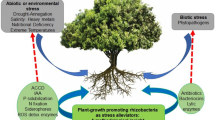
Abstract
One of the first mechanisms proposed to explain the plant growth promotion due to bacterial inoculation has been the nitrogen biological fixation; however, this mechanism has been of less agronomic significance than was initially expected. In counterpart, other mechanisms have been proposed to explain these inoculation effects, such as production of phytohormones and other plant growth regulators. Nowadays, we know that benefic effect of inoculation with rhizobacteria can be correlated with production and metabolism of chemically defined compounds such as auxins, cytokinins, gibberellins, and other plant growth regulators, such as abscisic acid and nitric oxide. In this chapter, we highlight the analytical and biological methods used to identify and quantify the most relevant phytohormones and plant growth regulators produced by Azospirillum sp. and other benefic rhizobacteria.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Salicylic Acid
- Jasmonic Acid
- Jasmonic Acid Biosynthesis
- Define Culture Medium
- Aminocyclopropane Carboxylic Acid
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Growth and development of all living organisms is determined by the interactions between their genomes and the growing environment. Consequently, they evolved a multitude of complex signaling systems to respond to external and internal cues to regulate these processes. Hormones play critical regulatory roles in this process. Research on plant hormones has resulted in the discovery of a wide variety, which include auxins, gibberellins (GAs), cytokinins (CKs), abscisic acid (ABA), ethylene (ET), jasmonic acid (JA), salicylic acid (SA), brassinosteroids (BRs), nitric oxide (NO), and strigolactones (SLs) (Santner and Estelle 2009). They control various developmental events throughout the plant life cycle ranging from patterning, cell identity, and differentiation as well as coordinated growth. Also, the hormones act as regulators of plant responses to biotic or abiotic stress conditions (Feussner and Wasternack 2002). The hormone activity is determined by its availability, which is controlled at the level of metabolism and distribution, and by the efficiency of the hormonal signal perception and transduction, which could be an impact in gene expression or protein activity control. Furthermore, an additional level of complexity in the regulation of developmental process took place when interactions occur between hormones or between hormone producer organisms. Many genera of bacteria, including Azospirillum, Bacillus, Achromobacter, Acetobacter, Azotobacter, Burkholderia, Herbaspirillum, Pseudomonas, Serratia, Xanthomonas, Klebsiella, Agrobacterium Enterococcus, and Enterobacter (Tsavkelova et al. 2006), are producers of diverse phytohormones as auxins, CKs, GAs, ABA, and JA among others (Ping and Boland 2004). The aim of this chapter is to present the current knowledge on phytohormone metabolism and production as well as different extraction, purification, and quantification protocols in bacteria, particularly those belonging to genus Azospirillum.
1.1 Auxins
Auxin is the generic name that represents a group of chemical compounds characterized by their ability to induce cell elongation in the subapical region of the stem and to reproduce the physiological effect of the most abundant, naturally occurring auxin molecule, indole-3-acetic acid (IAA). These compounds have been associated with different plant processes such as (a) gravitropism and phototropism, (b) vascular tissue differentiation, (c) apical dominance, (d) lateral and adventitious root initiation, (e) stimulation of cell division, and (f) stem and root elongation (Tale et al. 2006).
1.1.1 Biosynthesis and Metabolism of Auxins in Plants and Bacteria
IAA is structurally related to the amino acid tryptophan (Trp), and the first studies on auxin biosynthesis hundreds of species proposed this compound as the main precursor in nature. However, this fact has proved difficult to demonstrate in plants. Nevertheless four pathways have been postulated in plant convert Trp to IAA including the indole-3-pyruvic acid (IPA), the tryptamine (TAM), and the indole-3-acetonitrile (IAN) pathway, but there is an amino acid-independent pathway from indole or indole-3-glycerol phosphate. In the case of bacteria, at least six metabolic routes for IAA biosynthesis have been proposed and most of them use Trp as a precursor, as the IPA, TAM, IAN, indole acetamide (IAM), and Trp side-chain oxidase (TSO) pathways. In addition, a tryptophan-independent pathway has been suggested. Despite this diversity of pathways to produce the active phytohormone, prokaryotic IAA biosynthesis seems to follow predominantly two major routes: the IAM and the IPA (Lambrecht et al. 2000). Until now, at least four different pathways have been proposed/described for Azospirillum sp. three Trp-dependent pathways (via IPyA, IAM, and TAM) and one Trp-independent pathway (Cassán et al. 2014). A. brasilense Sp245 is one of the most studied strains world-wide and is considered as a type strain for this species, especially for its auxins biosynthesis. IAA production by A. brasilense Sp245 has extensively been studied with the IPA pathway, the main route for IAA biosynthesis. All genes involved in this pathway are found in the genome sequence of A. brasilense Sp245. Additionally, a nitrilase was also identified in the Sp245 genome sequence, which can catalyze the conversion of IAN to IAA.
1.2 Biosynthesis and Metabolism of Gibberellins
GAs, a large family of tetracyclic diterpenoid plant hormone, regulate many aspects of both vegetative and reproductive growth in plants, including seed germination, stem elongation, and leaf expansion; transitions from meristematic to shoot growth, and from vegetative growth to flowering, promotion of fruit set and subsequent growth, diseased resistance in rice and in response to different abiotic stress (Seo et al. 2009; Yang et al. 2013; Colebrook et al. 2014). Currently, approximately >130 GAs have been identified in plants, fungi, and bacteria, but only a small number are biologically active: GA1, GA3, GA4, and GA7. Many of the other GAs are biosynthetic intermediate or catabolites of bioactive GAs. They are biosynthesized from geranylgeranyl diphosphate (GGDP), a common C-20 precursor for diterpenoids and the plant biosynthesis can be divided into three steps according to the characteristics of the biosynthetic enzymes. In the first step, ent-kaurene, a tetracyclic hydrocarbon intermediate, is synthesized from GGDP by the ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS) in proplastid. In the second step, ent-kaurene is converted to GA12 by two microsomal cytochrome P450 monooxygenase in the endoplasmic reticulum. The ent-kaurene oxidase (KO) catalyzes the sequential oxidation on C-19 to produce ent-kaurenoic acid, which is subsequently converted to GA12 by the ent kaurenoic acid oxidase (KAO). The reactions in the final step involved the formation of C-20 and C-19 GAs in the cytoplasm. Thereby, GA12 is converted to GA4 (13 non-hydroxylated pathway), through oxidations on C-20 and C-3 by GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox), respectively. GA12 is also a substrate for GA13ox for the production of GA53, which is a precursor for GA1 in the 13 hydroxylated pathways. GA20ox catalyzes the sequential oxidation of C-20, including the loss of C-20 as CO2 and the formation of γ-lactone and thereby, C19-GAs are produced. The bioactive GAs are formed by the introduction of a 3β-hydroxyl group in the inactive precursors. On the other hand, GAs are metabolically deactivated in several diversity of mechanisms; being the best-characterized deactivation reaction catalyzed by GA 2-oxidases (GA2ox) (Yamaguchi 2008). Also, the GAs can be converted into conjugates by conjugation with glucose through a hydroxyl group of GA to give a GA-O-glucosyl ether or via the 6-carboxyl group to give a GA-glucosyl ester. Recently, two novel deactivation mechanisms were identified: epoxidation of non-13-hydroxylated GAs in rice and methylation of GAs in Arabidopsis.
1.2.1 Biosynthesis of Gibberellins in Azospirillum sp. and Other PGPR
Morrone et al. (2009) demonstrated that Bradyrhizobium japonicum encodes separate ent-copalyl diphosphate and ent-kaurene synthases. Thus, they provide the first evidence that this operon might indeed be involved in GAs biosynthesis in bacteria. Bottini et al. (1989) were the first to confirm the ability of Azospirillum sp. to produce GAs in chemically defined culture medium. They report the production of GA1 and GA3 on culture medium-free nitrogen of A. lipoferum Op33 by gas chromatography–mass spectrometry (GC-MS). Similar results were reported in A. brasilense by Janzen et al. (1992) and A. lipoferum AZm5 and A. brasilense VS9 by Esquivel-Cote et al. (2010). Additionally, it has been reported production of GA19 and GA9 in chemically defined medium of A. lipoferum Op33, suggesting the existence of different routes of synthesis in those bacteria. The first pathways presumably include the metabolism of GA19 (and its metabolite, GA20) to GA1. On the other hand, the second path where GA9 is (presumably) precursor of GA3. These pathways were confirmed in subsequent reports for A. lipoferum Op33 by using minimal medium supplemented with deuterated precursors GA20 and GA9 and subsequent identification as GA1 and GA3 by GC-MS. On the other hand, A. lipoferum Op33 can produce GA20 and GA5 which suggest the existence of a second 13α-hydroxylation pathway for the metabolism of GA20 to GA5 and GA3 3β-hydroxylation; however, this route has not been confirmed unequivocally. GAs production, metabolism, and hydrolysis of conjugates by Azospirillum sp. were summarized by Bottini et al. (2004).
