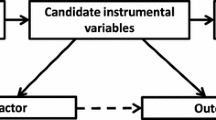
Abstract
The central aim of preventative care is to manage or avoid entirely life-threatening and costly disease endpoints. Success in this broad goal requires researchers and clinicians to correctly distinguish between biomarkers that cause disease from those that are simply correlated with outcome. The randomized controlled trial is a scientifically valid approach to assess causal relationships, but is time-consuming and expensive, and success is not a guaranteed endpoint. Recently, a statistical approach has been translated from the econometrics literature, a strategy which utilizes genetic information identified from human subjects as “instruments” to generate an assessment of causality between biomarker and disease. This methodology, dubbed Mendelian Randomization, is directly analogous to that of the controlled trial, circumventing the issues of confounding and reverse causation that precludes conventional epidemiological studies from making causal assessments. Owing to the growing dissection of genetically heritable traits in the literature, Mendelian Randomization has emerged as a high-value tool for efficient translation of genetics research to the bedside. In the following chapter, we present the framework of Mendelian Randomization and motivation for causal assessment, the analogy of Mendelian Randomization to the randomized controlled trial, discuss general considerations for study design and assumptions of the approach, and exemplify case studies from the literature of applications of MR to type 2 diabetes and other clinical endpoints.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Overview and Scope
The central aim of preventative care is to manage at an early stage—or avoid entirely—life-threatening and costly disease endpoints, like type 2 diabetes (T2D), coronary heart disease (CHD), or stroke. Precise and early diagnosis of pre-disease states with subsequent monitoring is a critical first step in this process, achieved through predictive strategies to identify high-risk patient populations. Once high-risk groups have been identified, a second step applies an intervention to a modifiable exposure (e.g., diet, exercise, lifestyle, etc.) or by other means (e.g., pharmacologic) at early stages, to ameliorate or avoid significant morbidity in later stages.
For success to be achieved, knowledge of the biological, physiological, and molecular factors that cause disease is required, as well as possession of a suitable, therapeutically beneficial intervention for the established cause. Thus, two key questions must be addressed: First, which exposures are causal for—as distinct from those merely correlated with—a disease state? Second, does an intervention for that exposure modify disease risk in a beneficial way? Addressing these two questions are active areas of research, and in human subjects this is achieved through randomized controlled trials, an expensive and time-consuming experiment to undertake.
Recently, an approach analogous to the study design of the controlled trial but that utilizes genetic information from human subjects to make causal inference has received much deserved attention (Katan 1986; Smith and Ebrahim 2003; Sheehan et al. 2008). Termed Mendelian Randomization (MR), the approach is intrinsically translational, owing to the increasing abundance of genetic information in large numbers of individuals; the increasing study of genetically heritable, but modifiable, biomarkers prognostic for disease endpoints; and the relative efficiency of the approach to evaluate causality, compared to a controlled trial. As such, we posit that MR studies will be an important tactic deployed in an overall strategy to comprehensively understand the biology of human disease, one that maximizes the pace in which new beneficial interventions successfully reach the bedside.
In this chapter, we present the framework of MR first with a discussion of the limits of epidemiology and motivation for causal assessment, the model underlying causal inference, and an analogy of MR to the randomized controlled trial (24.2). Then, we discuss general considerations for study design and assumptions of the approach (24.3). Finally, we exemplify case studies from the literature of applications of MR to T2D and other clinical endpoints (24.4). Awareness and understanding of the MR design, approach, assumptions, and methodology will be a useful keystone for translational scientists keen on pursuing interventional studies in humans in the near future.
2 Causal Inference: Rationale, Models, and Mendelian Randomization
To achieve the promise of preventive care, one must begin with knowledge of exposures that are predictive for the disease endpoint in question. These factors are generally obtained from epidemiological studies, whose primary purpose is to measure the correlation between an exposure and hazard to disease over time. These observations are typically made from longitudinal studies of population cohorts such as the Framingham Heart Study (Splansky et al. 2007) and numerous studies beyond this are ongoing. Classic prognostic biomarkers obtained from such studies include (not limited to) blood pressure and cholesterol levels and risk to coronary heart disease (Kannel et al. 1964), C-reactive protein for CHD (Danesh et al. 1998), as well as many others. For research primarily interested in predicting the hazard of disease, any biomarkers, variables, and environmental exposures are relevant.
