JV, Motulsky AG (1964) Developmental hemoglobin anomalies in a chromosomal triplication: D1 trisomy syndrome. Proc Natl Acad Sci U S A 51:89–97
Ingram VM (1957) Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature 180(4581):326–328
Jawaid K, Wahlberg K, Thein SL, Best S (2010) Binding patterns of BCL11A in the globin and GATA1 loci and characterization of the BCL11A fetal hemoglobin locus. Blood Cells Mol Dis 45(2):140–146
Jiang J, Best S, Menzel S et al (2006) cMYB is involved in the regulation of fetal hemoglobin production in adults. Blood 108(3):1077–1083
Joishy SK, Griner PF, Rowley PT (1976) Sickle b-thalassemia: identical twins differing in severity implicate nongenetic factors influencing course. Am J Hematol 1(1):23–33
Kutlar A, Kutlar F, Turker I, Tural C (2001) The methylene tetrahydrofolate reductase (C677T) mutation as a potential risk factor for avascular necrosis in sickle cell disease. Hemoglobin 25(2):213–217
Labie D, Pagnier J, Lapoumeroulie C et al (1985) Common haplotype dependency of high G gamma-globin gene expression and high Hb F levels in beta-thalassemia and sickle cell anemia patients. Proc Natl Acad Sci U S A 82(7):2111–2114
Lettre G, Sankaran VG, Bezerra MA et al (2008) DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci U S A 105(33):11869–11874
Machado RD, Pauciulo MW, Thomson JR et al (2001) BMPR2 haploinsufficiency as the inherited molecular mechanism for primary pulmonary hypertension. Am J Hum Genet 68(1):92–102
Magor GW, Tallack MR, Gillinder KR et al (2015) KLF1 null neonates display hydrops fetalis and a deranged erythroid transcriptome. Blood 125:2405–2417
Maier-Redelsperger M, Levy P, Lionnet F et al (2010) Strong association between a new marker of hemolysis and glomerulopathy in sickle cell anemia. Blood Cells Mol Dis 45(4):289–292
Makani J, Menzel S, Nkya S et al (2011) Genetics of fetal hemoglobin in Tanzanian and British patients with sickle cell anemia. Blood 117(4):1390–1392
Manolio TA (2013) Bringing genome-wide association findings into clinical use. Nat Rev Genet 14(8):549–558
Mendonça TF, Oliveira MC, Vasconcelos LR et al (2010) Association of variant alleles of MBL2 gene with vasoocclusive crisis in children with sickle cell anemia. Blood Cells Mol Dis 44(4): 224–228
Menzel S, Garner C, Gut I et al (2007a) A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet 39(10):1197–1199
Menzel S, Jiang J, Silver N et al (2007b) The HBS1L-MYB intergenic region on chromosome 6q23.3 influences erythrocyte, platelet, and monocyte counts in humans. Blood 110(10): 3624–3626
Menzel S, Garner C, Rooks H, Spector TD, Thein SL (2013) HbA2 levels in normal adults are influenced by two distinct genetic mechanisms. Br J Haematol 160(1):101–105
Menzel S, Rooks H, Zelenika D et al (2014) Global genetic architecture of an erythroid quantitative trait locus, HMIP-2. Ann Hum Genet 78:434–451
Miller IJ, Bieker JJ (1993) A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol Cell Biol 13(5):2776–2786
Miller ST, Sleeper LA, Pegelow CH et al (2000) Prediction of adverse outcomes in children with sickle cell disease. N Engl J Med 342(2):83–89
Milton JN, Sebastiani P, Solovieff N et al (2012) A genome-wide association study of total bilirubin and cholelithiasis risk in sickle cell anemia. PLoS One 7(4):e34741
Mtatiro SN, Singh T, Rooks H et al (2014) Genome wide association study of fetal hemoglobin in sickle cell anemia in Tanzania. PLoS One 9(11):e111464