1.3 Biosynthesis and Metabolism of Cytokinins
CKs play an important role at all phases of plant development from seed germination to senescence such as maintenance of stem cell systems in shoots and roots, nodule organogenesis, leaf senescence, root vascular development, control of shoot branching in interaction with auxin and in the response to abiotic and biotic stress (Kouchi et al. 2010; O’Brien and Benková 2013). Structurally, CKs are adenine derivatives substituted in the N6-position and they can be divided into two groups based on their side chain: those with isoprene derived side chains, which are predominant in plants; and those with aromatic side chains that are found in plants at a lower abundance. In higher plants, the predominant CKs are isopentenyladenine (iP), trans-zeatin (tZ)-, cis-zeatin (cZ), or dihydrozeatin-type derivatives and they can exist as free bases and also in the form of nucleosides and nucleotides. In the past decade, CKs biosynthesis has greatly progressed due in large part to the identification of key pathway. A key enzyme in isoprenoid CKs biosynthesis in plants is adenosine phosphate-isopentenyl transferase (IPT), which catalyzes N-prenylation of the adenosine 5′-phosphates (ATP, ADP, and AMP). The preferred donor of the isoprenoid side chain is dimethylallyl pyrophosphate (DMAPP) and the preferred acceptors are ATP and ADP. The primary products of synthesis are therefore either iP riboside 5′-triphosphate (iPRTP) or iP ribose 5′-diphosphate. The iP nucleotide is converted into tZ nucleotide by cytochrome P450 monooxygenases CYP735A1 and CYP735A2. To become biologically active, iP- and tZ-nucleotides are converted to nucleobase form by dephosphorylation and deribosylation. Recently, the existence of specific phosphoribohydrolase called LOG has been found in rice and Arabidopsis (Kuroha et al. 2009). An indirect pathway of CKs production involves the release of CKs by turnover of tRNA. CKs may be reversible or irreversible conjugated with sugars and amino acid, and they seem to serve as storage. Another way to control CKs content within tissue is by their irreversible degradation by the cytokinin oxidase/dehydrogenase action.
1.3.1 Biosynthesis of Cytokinins in Azospirillum sp. and Other Bacteria
CKs can be produced by bacteria, and they are used mainly as chemical signal either for communicating with the plant or as a tool for invading the plant host. In fact, CKs are also synthesized by phytopathogenic bacteria, such as Agrobacterium tumefaciens and Pseudomonas savastanoi, and is as a key factor in the formation of plant tumors. Barea et al. (1976) found that at least 90 % of the bacteria isolated from the rhizosphere of crops of agricultural interest were able to produce compounds type-CKs in defined culture medium. Horemans et al. (1986) modified the analytical procedure and were able to demonstrate that A. brasilense produces isopentenyl adenine (iP), isopentenyl adenine riboside (iPR), and zeatin (Z) in chemically defined culture medium. The most significant references CKs production by Azospirillum sp. was published by Strzelczyk et al. (1994), using a culture medium supplemented with different sources of carbon. They reported the production of isopentenyl adenine (iP), isopentenyl adenine riboside (iPR), trans-zeatin riboside (trans-Z), and zeatin (Z) in three strains of Azospirillum sp. isolated from sporocarps of ectomycorrhizal fungi Rhizopogon vinicolor, Laccaria laccata y Hebeloma crustuliniforme; however, they could just confirm iPR production in one of the three strains by gas chromatography (GC). An interesting case of synergism has been described for a mixed culture of A. brasilense and Arthrobacter giacomelloi showing a higher content of CKs, compared to those found in individual cultures of each microorganism (Cacciari et al. 1989). Tien et al. (1979) using different types of chromatography (HPLC and TLC) and a bioassay inoculation of pearl millet (Pearl millet) were the first to demonstrate the ability of A. brasilense to produce molecules of type-CKs; however, the partially purified compounds were not characterized. They reported that inoculation caused significant changes in the morphology of the root by increasing the number of lateral roots and root hairs density were similar to those obtained by the exogenous application of CKs. Similar results were found by Muralidhara and Rai (1986) in A. lipoferum.
1.4 Biosynthesis and Metabolism of Jasmonates
JA and its cyclic precursors and derivatives, collectively referred to as jasmonates (JAs), constitute a family of bioactive oxylipins. They function primarily in plant response towards biotic (insects, herbivores, and microbial pathogens) and abiotic (drought, salinity, ozone, UV light) stress, and a variety of plant growth and development processes, including flowering, fruit ripening, senescence, root growth, among others. JAs are generated via one specific branch of oxylipin biosynthesis, the AOS branch of the so-called LOX pathway, in an enzymatic process occurring in different subcellular compartments: plastid, peroxisome, and cytosol. The biosynthesis of JA has been extensively reviewed in recent years (Wasternack and Kombrink 2010; Kombrink 2012). The first step occurs in the membranes of chloroplasts, where the initial substrates α-linolenic acid (α-LeA; C18:3) or hexadecatrienoic acid (C16:3) are released from plastidial galactolipids by phospholipases such as DAD1 or GDL. In plants, generally, JAs synthesis occurs mainly from the C18:3 precursors through the octadecanoid pathway. Oxygenation of α-LeA is the initial step in JA biosynthesis. The oxygen has to be inserted in the C-13 position by the action of a chloroplastic 13-lipoxygenase (13-LOX) generating the 13-hydroperoxy derivative of linolenic acid (13-HPOT). The following step is the dehydration of the 13-HPOT into the allene oxide 12,13(S)-epoxyoctadecatrienoic acid (12,13(S)-EOT) by allene oxide synthase (AOS). This unstable allylic epoxide can decompose to different products, such as α- and γ-ketols, or spontaneously rearrange to racemic 12-oxophytodienoic acid, or it can be enzymatically cyclized by allene oxide cyclase (AOC) to optically pure cis-(+)-12-oxophytodienoic acid ((9S,13S)-OPDA). A small amount of hexadecatrienoic acid (16:3) can be converted by the same enzymes to dinor-12-oxo fitodienoic acid (dnOPDA), an OPDA structural homolog. Translocation of OPDA into peroxisomes, where the second half of JA biosynthesis occurs, has not been completely understood even though it depends on the carrier COMATOSE1/PEROXIMAL 1/PEROXISOME ABC TRANSPORTER (ABC CTS1/PXA1/PED3) and/or an ion trapping mechanism. OPDA reductase 3 (OPR3) catalyzes the reduction of OPDA and dnOPDA to 3-oxo-2-(2=[Z]-pentenyl)-cyclopentan-1-octanoic acid (OPC-8) and 3-oxo-2-(2=[Z]-pentenyl)-cyclopentan-1-hexanoic acid (OPC-6), respectively. JA synthesis proceeds with three rounds of β-oxidation that shorten the carbon side chain from the precursor molecule. Once synthesized, the (3R,7R)-JA is released into the cytoplasm from peroxisome by an unknown mechanism. JA can be further metabolized to different derivatives that take place mainly in the cytosol. The development of new, sensitive analytical techniques and isolation methods has led to an ever-increasing number of identified JA metabolites (Glauser et al. 2009). Among these compounds are the methyl ester and amino acid conjugates of JA, cucurbic acid, cis-jasmone, the glucose ester of JA, the 12-OH-JA and its sulfated, glucosylated and amino acid-conjugated derivatives; even the conjugate with the ethylene precursor aminocyclopropane carboxylic acid (ACC) and the glucose ester of OPC-4 were found. The conjugation of JA with Isoleucine (Ile) is an important step because jasmonoyl-Ile (JA-Ile) has been identified as a biologically active jasmonate. Some plant pathogenic strains of Pseudomonas syringae produce a virulence factor called coronatine that is structurally similar to JA-Ile. OPDA is also biologically active without conversion to JA derivatives. So far differences have not been reported in the JA biosynthesis pathway between plants and bacteria.