2.1 Correlation Does Not Imply Causation: Limits of Epidemiology
The underlying hypothesis tested in prospective cohort studies is that an identified marker is correlated, or associated, with hazard to a disease endpoint. An inappropriate conclusion from a significant correlation—particularly those with prior biological significance—is that the exposure also causes disease. Unfortunately, the causation assumption is a common one. Most (if not all) epidemiological monitoring studies do not provide a hypothesis test of causality, because most studies lack a formal intervention required for causality to be tested.
But why does one require an intervention to provide determination of causality? First off, determination of the direction of the associated variables—which is the cause and which is the effect—may be challenging (Fig. 24.1). In a prospective cohort study where a risk factor is measured years before disease onset, one might argue that reverse causation from the disease or disease treatment might be controlled. However, for many diseases, and particularly metabolic diseases, the disease process may have started many years before the disease is diagnosed, even as early as age 10 in the case of raised lipid levels (Whincup et al. 2002). In another example, increasing serum levels of an inflammatory biomarker may appear to precede a coronary event and, thus, correctly predict such events. But instead, unfavorable conditions in arterial walls (e.g., coronary plaques or calcification) could instead produce such an inflammatory marker, as damage to the coronary artery progresses over time. Specific inflammatory markers may not actually be the underlying cause of such events, even though such markers are substantially prognostic. A classic example of such confusion with an inflammatory marker is the much-studied association between raised high-sensitivity C-reactive protein (hsCRP) and CHD. MR using genetic variants that alter circulating hsCRP provided robust evidence, in several large studies, that hsCRP is not causal for CHD (Casas et al. 2006, CCGC et al. 2011).
The challenges in performing causal inference. Imagine one wants to determine if a specific exposure is causally related to a disease state (black arrow). Epidemiologic studies often cannot address the issue of reverse causation, where the disease leads to a change in the exposure, or confounding, where a second factor which is correlated to the exposure is related to the disease
Second, epidemiological studies may not fully capture—or control for—all exposures related to the endpoint of interest (Fig. 24.1). This is problematic only if such exposures are also associated with the marker of study. In this situation, our inference is confounded by that unmeasured factor. For metabolic diseases like type 2 diabetes, many variables are often correlated with the disease, e.g., body mass index (BMI) . Changes in BMI result in many perturbations in serum biomarkers and anthropometric traits; but correcting for BMI may not fully account for all of these additional effects. For example, correcting for BMI does not fully correct for adiposity and, in particular, visceral adiposity including fatty liver that is associated with and likely the cause of many adverse metabolic features. Without precise control of the direction of the prognostic marker, as well as addressing potential unmeasured factors that may confound, we are limited in our abilities to correctly interpret the manner in which an exposure impacts an important clinical outcome.
While epidemiology yields a large, baseline collection of biomarkers or exposures that are worth testing, correlational observation does not constitute etiologic evidence. In some cases, intervention based on epidemiologic evidence alone was successful. That said, we should temper these examples with the larger number of cases where causality was strongly refuted after controlled trial (Tatsioni et al. 2007).
2.2 Approach to Causal Inference: The Randomized Clinical Trial
In order to directly test for causality, any approach needs to address the issues of reverse causality and confounding as far as possible within feasible design parameters. Once established, the general framework in which these factors are controlled—and causality evaluated—is the randomized controlled trial (RCT ). For the purpose of exposition, imagine a pharmacologic therapeutic which lowers a particular biomarker, in which previous epidemiological study has correlated reproducibly with hazard to a disease endpoint (Fig. 24.2a). Here, we are interested in testing the hypothesis that lowering the biomarker through drug intervention reduces hazard to the disease endpoint of study. Patients are collected and randomized for the intervention who are disease-free at baseline, with biomarker and outcomes over time measured and compared between groups—this is the general design for trials used to determine clinical benefit (Gray et al. 2006). This design controls for reverse causality, as the intervention occurs before measurement of the outcome. This design also controls for confounding, with the assumption that the groups (intervention and placebo) are indistinguishable from one another for measured (as well as potentially unmeasured) factors.
Mendelian randomization is “nature’s clinical trial.” (a) A summarized depiction of a placebo-based clinical trial to evaluate efficacy of a therapeutic agent. A population is divided at random into two groups that are disease-free at the start of the study, which are indistinguishable from each other, except that one group is administered a drug which reduces a biomarker and the other is given a placebo. The prediction is that dosage of drug lowers the biomarker, which in turn lowers hazard to the endpoint if the biomarker is causally related to the endpoint. (b) A summarized version of the Mendelian Randomization design. A genetic variant is selected with a strong impact on the biomarker of interest. As a result of meiosis, a sampled population is “naturally” divided into two groups: carriers and noncarriers of the gene. These two groups are equivalent to having experienced reduced or elevated levels of the biomarker—analogous to having received a “weak drug” since birth. Association between variant and endpoint rejects the null hypothesis that the biomarker is not causally related to the disease endpoint
The barriers to activate this line of experimental inquiry are daunting. First, the approach relies upon possession a suitable, proven intervention for the biomarker of interest, and such an intervention might not actually be readily available. For drug intervention trials, a great deal of time and expense are required for any drug to enter this stage of testing (i.e., Phase I and II trials). However, even with an intervention in hand, this trial takes years to deploy and a great deal of cost to complete, in order to obtain a final result. The low-throughput and increasingly abundant prognostic biomarkers that await formal causal evaluation now represent a significant barrier to progress in translational science.