Nagel RL, Fabry ME, Pagnier J et al (1985) Hematologically and genetically distinct forms of sickle cell anemia in Africa. The Senegal type and the Benin type. N Engl J Med 312(14):880–884
Nagel RL, Rao SK, Dunda-Belkhodja O et al (1987) The hematologic characteristics of sickle cell anemia bearing the Bantu haplotype: the relationship between Gg and Hb F level. Blood 69:1026–1030
Nagel RL, Erlingsson S, Fabry ME et al (1991) The Senegal DNA haplotype is associated with the amelioration of anemia in African-American sickle cell anemia patients. Blood 77: 1371–1375
Nath KA, Hebbel RP (2015) Sickle cell disease: renal manifestations and mechanisms. Nat Rev Nephrol 11(3):161–171
National Institutes of Health: National Heart Lung and Blood Institute (2014) Evidence-based management of sickle cell disease NIH Expert Panel Report
Nebor D, Broquere C, Brudey K et al (2010a) Alpha-thalassemia is associated with a decreased occurrence and a delayed age-at-onset of albuminuria in sickle cell anemia patients. Blood Cells Mol Dis 45(2):154–158
Nebor D, Durpes MC, Mougenel D et al (2010b) Association between Duffy antigen receptor for chemokines expression and levels of inflammation markers in sickle cell anemia patients. Clin Immunol 136(1):116–122
Neonato MG, Lu CY, Guilloud-Bataille M et al (1999) Genetic polymorphism of the mannose-binding protein gene in children with sickle cell disease: identification of three new variant alleles and relationship to infections. Eur J Hum Genet 7(6):679–686
Nishank SS, Singh MP, Yadav R et al (2013) Endothelial nitric oxide synthase gene polymorphism is associated with sickle cell disease patients in India. J Hum Genet 58(12):775–779
Noguchi CT, Torchia DA, Schechter AN (1983) Intracellular polymerization of sickle hemoglobin. Effects of cell heterogeneity. J Clin Invest 72(3):846–852
Noguchi CT, Rodgers GP, Serjeant G, Schechter AN (1988) Levels of fetal hemoglobin necessary for treatment of sickle cell disease. N Engl J Med 318(2):96–99
Nolan VG, Baldwin C, Ma Q et al (2005) Association of single nucleotide polymorphisms in klotho with priapism in sickle cell anaemia. Br J Haematol 128(2):266–272
Nolan VG, Adewoye A, Baldwin C et al (2006) Sickle cell leg ulcers: associations with haemolysis and SNPs in Klotho, TEK and genes of the TGF-beta/BMP pathway. Br J Haematol 133(5):570–578
Ofori-Acquah SF, Lalloz MR, Layton DM (2001) Nucleotide variation regulates the level of enhancement by hypersensitive site 2 of the beta-globin locus control region. Blood Cells Mol Dis 27(5):803–811
Oliveira MC, Mendonca TF, Vasconcelos LR et al (2009) Association of the MBL2 gene EXON1 polymorphism and vasoocclusive crisis in patients with sickle cell anemia. Acta Haematol 121(4):212–215
Passon RG, Howard TA, Zimmerman SA, Schultz WH, Ware RE (2001) Influence of bilirubin uridine diphosphate-glucuronosyltransferase 1A promoter polymorphisms on serum bilirubin levels and cholelithiasis in children with sickle cell anemia. J Pediatr Hematol Oncol 23(7):448–451
Pauling L, Itano HA et al (1949) Sickle cell anemia, a molecular disease. Science 109(2835):443
Platt OS, Thorington BD, Brambilla DJ et al (1991) Pain in sickle cell disease: rates and risk factors. N Engl J Med 325(1):11–16
Platt OS, Brambilla DJ, Rosse WF et al (1994) Mortality in sickle cell disease: life expectancy and risk factors for early death. N Engl J Med 330(23):1639–1644
Powars DR (1991) Beta s-gene-cluster haplotypes in sickle cell anemia. Clinical and hematologic features. Hematol Oncol Clin North Am 5(3):475–493