1.4.1 Biosynthesis and Metabolism of Jasmonates in Bacteria
JAs have not been extensively investigated in bacteria. The presence of JA was identified in culture medium of the bacterium E. coli RC424 and RC-7 strains (Abdala et al. 1999). Also, JA was reported as produced by isolated endophytic bacteria from roots of Prosopis strombulifera (Piccoli et al. 2011). Achromobacter xylosoxidans (SF2) and Bacillus pumilus (SF3 and SF4) were selected as PGPR, among other characteristics, by the production of JA and OPDA (Forchetti et al. 2007).
1.5 Biosynthesis and Metabolism of ABA
ABA is an important phytohormone playing many physiological roles in plants including seed dormancy, stomatal movement, growth, and various environmental stresses (abiotic and biotic) (Jiang and Hartung 2008; Sreenivasulu et al. 2010). It is produced primarily in vascular tissues of roots and leaves, in guard cells and in seeds. Major steps of ABA biosynthesis take place in plastids, while the last two steps occur in the cytosol. ABA is an apocarotenoid synthesized via oxidative cleavage of epoxy-carotenoids in plants (Zeevaart and Creelman 1988). The epoxidation of zeaxanthin and antheraxanthin to form violaxanthin and neoxanthin is catalyzed by zeaxanthin epoxidase (ZEP/AtABA1). The products are isomerized to produce 9-cis isomers which are cleaved by nine-cis-epoxycarotenoid dioxygenase (NCED) to form xanthoxin. The later is subsequently converted to ABA by two oxidases, a short-chain dehydrogenase/reductase (SDR) and aldehyde oxidase 3 (AAO3). The level of active ABA is determined by its rate of biosynthesis, catabolism, transport between different organs and sensitivity of the cell/tissue to the hormone. When compared with biosynthesis pathway, ABA catabolism is much simpler. ABA can be hydroxylated at three different methyl groups in the ring structure (C-7′, C-8′, and C-9′), which leads to three pathways for ABA hydroxylation and produces three substantial biological activities metabolites. Although the hydroxylation does not reduce the biological activity of ABA thoroughly, it can trigger further inactivation steps. Among the hydroxylated products, only the 8′-hydroxy ABA can be changed into (phaseic acid) PA by spontaneous cyclization and then into dihydophaseic acid (DPA) by further reduction. ABA is inactivated at the C-1 hydroxyl group by different chemical which form different conjugates and accumulate in vacuoles or apoplastic space. Among them, ABA glucosyl ester (ABA-GE) is the most widespread conjugate which is catalyzed by ABA glucosyltransferase.
1.5.1 Biosynthesis of ABA in Azospirillum sp. and PGPR
In certain bacteria, ABA is synthesized as a C-15 terpenoid (sesquiterpene); however, more nothing is known about the biochemistry of ABA (biosynthesis and metabolism) or about a possible function of this molecule in bacteria. There are only a few reports on ABA production by Azospirillum sp. in chemically defined culture medium and in inoculated plants. Kolb and Martin (1985) were the first to report on ABA production by A. brasilense Ft326 in defined culture medium. A. brasilense Az39 and Cd have the capacity to produce ABA (75 and 6.5 ng mL−1 medium, respectively) in chemically defined medium as identified Perrig et al. (2007) by GC-MS. ABA was also characterized by gas chromatography with electron impact mass spectrometry (GC-EI-MS) in the supernatant of the model strain A. brasilense Sp245 from chemically defined media by Cohen et al. (2008). B. pumilus (SF3 and SF4) and A. xylosoxidans (SF2) produced JA and ABA in Luria Bertani (LB) culture media and increased the concentration when cultures were subjected to abiotic stress (Forchetti et al. 2007).
1.6 Biosynthesis and Metabolism of Salicylic Acid
SA is a phenolic compound, which is produced by a wide range of prokaryotic and eukaryotic organisms. Indeed, SA has been found to play a key role in various physiological and biochemical activities of plants such as thermogenesis, photosynthesis, stomatal closure, seed germination, plant growth (Abreu and Munné-Bosch 2009), response to local and systemic against pathogens, and response to abiotic stresses such as drought, chilling, heavy metal toxicity, heat, and osmotic stress (Hayat et al. 2010; Ng et al. 2011). SA is synthesized in plants through two distinct enzymatic pathways: the phenylalanine ammonia lyase (PAL)-mediated phenylalanine pathway in the cytoplasm, and the isochorismate synthase (ICS)-mediated isochorismate pathway in the chloroplast. Both of these pathways originate from chorismate, which is an intermediate of plant phenylpropanoid pathway. The common and initial enzymatic step of PAL pathway is the conversion of chorismate-derived l-phenylalanine to trans-cinnamic acid by phenylalanine ammonia-lyase. Subsequently, trans-cinnamic acid is hydroxylated to form O-coumarate followed by oxidation of the side chain to yield SA. Alternatively, the side chain of trans-cinnamic acid can be initially oxidized to give benzoic acid, which is then hydroxylated to produce SA. Thus, the difference between the two routes is the hydroxylation of the aromatic ring before or after the chain-shortening reactions. Most of the SA synthesized may undergo a number of biologically relevant chemical modifications including glucosylation, methylation, and amino acid conjugation. Glucose conjugation catalyzed by cytosolic SA glucosyltransferases, results in the formation of SA glucoside [SA 2-O-b-d-glucoside] and SA glucose ester (Rivas-San Vicente and Plasencia 2011). The methylation converted SA to methyl salicylate (MeSA), this reaction is catalyzed by an SA carboxyl methyltransferase, and this volatile derivate is an important long-distance signal in tobacco and Arabidopsis systemic acquired resistance. Amino acid conjugation of SA is less well characterized, but may be involved in SA catabolism. Sulfonation is important for the activation or deactivation of various hormones and recently, SA has been shown to be sulfonated in vitro by members of the SOT family of sulphotransferases.
1.6.1 Biosynthesis and Metabolism of Salicylic Acid in Azospirillum sp. and PGPR
Several genera of bacteria such as Pseudomonas aeruginosa and P. fluorescens can synthesize SA from chorismate via two reactions catalyzed by ICS and isochorismate pyruvate lyase (IPL). ICS catalyzes the synthesis of isochorismate from chorismate and IPL catalyzes the conversion of SA from isochorismate (Mercado-Blanco et al. 2001). No gene encoding IPL has been cloned from plant species; whereby in plants, the conversion from IC to SA may be mediated by a route different from that characterized in bacteria. By contrast, SA synthesis in Yersinia enterocolitica and Mycobacterium tuberculosis is mediated by a single, bifunctional enzyme the SA synthase (SAS) that directly converts chorismate to SA via an isochorismate intermediate (Harrison et al. 2006). The presence of SA on Azospirillum sp. cultures was usually detected by not entirely suitable methodologies for SA determination, as silica TLC (Shah et al. 1992). Tortora et al. (2011) confirmed SA production by A. brasilense strains REC2 and REC3 under iron-starved conditions and isolated this compound from other catechols by the use of silica TLC coupled with fluorescence spectroscopy and GC-MS analysis. Siderophores produced by them showed in vitro antifungal activity against Colletotrichum acutatum M11. This fact was coincident with results obtained from phytopathological tests performed in plants, where a reduction of anthracnose symptoms on strawberry plants previously inoculated with A. brasilense was observed (Pedraza et al. 2010).
2 Methods
In the first part of the chapter, we offer several methods for the assessment (extraction and purification) of different hormones from bacterial cultures and plant tissues, with the aim that users can choose the most appropriate according to the availability of equipments and chemical reagents in their laboratories. The following section of the chapter describes analytical and biological methods for phytohormones identification and quantification. In addition, an introduction to Mass Spectrometry methodology is included as final remark.