2.3 The Case for Mendelian Randomization
MR studies will not replace RCTs, but rather serve as a complement to them. Beyond the technical feasibility of the MR study design in addressing reverse causality and nongenetic confounding (Fig. 24.3), application of the MR design offers other advantages. Perhaps the main advantage is cost and pace of discovery: multiple phases of RCT take years and millions of dollars to complete, and success is not guaranteed at the end of the trial (Arrowsmith and Miller 2013). Approaches that can help prioritize trials most likely to succeed would be of tremendous benefit in terms of cost saving and development of safe and effective therapeutics. Furthermore, high-throughput technologies developed in the last 10 years have increased the rate of assay and characterization of human phenotypes and molecular markers in genetic and epidemiologic studies. Such growth makes RCT on all such discoveries infeasible; thus, screening tools like MR can help to separate prognostic from high-priority biomarkers with etiologic support from human subjects, where the latter are moved into RCT.
The Mendelian randomization design in graphical terms. Here, we depict that a gene (in this case, PCSK9) influences the exposure (LDL-C). As a result, a polymorphism that strongly influences LDL-C which segregates randomly in the population can be used to test the hypothesis that LDL-C causally impacts risk to CHD (black arrow). Other variants which may associate with confounding factors, which impact the endpoint beyond LDL-C, violate the assumptions of MR and are not considered for analysis (gray arrows). Individually weak variants may be used in a combined score, if assumptions for each variant are met
MR studies are also uniquely powerful beyond the designs of the RCT. First, MR has the advantage that it usually tests the effects of lifelong exposure to a subtle change in a potential risk factor. In contrast, RCTs test the effects of much more acute, short-term, and stronger interventions. Second, MR also provides a strategy for scientific advance, whereby in a standard RCT, it might be unethical to provide such an intervention, for example, evaluating the causal relationship between developmental stunting and infection to strains of Plasmodium known to cause life-threatening malaria (one is ethically obligated to treat the infection!).
Owing to the efficiency of design, availability of data, and utility as a screening tool, MR is a powerful experimental approach, one which can help to clarify cases where an RCT has not been successful or to motivate new RCTs where evidence is altogether unclear.
2.4 The Analogy of MR to Randomized Clinical Trial
Despite their differences, the RCT is a useful analogy for the Mendelian Randomization design (Fig. 24.2b). Intuitively, MR is “nature’s randomized trial” (Thanassoulis and O’Donnell 2009) whereby individuals in the population have been randomly assigned into carrier and noncarrier genotype groups, at meiosis. Ideally, these two groups are indistinguishable from one another, except for genotype at the given locus of interest. The genetic variant that is used to stratify groups is carefully selected, such that carriage of a specific allele modifies the level of the trait exposure one wants to test a causal role for to the endpoint. For example, carriage of loss-of-function alleles at the proprotein convertase subtilisin/kexin type 9 (PCSK9) gene results in lower LDL cholesterol (LDL-C) compared to noncarriers, and these two genotype groups are otherwise indistinguishable from one another. Thus, a loss-of-function genotype would facilitate a formal test of the hypothesis that LDL-C is causality related to CHD or stroke (Fig. 24.3) and would make the prediction that carriage of PCSK9 loss-of-function would protect against hazard to these endpoints, via LDL-C lowering (Cohen et al. 2006). Intuitively, one can conceptualize allele carriage (dosage) as analogous to the intervention in a typical RCT, as a weak drug perturbation administered over a lifetime. And because genotypes are almost always going to be independent of nongenetic confounding and are unmodified by disease processes, reverse causality is controlled for appropriately.
3 Considerations and Approaches for Mendelian Randomization
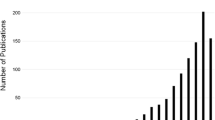
Given the increasing abundance of genetic information available today and the power of the approach, it is perhaps unsurprising that the number of MR publications has skyrocketed, with over 550 citations after 2006, compared to <30 before hand. MR studies can be initiated in many ways, varying in their approach to study design and selection of genetic variants, which may come with additional required assumptions. We turn our attention next to these details.