Premawardhena A, Fisher CA, Fathiu F et al (2001) Genetic determinants of jaundice and gallstones in haemoglobin E beta thalassaemia. Lancet 357(9272):1945–1946
Rees DC, Lambert C, Cooper E et al (2009) Glucose 6 phosphate dehydrogenase deficiency is not associated with cerebrovascular disease in children with sickle cell anemia. Blood 114(3):742–743, author reply 743–744
Rees DC, Williams TN, Gladwin MT (2010) Sickle-cell disease. Lancet 376(9757):2018–2031
Sankaran VG, Menne TF, Xu J et al (2008) Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322(5909):1839–1842
Sankaran VG, Xu J, Orkin SH (2010) Advances in the understanding of haemoglobin switching. Br J Haematol 149(2):181–194
Sankaran VG, Menne TF, Scepanovic D et al (2011) MicroRNA-15a and −16-1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc Natl Acad Sci U S A 108(4):1519–1524
Sankaran VG, Joshi M, Agrawal A et al (2013) Rare complete loss of function provides insight into a pleiotropic genome-wide association study locus. Blood 122(23):3845–3847
Sebastiani P, Nolan VG, Baldwin CT et al (2007) A network model to predict the risk of death in sickle cell disease. Blood 110(7):2727–2735
Sebastiani P, Solovieff N, Hartley SW et al (2010) Genetic modifiers of the severity of sickle cell anemia identified through a genome-wide association study. Am J Hematol 85(1):29–35
Serjeant GR, Serjeant BE (2001) Sickle cell disease, 3rd edn. Oxford University Press, Oxford
Serjeant GR, Higgs DR, Hambleton IR (2007) Elderly survivors with homozygous sickle cell disease. N Engl J Med 356(6):642–643
Sharan K, Surrey S, Ballas S et al (2004) Association of T-786C eNOS gene polymorphism with increased susceptibility to acute chest syndrome in females with sickle cell disease. Br J Haematol 124(2):240–243
Sharpe CC, Thein SL (2014) How I treat renal complications in sickle cell disease. Blood 123(24):3720–3726
Shiba HF, El-Ghamrawy MK, Shaheen IA, Ali RA, Mousa SM (2014) Glutathione S-transferase gene polymorphisms (GSTM1, GSTT1, and GSTP1) in Egyptian pediatric patients with sickle cell disease. Pediatr Dev Pathol 17(4):265–270
Siatecka M, Bieker JJ (2011) The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood 118(8):2044–2054
Smith-Whitley K, Pace BS (2007) Sickle cell disease: a phenotypic patchwork. In: Pace BS (ed) Renaissance of sickle cell disease research in the genome era, 1st edn. Imperial College Press, London, pp 45–67
Soranzo N, Spector TD, Mangino M et al (2009) A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet 41(11):1182–1190
Stadhouders R, Thongjuea S, Andrieu-Soler C et al (2012) Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J 31(4): 986–999
Stadhouders R, Aktuna S, Thongjuea S et al (2014) HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J Clin Invest 124(4):1699–1710
Stamatoyannopoulos G (2005) Control of globin gene expression during development and erythroid differentiation. Exp Hematol 33(3):259–271
Steinberg MH (2009) Genetic etiologies for phenotypic diversity in sickle cell anemia. ScientificWorldJournal 9:46–67
Steinberg MH, Embury SH (1986) Alpha-thalassemia in blacks: genetic and clinical aspects and interactions with the sickle hemoglobin gene. Blood 68(5):985–990
Steinberg MH, Sebastiani P (2012) Genetic modifiers of sickle cell disease. Am J Hematol 87(8):795–803
Steinberg MH, West MS, Gallagher D, Mentzer W (1988) Effects of glucose-6-phosphate dehydrogenase deficiency upon sickle cell anemia. Blood 71(3):748–752