2.1 Phytohormones Extraction and Purification
The methods described here are: (a) Ion exchange chromatography, which employs Mini-column Sephadex A-25; (b) Anion exchange chromatography including Mini-column Amino (NH 2 ); (c) Liquid-liquid partition which employs double partition with ethyl ether; and (d) Direct use of the sample by centrifugation and filtering of supernatants.
2.1.1 Sample Assessment for JA Determination
This methodology is described by Gidda et al. (2003) and Andrade et al. (2005). For that, homogenize the lyophilized culture medium [approximately 0.2 g dry weight (DW)] with 10 mL 100 % methanol and add 50 ng (5 μL) of the corresponding deuterated standard (JA, OPDA, JA-Ile, 11 and 12-OH-JA). Filter the homogenate under vacuum through a column with a cellulose filter and collect the eluate and dry at 40 °C under vacuum in rotary evaporator. For the acetylation of endogenous hydroxylated-JAs, add a 400 μL of pyridine and acetic acid (2:1) and incubate overnight at 20 °C. Dry the extract at 40 °C under vacuum in rotary evaporator and prepare each cartridge of the column with a wash of 3 mL 100 % methanol. Discard this fraction and dissolve the dried extract in 10 mL of 100 % methanol and to load on columns filled with 3 mL Diethyl-aminoethyl (DEAE) Sephadex A25 or similar. Wash with 3 mL 0.1 M acetic acid in methanol and discard the eluate. Wash with 3 mL 1 M acetic acid in methanol and discard the eluate. Wash with 3 mL 1.5 M acetic acid in methanol and collect and evaporate the eluate under vacuum. Reconstitute the dried extract in 50 μL 100 % methanol (HPLC grade) and vortex briefly. For further jasmonates identification and quantification, inject 15 μL into LC-MS-MS.
Note: This methodology can be used for simultaneous determination of JA from bacterial culture medium or inoculated plant tissues. In the second case, approximately of 200 mg dry weight plant material must be ground in a mortar with liquid nitrogen and add 10 mL 100 % methanol.
2.1.2 Samples Assessment for ABA Determination
This methodology is described by Luna et al. (1993). For that, homogenize the lyophilized culture medium (approximately 0.2 g dry weight, DW) with 20 mL imidazole buffer (pH 7) plus 2,6-di-tert-butyl-p-cresol as antioxidant and add 50 ng (5 μL) of the corresponding deuterated standard (ABA). Incubate each sample overnight at 4 °C to allow extraction and standard equilibration. Centrifuge the sample for 15 min at 1,530 × g, then collect the supernatant and evaporate it under vacuum until to eliminate the isopropanol. An amino anion exchange minicolumn or similar is conditioned with 100 % methanol, deionized water, and imidazole buffer 0.02 M. The aqueous fraction from step 4 is loaded onto the minicolumn. Wash sequentially the minicolumn with 6 mL each of hexane, ethyl acetate, and acetonitrile and discard these fractions. ABA is eluated with 6 mL of methanol:acetic acid (95:5, v/v) and the eluate fraction is evaporated to dryness under vacuum in rotary evaporator. For further ABA identification and quantification, inject 15 μL into high-performance liquid chromatography coupled to mass spectrometers (LC-MS-MS).
Note: This methodology can be used for simultaneous determination of ABA from bacterial culture medium or inoculated plant tissues. In the second case, approximately of 200 mg dry weight plant material must be ground in a mortar with liquid nitrogen and add 20 mL imidazole buffer (pH 7) plus 2,6-di-tert-butyl-p-cresol as antioxidant to the ground.
2.1.3 Samples Assessment for Simultaneous Determinations
For Simultaneous Determination of ABA, JA, IAA, and SA
This methodology is described by Durgbanshi et al. (2005). For that, centrifuge 20 mL of defined bacterial cultures, in exponential growth phase, separate at 8,000 rpm, 4 °C, for 15 min and add 50 ng (5 μL) of the corresponding deuterated standard (ABA, SA, GA3, JA, and IAA). Acidify the supernatants at pH 2.5–2.8 with acetic acid solution (15 %, v/v) and partitioned two times with 20 mL of ethyl ether. Evaporate ethyl ether to dryness at 36 °C and resuspend dried samples in 1,500 μL methanol. Filter through a 0.22 μm cellulose acetate filter and dry methanol in a Speed Vacuum. Resuspend in 5 μL methanol (100 %) and injected into the analytical system.
Note: This methodology can be used for simultaneous determination of ABA, JA, IAA, and SA from bacterial culture medium or inoculated plant tissues. In the second case, approximately of 200 mg dry weight plant material must be ground in a mortar with liquid nitrogen, then add 5 mL ultra-pure water to the ground material.
For Simultaneous Determination of IAA, ABA, GA3, and Z
This methodology is described by Perrig et al. (2007). For that, centrifuge 20 mL of defined bacterial cultures, in exponential growth phase, separate at 8,000 rpm, 4 °C, for 15 min. Add 100 ng of the corresponding deuterated standard (ABA, SA, GA3, JA, and IAA) and acidify the supernatants at pH 2.5–2.8 with acetic acid solution (15 %, v/v). Keep the sample at 4 °C for 2 h and partition four times with the same volume of acetic-acid-saturated ethyl acetate (1 %, v/v). Evaporate ethyl ether to dryness at 36 °C and dried samples are diluted in 100 μL acetic acid/methanol/water solution (1:30:70, v/v) for ABA determination, acetic acid/acetonitrile/water solution (1:15:85, v/v) for IAA determination, and methanol/water solution (30:70, v/v) for GA3. Inject into a reverse phase octadecyl carbon chain (C18) column (300 × 3.9 mm) in an HPLC system coupled to a UV–Vis diode-array spectrometer.
Note: This methodology can be used for simultaneous determination of IAA, ABA, GA3, and Z from bacterial culture medium or inoculated plant tissues. In the second case, approximately of 200 mg dry weight plant material must be ground in a mortar with liquid nitrogen, then add 5 mL ultra-pure water to the ground material.
Direct Use of the Bacterial Culture for IAA Determination
Centrifuge 1.5 mL of bacterial culture at 10,000 rpm for 15 min at room temperature and filter the supernatant using 0.22 μm cellulose acetate filters. Keep the sample in darkness conditions at 4 °C and use the clarified supernatant to colorimetric reaction with Salkowsky’s reagent.
Note: This methodology can be used for determination of IAA and other related auxins from bacterial culture medium.
2.2 Phytohormones Identification and Quantification
2.2.1 Biological Methods
Biological activity of microbial phytohormones depends on their concentrations in culture supernatants and microbial ability to produce these molecules (Cassán et al. 2009a). Phytohormones may influence early seed germination, early seedling growth, plant colonization, and bacterial establishment. The methodology proposed in this section allows you to estimate the bacterial ability to produce cytokinins, gibberellins, and/or auxins in inoculated plants or in chemically defined culture medium.
Determination of Gibberellic Acid
A sensitive, easy, and fast bioassay is described by Cassán et al. (2009b) for detection and quantification of active GAs from bacterial solutions. For that, maize (Zea mays L.) or rice (Oryza sativa L.) seeds are surface disinfected by soaking 3 min in 1 % NaClO, and then washed with sterile distilled water to eliminate traces of this compound. Seeds are pre-germinated for 48 h at 30 °C and 80 rpm shaking in an Erlenmeyer flask containing sterile 80 μM uniconazole [S-3307, (±) (E)-1-(4-chlorophenyl)-4,4-dimethyl-2-(1,2,4-triazol-1-yl)-1-penten-3-ol]. Almost five seeds are planted at 1 cm depth in individual pots, each one containing 200 mL of sterile perlite-sand (1:1) mixture. After full emergence, seedlings are thinned to three plants per pot. Plant pots are kept at field capacity by daily irrigation with sterile distilled water, and once a week fertilized with 50 % of Hoagland’s solution (Hoagland and Boyer 1936). Plants (three plants per pot, five replicates per treatment) are cultured in a growth chamber with daily cycles of 16 h at 25–30 °C with a light intensity of 450 μmol m−2 s−1 and 8 h at 20 °C in darkness for 7 days. Plants are grouped in the following treatments: (a) cell inoculated: each seedling is inoculated with 1 mL of bacterial culture medium with up to 109 cells mL−1; (b) supernatant inoculated: each seedling is inoculated with 1 mL of supernatant of culture medium (10,000 rpm × 10 min at room temperature); (c) GA3 treated: GA3 is dissolved in pure ethanol to obtain 0.1, 0.5, 1.0, 5.0, and 10.0 μg mL−1 solutions and then 10 μL are applied individually at the first leaf with microsyringe for every seedling for the same treatment; (d) Control: seedlings individually treated with pure ethanol. At the end of the experiment the plants are carefully pulled out from the pots and rinsed for 5 s in the potassium phosphate buffer. The following parameters are measured as per plant basis: first internodes length and total shoot length.
Note: There are some dwarf mutants deficient in the production of physiologically active gibberellins, like the dwarf-1 (d1) described in maize (Zea mays L.) by Fujioka et al. (1988) and the dwarf-x (dx or cv. Tan-ginbozu) described in rice (Oryza sativa L.) by Kobayashi et al. (1989). These mutants express the dwarf phenotype since young seedling stage, but the exogenous application of gibberellic acid (GA3) or the inoculation with gibberellins-producer bacteria, allows expression of the normal phenotype. For this reason, the use of the described dwarf mutants is strongly recommended.
Determination of Indole Acetic Acid
A sensitive, easy, and fast bioassay is described by Epel et al. (1987) for detection and quantification of IAA in bacterial cultures. The bioassay consists of measuring the weight increase of hypocotyl sections from etiolated cucumber seedlings incubated in a simple growth medium. The sensitivity of the test is high with a significant response at an IAA concentration as low as 1 ng mL. The bioassay requires at least 3 h to evaluate and is easy to perform. Some advantages of this methodology are listed below: (1) the slope of the log-linear concentration-response curve is moderately steep; (2) the assay is insensitive to the pH of the media between 4 and 7 and can be performed in the presence or absence of buffer; and (3) the growth of the etiolated hypocotyl sections is insensitive to GA and to kinetin.
Cucumber seeds (Cucumis sativus L.) are germinated and grown in vermiculite subtract under darkness and watery irrigation conditions at 25 °C. When the etiolated seedlings were 5–8 cm in length (generally 4–5 days, depending on freshness of seeds) hypocotyl sections 10 mm in length are excised from the hypocotyl, starting about 2 mm below the hook, unless stated otherwise. The excised sections are immediately placed in tap water until sufficient sections had been cut. The sections are then washed in running tap water for about 30 min. The seedlings should be protected of direct light during this procedure. Washed sections, gently blotted with filter paper, are weighed in groups of 10–12 with a tared electronic balance with mg precision (typical weight, about 250 mg) and placed in glass scintillation vials containing 3 mL standard growth medium (pH 7.0) containing 2 mM KCl, 0.1 mM CaCl and 10 mg L−1 chloramphenicol. IAA solutions are added individually into scintillation vials to produce a typical dose–response calibration curve. These solutions should contain a concentration of pure IAA between 0.1 and 100 μg mL−1. The bacterial supernatants are also added in specific vials as problem treatment. After the treatment, the covered vials are gently shaken (80 rpm min−1) at 30 °C between 3 and 12 h (linearity increases with the time of exposition) and sections removed, blotted, and grouped weighed. Each point should represent three replicates of 10–12 sections each. The growth response of IAA solutions is expressed as percentage of increase in weight [%] vs. log μg mL−1 IAA. The supernatant IAA concentration calculation is derived of that dose–response curve.
Determination of Cytokinins
The cucumber cotyledon greening bioassay reported by Fletcher et al. (1981) with modifications is used for detecting cytokinins from several natural sources, including bacterial cultures. The sensitivity of the methodology is extremely low and useful to detect a minimal of 0.0001 mg L−1 of N′-benzyladenine, zeatin, kinetin, or zeatin riboside in solution. Of the more frequent cytokinins tested, kinetin appeared to be the least active. Cucumber (Cucumus sativus L.) seeds are planted in vermiculite, in peat flats (20 × 15 × 7 cm) and germinated in the dark at 28 °C. The cotyledons from 5-days old plants are excised in the absence of green light, making certain that the hypocotyl hook is removed. The cotyledons are placed in 5-cm Petri dishes containing 3 mL of test solution which consisted of distilled H2O, cytokinins at various concentrations to generate a dose–response curve or bacterial supernatant, and 40 mM KCl. The dishes are returned to the dark at 28 °C for 20 h. After incubation period, they are exposed to fluorescent light with an intensity of 12.9 W m−2. After 3.5 h, cotyledons are homogenized, and the chlorophyll extracted in 8 mL 80 % acetone solution. The volume is brought up to 10 mL with acetone and then centrifuged at 2,500 × g for 10 min. The absorbance of the supernatants is read at 663 (chlorophyll A) and 645 nm (chlorophyll B) to obtain the partial and total concentrations according to Arnon (1949).
2.2.2 Analytical Methods
Determination of Indole-3-Acetic Acid by Spectrophotometry
The precise identification of auxin and related molecules can be obtained according to previous extraction, concentration, separation, and identification of the compounds by complex methodologies as high-performance liquid chromatography or GC-MS. Such methods, however, are time-consuming and cannot be used as routine assays. To solve these problems, various authors have used a colorimetric technique derived from that of Salkowski (1885) for indole detection. This method has been used for years because it is simple, rapid, and cheap and allows the daily analysis of numerous bacterial supernatants. Glickman and Dessaux (1995) examined the sensitivity and the specificity of three versions of the Salkowski colorimetric technique. Two of these allowed the detection of indoleacetic acid over a low range of concentrations (0.5–20 mg mL−1), while the third permitted the detection of IAA over a range of higher concentrations (5–200 mg mL−1). These methodologies are shown to be specific for IAA, IPA, and IAM. The first colorimetric technique was performed according to Pilet and Chollet (1970), using reagent R1, which consisted of 12 g of FeCl3 per liter in 7.9 M H2SO4. One milliliter of reagent R1 will be added to 1 mL of the sample solution, well mixed, deposited in a 3 mL spectrophotometer cuvette, and will be kept in the darkness conditions for 30 min at room temperature. The second and third colorimetric methods termed were derived from that of Tang and Bonner (1947). They used reagent R2, which consisted of 4.5 g of FeCl3 per liter in 10.8 M H2SO4. An aliquot of 1 or 2 mL of reagent R2 will be added to 1 mL of sample solution, and the mixture will be processed as indicated for first technique. In all cases, mixtures will be examined at OD530 for determination of indolic compounds. The quantification of IAA will be performed by the use of a calibration curve of pure IAA solution.
Simultaneous Determination of IAA, ABA, GA3 by HPLC
The chromatographic analysis is performed according to Kelen et al. (2004). For that chromatography system consists of pump with an auto injector and diode array detector, a column oven and a degasser system. The column used is C18 [250 mm × 4.6 I.D stainless steel analytical column with 5 μm particle size]. The electromotive force (e.m.f.) values used to evaluate the pH of the mobile phase are measured with a pH/ion analysis apparatus using an Ag/AgCl combination pH electrode. All solutions are externally thermostated at 25 ± 0.1 °C. Stabilize the electrode in appropriate acetonitrile water mixtures before the e.m.f. measurements. The mobile phases are acetonitrile-water (26:74 v/v) containing 30 mM p phosphoric acid with sodium hydroxide at PH 4.00. Equilibrate the C18 column for each mobile phase condition with a time limit of 30 min. Maintain the column temperature constant 25 ± 0.1 °C. The separation is carried out by isocratic elution at 0.8 mL min−1 of flow rate. An injection volume of 10 μL is used for each analysis. The standard solution of the individual acid are prepared in the mobile phase and chromatographed separately to determine the retention time for each molecule. The signal of the compounds is monitored at 208, 265, and 280 nm for GA3, ABA, and IAA, respectively. Calculate the capacity factors from k = (tR − to)/to, where to is the hold-up time, and tR is the retention time of each hormone for each mobile phase. In this equation the hold-up time, to, is established for every mobile phase composition using potassium bromide solution [0.01 % (w/v) in water, μmax = 200 nm]. Determinate the retention time and capacity factor of solutes from three different injections. The identification of different peaks is based on retention time and spiking of the sample.