Steinberg MH, Lu ZH, Barton FB et al (1997) Fetal hemoglobin in sickle cell anemia: determinants of response to hydroxyurea. Multicenter Study of Hydroxyurea. Blood 89(3):1078–1088
Styles LA, Hoppe C, Klitz W et al (2000) Evidence for HLA-related susceptibility for stroke in children with sickle cell disease. Blood 95(11):3562–3567
Sullivan KJ, Kissoon N, Duckworth LJ et al (2001) Low exhaled nitric oxide and a polymorphism in the NOS I gene is associated with acute chest syndrome. Am J Respir Crit Care Med 164(12):2186–2190
Suzuki M, Yamazaki H, Mukai HY et al (2013) Disruption of the Hbs1l-Myb locus causes hereditary persistence of fetal hemoglobin in a mouse model. Mol Cell Biol 33(8):1687–1695
Suzuki M, Yamamoto M, Engel JD (2014) Fetal globin gene repressors as drug targets for molecular therapies to treat the beta-globinopathies. Mol Cell Biol 34(19):3560–3569
Tallack MR, Perkins AC (2013) Three fingers on the switch: Kruppel-like factor 1 regulation of gamma-globin to beta-globin gene switching. Curr Opin Hematol 20(3):193–200
Tamouza R, Neonato MG, Busson M et al (2002) Infectious complications in sickle cell disease are influenced by HLA class II alleles. Hum Immunol 63(3):194–199
Tamouza R, Busson M, Fortier C et al (2007) HLA-E*0101 allele in homozygous state favors severe bacterial infections in sickle cell anemia. Hum Immunol 68(10):849–853
Tang DC, Prauner R, Liu W et al (2001) Polymorphisms within the angiotensinogen gene (GT-repeat) and the risk of stroke in pediatric patients with sickle cell disease: a case–control study. Am J Hematol 68(3):164–169
Tantawy AA, Adly AA, Ismail EA, Aly SH (2015) Endothelial nitric oxide synthase gene intron 4 VNTR polymorphism in sickle cell disease: relation to vasculopathy and disease severity. Pediatr Blood Cancer 62(3):389–394
Taylor JG, Tang DC, Savage SA et al (2002) Variants in the VCAM1 gene and risk for symptomatic stroke in sickle cell disease. Blood 100(13):4303–4309
Taylor JG, Nolan VG, Mendelsohn L et al (2008) Chronic hyper-hemolysis in sickle cell anemia: association of vascular complications and mortality with less frequent vasoocclusive pain. PLoS One 3(5):e2095
Thangarajh M, Yang G, Fuchs D et al (2012) Magnetic resonance angiography-defined intracranial vasculopathy is associated with silent cerebral infarcts and glucose-6-phosphate dehydrogenase mutation in children with sickle cell anaemia. Br J Haematol 159(3):352–359
Thein SL (2011) Genetic modifiers of sickle cell disease. Hemoglobin 35(5–6):589–606
Thein SL, Craig JE (1998) Genetics of Hb F/F cell variance in adults and heterocellular hereditary persistence of fetal hemoglobin. Hemoglobin 22:401–414
Thein SL, Menzel S (2009) Discovering the genetics underlying foetal haemoglobin production in adults. Br J Haematol 145(4):455–467
Thein SL, Menzel S, Peng X et al (2007) Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci U S A 104(27):11346–11351
Thein SL, Menzel S, Lathrop M, Garner C (2009) Control of fetal hemoglobin: new insights emerging from genomics and clinical implications. Hum Mol Genet 18(R2):R216–R223
Thomas PW, Higgs DR, Serjeant GR (1997) Benign clinical course in homozygous sickle cell disease: a search for predictors. J Clin Epidemiol 50(2):121–126
Uda M, Galanello R, Sanna S et al (2008) Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A 105(5):1620–1625
Ulug P, Vasavda N, Awogbade M et al (2009) Association of sickle avascular necrosis with bone morphogenic protein 6. Ann Hematol 88(8):803–805