Mass Spectrometry
This technique works by ionization of compounds to generate charged molecules or molecule fragments. The mixture of electrically charged molecules is accelerated by a magnetic field, all in a high vacuum, this in order to prevent reactions occurring with other particles. The fragments of molecules which are obtained by mass spectrometry are the “fingerprint” of each analyte being tested; it can obtain identification with a very high precision. Compounds are separated by chromatographic methods prior to entering the mass spectrometer when hormones are determined. The gas chromatograph (GC) is a common combination with mass spectrometry (MS), where the molecules must be volatile and tolerate high temperatures. The ionization of molecules associated with gaseous chromatographer generally is produced by electron impact, where electrons are generated by through thermionic emission. The disadvantage of this technique of ionization is that they produce a substantial fragmentation of the molecules of the compound in evaluation. In general, the organic molecules are labile at high temperatures and in many cases non-volatile, by which such molecules must be modified by derivatization to improve their volatility, which not only is annoying to perform but also it is different according to family of plant hormones. The solution to such problem occurred after the appearance of high-performance liquid chromatography coupled to mass spectrometers (LC-MS), which allowed the separation of organic compounds without prior modification of such molecules at the entrance of the mass spectrometer (Fig. 7.1), and also the use of electrospray ionization (ESI), developed by Fenn et al. (1989). Micro-volume of solvent are required for the ESI technique. The HPLCs used possess flows in the order of microliters of solvent per minute (μL min−1) or nano liter (nL m−1) in the case of liquid chromatographs ultra performance (UPLC), which improves the definition of chromatographic peaks and ESI ionization. ESI is a soft ionization with very little fragmentation. The solvent with the molecules of interest pass through a capillary in an electric potential of 3–4 kV is applied. As a result highly charged droplets are produced, which are dried by the interface temperature and the gas stream nitrogen. The droplets are electrically overloaded and suffer a “coulomb explosion” and individual molecules charged “ionic form” positively or negatively, depending on the filler used in the ionization, are generated (Fig. 7.1). Analysis of these charged molecules is performed by different analyzers, for example, quadrupole (Q), ion trap (ionic tramp), or time of flight (TOF) and/or the combination of them (Fig. 7.2). Analyzers perform the separation of each molecule or fragment analysis by mass/charge ratio (m/z). When more than one analyzer is used, this is called tandem mass spectrometry MS/MS or MSn. In mass spectrometers in tandem, the parent ions with a specific m/z are selected in the first analyzer, after they are fragmented in the collision cell and the daughter ions with a specific m/z are selected in the second analyzer. A parent/daughter ions pair is often called to as a “transition” for example in salicylic acid: m/z 137/93. The identification and quantification in this case is performed by fragment molecular compound in the operating mode of Multiple Reaction Monitoring (MRM). In the mass spectrum, we can observe the relative abundance of ionized fragments and the m/z, where we can identify the “molecular ion” which is the molecular weight of the molecule in ionic form, and the other peak is called “Peak base,” which is the analyte’s relative abundance (100 %) and then find the different fragments obtained. Quantification is performed from areas that generate each of the molecules or fragments in a chromatogram that appear in the retention time (RT) for each compound. Such areas are associated with different concentrations generated by the endogenous and the internal standard (deuterated compounds), which have the same structure and different weight that the analyte being evaluated. These areas are associated with different concentrations generated by the synthetic and the internal standard (deuterated compound). For example, in the chromatogram of the ABA (Fig. 7.3) and (2H6)-ABA (Fig. 7.4) and their RT and related areas are observed. Such quantification of the endogenous compound can be done by direct relation: (peak area of endogenous compound/peak area of the deuterated compound) × amount of deuterated compound or through the use of a calibration curve between the area of the endogenous and deuterated compound and their concentrations.
Schematic representation of the electrospray ionization published by Banerjee and Mazumdar (2012)
Determination of Cytokinins by GC-MS-SIM
The chromatographic analysis is performed according to Timmusk et al. (1999). For that, HPLC system consists of a pump with a low-pressure gradient mixer (a 6-mL mixing volume between the pump and the sample injector), an online scanning spectrophotometric UV detector, and a 125 × 4 mm and a 75 × 4 mm column connected in series. The gradient profile is: 0 min, 1.0 %; 8 min, 3.0 %; 21 min, 16.6 %; and 25 min, 40.0 % acetonitrile in 2 % acetic acid and the low rate 2 mL min−1. A standard run includes a mixture of the authentic cytokinins, zeatin, dihydrozeatin, and iP; their nucleosides, nucleotides, and 9-glucosides; and KR. Added dideoxyadenosine (ddAR, 100 pmol) before injection as a means to check the retention time. Store the fractions with putatives a cytokines at −20 °C. Evaporate the fractions with putative cytokinins to dryness and dissolve in 0.5 mL of methanol. Permethylation of standards and samples are performed according to Kovac (1993), except that chloroform that is used instead of dichloromethane and the organic phase are backwashed twice with water. After evaporation to dryness in a stream of N2 at 60 °C, the samples are dissolved in 3 mL of chloroform and a 1 mL aliquot is injected into a g.c. coupled to a mass spectrometer. The mass spectrometer conditions are: EI ionizing voltage, 70 eV; source temperature, 300 °C; and interface temperature, 290 °C. Chromatograms are obtained by Selected Ion Monitoring (SIM) with a well time of 0.05 s and a mass range span of 0.3 amu. The gas chromatography system has a splitless injection port at 300 °C and a 15 m × 0.25 mm × 0.1 mm DB-5 ms column with He as the carrier gas at 27.6 kPa. The temperature program is started at 80 °C for 4 min, then is increased 10 °C min−1 to 290 °C, and finally is maintained for 10 min.
Simultaneous Determination of ABA, JA, IAA, and SA by HPLC-LC-MS
The chromatographic analysis is performed according to Durgbanshi et al. (2005) using an autosampler and a quaternary pump system. Inject aliquots (20 μL) on a Nucleosil ODS reversed-phase column (100 × 2 mm i.d., 5 μm). Elute the phytohormones with a gradient of methanol and 0.01 % CH3COOH in water that started from 10:90 (v/v) and linearly reached 60:40 (v/v) in 10 min. In the following 4 min, increase the gradient to 80:20 (v/v). Retain isocratic conditions of 80:20 during the last 2 min of the run, restore the initial conditions, and allow equilibrating for 5 min, giving a total time of 21 min per sample. The solvent flow rate is 0.3 mL min−1 with working pressures around 70–100 bar. Introduce the effluents from the HPLC into a triple-quadrupole mass spectrometer. Drying gas, as well as nebulizing gas, is nitrogen generated from pressurized air in a nitrogen generator. The nebulizer gas flow is set to 80 L h−1 and the desolvation gas flow to 800–900 L h−1. The collision gas for operation in the tandem MS (MS/MS) is 99.995 % pure argon mode, with a pressure of 2 × 10−3 mbar in the collision cell. The desolvation gas temperature is 350 °C, the source temperature 120 °C, and the capillary voltage 3 kV. The mass spectrometer is operated in multiple reactions monitoring (MRM) mode.
Simultaneous Determination of IAA, ABA, GA3 by GC-MS-SIM
The chromatographic analysis is performed according to Perrig et al. (2007). For that, separate the bacterial cultures into several 20 mL fractions for determination of IAA, ABA, and GA3. Centrifuge the culture fractions at 8,000 rpm for 20 min at 4 °C, acidify supernatants at pH 2.5 with acetic acid solution (1 %, v/v), and add 100 ng of corresponding deuterated internal standard (ABA, IAA, or GA3) at the samples. Then keep the sample at 4 °C for 2 h. Partition each sample four times with the same volume of acetic-acid-saturated ethyl acetate (1 %, v/v). After the last partition, evaporate acidic ethyl acetate to dryness at 36 °C. Dilute the dried samples in 100 μL acetic acid/methanol/water (1:30:70) for ABA determination, acetic acid/acetonitrile/water (1:15:85, v/v) for IAA determination, and methanol/water (30:70 v/v) for GA3 and Z determination. Inject into a reverse phase C18 HPLC column (300 × 3.9 mm) coupled to a UV-diode-array spectrometer. Elute each sample at 1 mL min−1 flow rate, fractions eluting at the retention time corresponding to each pure standard. IAA, ABA, and GA3 are identified and quantified by GC-MS with selective ion monitoring (GC-MS-SIM). UV-absorbing fractions at 254, 262, and 220 nm are grouped for IAA, ABA, and GA3 determination, respectively, and then methylated with ethereal diazomethane and silylated with 1:1 pyridine/BSTFA [bis (trimethylsilyl) trifluoroacetamide] plus 1 % trimethyl-chlorosilane to obtain methyl-trimethylsilyl derivatives of IAA, ABA, and GA3. Inject aliquots of each sample directly into a capillary column (15 m × 0.25 mm, 0.25 μM methyl silicone) fitted in a GC system with a capillary direct interface to a 5970B Mass Selective Detector. The GC temperature program is 60–195 °C at 20 °C min−1, then 4 °C min−1 to 260 °C. Carrier gas (He) flow rate is 1 mL min−1, interface temperature is 280 °C, and data acquisition is controlled by an HP 300 Series computer. The amount of free ABA is calculated by comparison of peak areas of the ion at a mass/charge (m/z) 196 (molecular ion for [2H6] ABAMeTMSi) and the ion at m/z 190 (molecular ion for [1H] ABAMeTMSi) at the corresponding time (Kovats 1958). Similarly, the amount of free IAA is calculated by comparison of peak areas for the parent ion (m/z) 194 and (m/z) 189 and amount of free GA3 by comparison of peak areas for parent ion (m/z) 506 and (m/z) 504.
Simultaneous Determination of ABA, JA, IAA, GA3, and SA by LC-ESI-MS-MS
The hormones are determined by liquid chromatography with electrospray ionization (LC-ESI). For that, perform the liquid chromatography and mass spectrometry analyses using a quaternary pump equipped with auto-sampler. A C18 column (2.1 mm × 100 mm, 5 m) is used at 28 °C, with injected volume 10 μL. The binary solvent system used for elution gradient consists of 0.2 % acetic acid in H2O (solvent B), and MeOH (solvent A), at a constant flow rate of 200 μL min−1. Apply a linear gradient profile with the following proportions (v/v) of solvent A [t (min), % A]: (0, 40), (25, 80), with 7 min for re-equilibration. Mass spectrometry/mass spectrometry (MS/MS) experiments are performed on a double quadrupole mass spectrometer. All analyses are performed using turboion spray source in negative ion mode with the following settings for each hormone: capillary voltage −3,000 V, energy cone 35 V, RF Lens 1 (20), RF Lens 2 (0.3), source temperature 100 °C, solvation temperature 380 °C, gas cone 100 L h−1, collision (50), and multiplier (650). Optimize the MS/MS parameters in infusion experiments using individual standard solutions of each hormone at a 50 ng μL−1 diluted in mobile phase A/B (40:60 v/v). MS/MS product ions are produced by collision-activated dissociation of selected precursor ions in the collision cell of the double quadrupole mass spectrometer, and mass is analyzed using the second analyzer of the instrument. For quantifying, inject the samples in MRM modes, since many compounds could present the same nominal molecular mass. The combination of parent mass and unique daughter fragment ions is used to selectively monitor hormones in plants extracts. MRM acquisition is performed using the 137/93 and 141/97 transitions for SA and (2H4)-SA; 263/153 and 269/159 for ABA and (2H6)-ABA; 209/59 and 215/59 for JA and (2H6-JA); and 174/130 and 179/135 for IAA and (2H5)-AIA, respectively, with dwell 1,000 ms for each transition. Acquire and analyze data using software, for example, MassLynx™ 4.1 and QuanLynx™ 4.1. For quantification, obtain the values from a calibration curve previously constructed using known amounts of each hormone and their pure standard/deuterated internal standard ratio.
References
Abdala G, Miersch O, Correa N, Rosas S (1999) Detection of jasmonic acid in cultures of Escherichia coli and Saccharomyces cerevisiae. Nat Prod Lett 14(1):55–63
Abreu M, Munné-Bosch S (2009) Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana. J Exp Bot 60(4):1261–1271
Andrade A, Vigliocco A, Alemano S, Miersch O, Botella M, Abdala G (2005) Endogenous jasmonates and octadecanoids in hypersensitive tomato mutants during germination and seedling development in response to abiotic stress. Seed Sci Res 15(4):309–318
Arnon D (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in β vulgaris. Plant Physiol 24:1–15
Banerjee S, Mazumdar S (2012) Electrospray ionization mass spectrometry: a technique to access the information beyond the molecular weight of the analyte. Int J Anal Chem. doi: 10.1155/2012/282574
Barea J, Navarro E, Montoya E (1976) Production of plant growth regulators by rhizosphere phosphate-solubilizing bacteria. J Appl Bacteriol 40:129–134
Bottini R, Fulchieri M, Pearce D, Pharis RP (1989) Identification of gibberellins A1, A3 and iso-A3 in culture of Azospirillum lipoferum. Plant Physiol 89:1–3
Bottini R, Cassán F, Piccoli P (2004) Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl Microbiol Biotechnol 65(5):497–503
Cacciari I, Lippi D, Pietrosanti T (1989) Phytohormone-like substances produced by single and mixed diazotrophic cultures of Azospirillum spp. and Arthrobacter. Plant Soil 115:151–153
Cassán F, Perrig D, Sgroy V, Masciarelli O, Penna C, Luna V (2009a) Azospirillum brasilense Az39 and Bradyrhizobium japonicum E 109 promote seed germination and early seedling growth, independently or co-inoculated in maize (Zea mays L.) and soybean (Glycine max L.). Eur J Soil Biol 45:28–35
Cassán F, Masciarelli O, Luna V (2009b) Plant growth promoting Rhizobacteria assays. In: Narwal SS, Catalán CA, Sanprieto DA, Vattuone MA, Polyticka B (eds) Plant bioassays. Studium Press LLC, Houston, pp 157–186
Cassán F, Vanderleyden J, Spaepen S (2014) Physiological and agronomical aspects of phytohormone production by model plant growth promoting rhizobacteria (PGPR) belonging to the genus Azospirillum. J Plant Growth Regul 33:440–459
Cohen A, Bottini R, Piccoli P (2008) Azospirillum brasilense Sp 245 produces ABA in chemically-defined culture medium and increases ABA content in Arabidopsis plants. Plant Growth Regul 54:97–103
Colebrook E, Thomas S, Phillips A, Hedden P (2014) The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol 217:67–75
Durgbanshi A, Arbona V, Pozo O, Miersch O, Sancho J, Gomez-Cardenas A (2005) Simultaneous determination of multiple phytohormones in plants extracts by liquid chromatography-electrospray tandem mass spectrometry. J Agric Food Chem 53:8437–8442
Epel B, Erlanger M, Yahalom A (1987) The etiolated cucumber hypocotyl weight-growth test: a sensitive, easy and ultrafast bioassay for IAA. Plant Growth Regul 5(1):3–14
Esquivel-Cote R, Ramírez-Gama R, Tsuzuki-Reyes G, Orozco-Segovia A, Huante P (2010) Azospirillum lipoferum strain AZm5 containing 1-aminocyclopropane-1-carboxylic acid deaminase improves early growth of tomato seedlings under nitrogen deficiency. Plant Soil 337:65–75
Fenn J, Mann M, Meng C, Wong S, Whitehouse C (1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246(4926):64–71
Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53:275–297
Fletcher A, Kallidumbil V, Steele P (1981) An improved bioassay for cytokinins using cucumber cotyledons. Plant Physiol 69:675–677
Forchetti G, Masciarelli O, Alemano S, Alvarez D, Abdala G (2007) Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, production of jasmonates and abscisic acid in culture medium. Appl Microbiol Biotechnol 76(5):1145–1152