van der Harst P, Zhang W, Mateo Leach I et al (2012) Seventy-five genetic loci influencing the human red blood cell. Nature 492(7429):369–375
Vasavda N, Menzel S, Kondaveeti S et al (2007) The linear effects of alpha-thalassaemia, the UGT1A1 and HMOX1 polymorphisms on cholelithiasis in sickle cell disease. Br J Haematol 138(2):263–270
Vasavda N, Badiger S, Rees D et al (2008) The presence of alpha-thalassaemia trait blunts the response to hydroxycarbamide in patients with sickle cell disease. Br J Haematol 143(4): 589–592
Vicari P, Adegoke SA, Mazzotti DR et al (2015) Interleukin-1beta and interleukin-6 gene polymorphisms are associated with manifestations of sickle cell anemia. Blood Cells Mol Dis 54(3): 244–249
Viprakasit V, Ekwattanakit S, Riolueang S et al (2014) Mutations in Kruppel-like factor 1 cause transfusion-dependent hemolytic anemia and persistence of embryonic globin gene expression. Blood 123:1586–1595
Wang WC, Pavlakis SG, Helton KJ et al (2008) MRI abnormalities of the brain in one-year-old children with sickle cell anemia. Pediatr Blood Cancer 51(5):643–646
Ware RE (2010) How I use hydroxyurea to treat young patients with sickle cell anemia. Blood 115(26):5300–5311
Ware RE, Eggleston B, Redding-Lallinger R et al (2002) Predictors of fetal hemoglobin response in children with sickle cell anemia receiving hydroxyurea therapy. Blood 99(1):10–14Weatherall MW, Higgs DR, Weiss H, Weatherall DJ, Serjeant GR (2005) Phenotype/genotype relationships in sickle cell disease: a pilot twin study. Clin Lab Haematol 27(6): 384-390
Welch JJ, Watts JA, Vakoc CR et al (2004) Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104(10):3136–3147
Wilber A, Nienhuis AW, Persons DA (2011) Transcriptional regulation of fetal to adult hemoglobin switching: new therapeutic opportunities. Blood 117(15):3945–3953
Wonkam A, Ngo Bitoungui VJ, Vorster AA et al (2014) Association of variants at BCL11A and HBS1L-MYB with hemoglobin F and hospitalization rates among sickle cell patients in Cameroon. PLoS One 9(3):e92506
Xu J, Sankaran VG, Ni M et al (2010) Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev 24(8):783–798
Xu J, Peng C, Sankaran VG et al (2011) Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science 334(6058):993–996
Xu J, Bauer DE, Kerenyi MA et al (2013) Corepressor-dependent silencing of fetal hemoglobin expression by BCL11A. Proc Natl Acad Sci U S A 110(16):6518–6523
Yawn BP, Buchanan GR, Afenyi-Annan AN et al (2014) Management of sickle cell disease: summary of the 2014 evidence-based report by expert panel members. JAMA 312(10):1033–1048
Zhang X, Zhang W, Ma SF et al (2014) Hypoxic response contributes to altered gene expression and precapillary pulmonary hypertension in patients with sickle cell disease. Circulation 129(16):1650–1658
Zhou D, Liu K, Sun CW, Pawlik KM, Townes TM (2010) KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat Genet 42(9):742–744
Zimmerman SA, Ware RE (1998) Inherited DNA mutations contributing to thrombotic complications in patients with sickle cell disease. Am J Hematol 59(4):267–272
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Gardner, K., Thein, S.L. (2016). Genetic Factors Modifying Sickle Cell Disease Severity. In: Costa, F., Conran, N. (eds) Sickle Cell Anemia. Springer, Cham. https://doi.org/10.1007/978-3-319-06713-1_15
Download citation
DOI: https://doi.org/10.1007/978-3-319-06713-1_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-06712-4
Online ISBN: 978-3-319-06713-1
eBook Packages: MedicineMedicine (R0)