Fujioka S, Yamame H, Spray S, Phinney B, Gaskin P, Mac Millan J, Takahashi N (1988) Qualitative and quantitative analyses of gibberellins in vegetative shoots of normal, dwarf1, dwarf2, dwarf3 and dwarf5 seedlings of Zea mays L. Plant Physiol 88:1367–1372
Gidda S, Miersch O, Levitin A, Schmidt J, Wasternack C, Varin L (2003) Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J Biol Chem 278:17895–17900
Glauser G, Dubugnon L, Mousavi SAR, Rudaz S, Wolfender J-L, Farmer E (2009) Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. J Biol Chem 284:34506–34513
Glickman E, Dessaux Y (1995) A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl Environ Microbiol 61:793–796
Harrison A, Yu M, Gårdenborg T, Middleditch M, Ramsay J, Baker E, Lott J (2006) The structure of Mbtl from Mycobacterium tuberculosis, the first enzyme in the biosynthesis of the siderophore mycobactin, reveals it to be a salicylate synthase. J Bacteriol 188:6081–6091
Hayat Q, Hayat S, Irfan M, Ahmad A (2010) Effect of exogenous salicylic acid under changing environment: a review. Environ Exp Bot 68:14–25
Hoagland D, Boyer T (1936) General nature and process of salt accumulation by roots with description of experimental methods. Plant Physiol 11:471–507
Horemans S, Koninck K, Neuray J, Hermans R, Vlassak K (1986) Production of plant growth substances by Azospirillum sp. and other rhizosphere bacteria. Symbiosis 2:341–346
Janzen R, Rood S, Dormar J, McGill W (1992) Azospirillum brasilense produces gibberellins in pure culture and chemically-medium and in co-culture on straw. Soil Biol Biochem 24:1061–1064
Jiang F, Hartung W (2008) Long-distance signalling of abscisic acid (ABA): the factors regulating the intensity of the ABA signal. J Exp Bot 59:37–43
Kelen M, Demiralay E, Sen S, Ozkan G (2004) Separation of abscisic acid, indole-3-acetic acid, gibberellic acid in 99 R (Vitis berlandieri x Vitis rupestris) and rose oil (Rosa damascena Mill.) by reversed phase liquid chromatography. Turk J Chem 28:603–610
Kobayashi M, Sakurai A, Saka A, Takahashi N (1989) Quantitative analysis of endogenous gibberellins in normal and dwarf cultivars of rice. Plant Cell Physiol 30:963–969
Kolb W, Martin P (1985) Response of plant roots to inoculation with Azospirillum brasilense and to application of indoleacetic acid. In: Klingmüller W (ed) Azospirillum III: genetics, physiology, ecology. Springer, Berlin, pp 215–221
Kombrink E (2012) Chemical and genetic exploration of jasmonate biosynthesis and signaling paths. Planta 236:1351–1366
Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M (2010) How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol 51:1381–1397
Kovac K (1993) Alkylation. In: Blau K, Halket JM (eds) Handbook of derivatives for Chromatography. Jhon Wiley. New York
Kovats E (1958) Gas chromatographische charakteriserung organischer verbindungen I. Retentions indices aliphatischer halogenide, alkohole, aldehyde und ketone. Helv Chim Acta 41:1915–1932
Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H (2009) Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21:3152–3169
Lambrecht M, Okon Y, Vande Broek A, Vanderleyden J (2000) Indole-3-acetic acid: a reciprocal signalling molecule in bacteria-plant interactions. Trends Microbiol 8:298–300
Luna V, Soriano M, Bottini R, Sheng C, Pharis R (1993) Levels of endogenous gibberellins, abscisic acid, indol 3 acetic acid and naringenin during dormancy of peach flower buds. Acta Horticult 329:265–267
Mercado-Blanco J, Rodríguez-Jurado D, Pérez-Artés E, Jiménez-Díaz R (2001) Detection of the nondefoliating pathotype of Verticillium dahliae in infected olive plants by nested PCR. Eur J Plant Pathol 50:609–619
Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, Peters R (2009) Gibberellin biosynthesis in bacteria: separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett 583:475–480
Muralidhara R, Rai P (1986) Plant growth regulators produced by diazotrophic bacteria. In: National seminar on microbial ecology, pp 18–23
Ng G, Seabolt S, Zhang C, Salimian S, Watkins TA, Lu H (2011) Genetic dissection of salicylic acid-mediated defense signaling networks in Arabidopsis. Genetics 189:851–859
O’Brien JA, Benková E (2013) Cytokinin cross-talking during biotic and abiotic stress responses. Front Plant Sci 4:451
Pedraza R, Motok J, Salazar S, Ragout A, Mentel M, Tortora M, Guerrero Molina M, Winik B, Díaz Ricci J (2010) Growth-promotion of strawberry plants inoculated with Azospirillum brasilense. World J Microbiol Biotechnol 26(2):265–272
Perrig D, Boiero M, Masciarelli O, Penna C, Ruiz O, Cassan F, Luna V (2007) Plant-growth-promoting compounds produced by two agronomically important strains of Azospirillum brasilense, and implications for inoculant formulation. Appl Microbiol Biotechnol 75:1143–1150
Piccoli P, Travaglia C, Cohen A, Sosa L, Cornejo P, Masuelli R, Bottini R (2011) An endophytic bacterium isolated from roots of the halophyte Prosopis strombulifera produces ABA, IAA, gibberellins A1 and A3 and jasmonic acid in chemically-defined culture medium. Plant Growth Regul 64:207–210
Pilet P, Chollet R (1970) Sur le dosage colorimétrique de l’acide indolylacétique. C R Acad Sci Ser D 271:1675–1678
Ping L, Boland W (2004) Signals from the underground: bacterial volatiles promote growth in Arabidopsis. Trends Plant Sci 9(6):263–266
Rivas-San Vicente M, Plasencia J (2011) Salicylic acid beyond defense: its role in plant growth and development. J Exp Bot 26:3321–3338
Salkowski E (1885) Ueber das Verhalten der Skatolcarbonsa¨ure im Organismus. Z Physiol Chem 9:23–33
Santner A, Estelle M (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459:1071–1078
Seo M, Nambara E, Choi G, Yamaguchi S (2009) Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69:463–472
Shah S, Karkhanis V, Desai A (1992) Isolation and characterization of siderophore, with antimicrobial activity, from Azospirillum lipoferum. Curr Microbiol 25:347–351
Sreenivasulu N, Radchuk V, Alawady A, Borisjuk L, Weier D, Staroske N, Fuchs J, Miersch O, Strickert M, Usadel B, Wobus U, Grimm B, Weber H, Weschke W (2010) De-regulation of abscisic acid contents causes abnormal endosperm development in the barley mutant seg8. Plant J 64:589–603
Strzelczyk E, Kamper M, Li C (1994) Cytocinin-like-substances and ethylene production by Azospirillum in media with different carbon sources. Microbiol Res 149:55–60
Tale W, Paponov I, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7:847–859
Tang Y, Bonner J (1947) The enzymatic inactivation of indoleacetic acid. I. Some characteristics of the enzyme contained in pea seedlings. Arch Biochem 13:11–25
Tien T, Gaskins M, Hubbell D (1979) Plant growth substances produced by Azsopirillum brasilense and their effect on the growth of pearl millet (Pennisetum americanum L.). Appl Environ Microbiol 37:1016–1024
Timmusk S, Nicander B, Granhall U, Tillberg E (1999) Cytokinin production by Paenibacillus polymyxa. Soil Biol Biochem 31:1847–1852
Tortora M, Diaz Ricci J, Pedraza R (2011) Azospirillum brasilense siderophores with antifungal activity against Colletotrichum acutatum. Arch Microbiol 193:275–286
Tsavkelova E, Klimova S, Cherdyntseva T, Netrusov A (2006) Microbial producers of plant growth stimulators and their practical use: a review. Appl Biochem Microbiol 42:117–126
Wasternack C, Kombrink E (2010) Jasmonates: structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem Biol 5:63–77
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251
Yang D, Dong W, Zhang Y, He Z (2013) Gibberellins modulate abiotic stress tolerance in plants. Sci Sinica Vitae 43:1119–1126
Zeevaart JAD, Creelman RA (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol 39:439–473
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Castillo, P., Molina, R., Andrade, A., Vigliocco, A., Alemano, S., Cassán, F.D. (2015). Phytohormones and Other Plant Growth Regulators Produced by PGPR: The Genus Azospirillum . In: Cassán, F., Okon, Y., Creus, C. (eds) Handbook for Azospirillum. Springer, Cham. https://doi.org/10.1007/978-3-319-06542-7_7
Download citation
DOI: https://doi.org/10.1007/978-3-319-06542-7_7
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-06541-0
Online ISBN: 978-3-319-06542-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)