3.1 Study Design Considerations for Mendelian Randomization
A primary study design for MR can be initiated in a large prospective cohort, where each variable of interest is measured directly in all participants. These measures include the monitored endpoint, the intermediate trait or biomarker, genetic variables which influence the tested intermediate trait, and any additional confounding variables (Fig. 24.3). This design has the advantage of the availability of individualized genotypes and phenotypes and, thus, allows the most direct control over the analysis and interpretation. The main limitation of this design is low statistical power owing to small sample sizes. The probability of obtaining a meaningfully interpretable result is proportional to the number of participants surveyed, particularly the number who have suffered an endpoint. Prospective cohort studies monitor thousands of individuals, of which only hundreds of whom progress to a disease condition. One important question to determine is if observed correlation between trait and endpoint—estimated from epidemiologic studies—is the same as a causal estimate through a genetic instrument. While it is plausible to combine evidence across multiple studies through meta-analysis, the numbers of disease endpoints may still not be sufficient to generate a conclusive result for even this straightforward comparison.
To overcome this limitation, a class of MR statistical approaches have been developed that take advantage of “summary-level” association data. Summary-level data are distinct from individual-level genotype or phenotype data, in that the estimates of effects between SNP and traits that are used for the MR study are pooled across a large number of individuals (often hundreds of thousands), though no individual relationships are identifiable from such data. Summary level data are available for a large number of traits and disease endpoints, with traits that may potentially confound. This "two-stage" approach generates a substantial boost in statistical power to estimate causal relationships, but at the costs of some control over the testing procedure. Importantly, the statistical test for causality is valid for this data type (Burgess and Thompson 2013), though the standard assumptions still must be met.
3.2 Development of Genetic Instruments for Mendelian Randomization
The most easily interpretable MR analyses begin with a single genetic instrument, one with a profound impact on the intermediate trait of interest. Often, these are coding mutations that result in partial or complete loss of function, i.e., see examples described below in PCSK9 or LIPG for LDL and HDL cholesterol, respectively (Cohen et al. 2006; Voight et al. 2012). While it is advantageous to select instruments where the mechanism between genotype and phenotype has been established, this is not strictly required for MR to provide a meaningful result. Individual genetic instruments that stratify populations into two groups (carriers vs. noncarriers) allow for a straightforward evaluation of effects, particularly confounding associations, and fit readily into the controlled trial analogy.
Unfortunately, individual genetic instruments that have a strong impact on the intermediate trait are the exception, rather than the rule. Often, genetic variants with a strong trait effect tend to be rare in the population. Catalogs of low-frequency human genetic variation, also tending toward rare, have only been made available recently (1000 Genomes Project Consortium et al. 2012), and much work remains to relate this spectrum of variation to human phenotypes. That said, even if rare variants and their association to disease were known, owing to their frequency, the statistical power to make causal inference would still be challenging; one might still need to combine evidence across multiple variants into a single statistical test to make appropriate inference.
It should also be noted that an individual, specific genetic instrument tests only a single mechanism and trait/disease pathway. In some cases, this might be precisely what one requires. In other cases, a stronger argument can be made through examination of multiple gene targets and mechanisms of action (Voight et al. 2012). Consequently, to address both of these concerns, statistical approaches that aggregate multiple genetic instruments into a single assessment of causality have been developed (Johnson 2012; Burgess and Thompson 2013), resulting in a valid assessment if certain assumptions are satisfied. While this approach is typically based on summary-level data, it can also be applied in the individual-level data types, via a genetic risk score, a variable that linearly combines allele dosage with effect over a collection of genetic variants (Voight et al. 2012; Johnson 2012).
3.3 Assumptions Underlying Mendelian Randomization
For all MR study designs, there are three primary assumptions that must be held in order for the test to be valid (Smith and Ebrahim 2003; Hernán and Robins 2006; Sheehan et al. 2008):
-
1.
Confounding . The genetic instrument is not related to another, causal variable which has a measurable effect on the endpoint (confounder). A confounding variable could be another trait or other factor that associated with the trait, SNP, and/or endpoint measurements. This is often referred to as pleiotropy in the genetics literature—meaning a genetic variant influences two or more independent traits.
-
2.
Potency . The genetic instrument selected must have a strong and reliable impact on the intermediate trait tested.
-
3.
Exclusivity . The effect on the endpoint must occur through the intermediate trait of interest, with no additional impact outside the two variables measured. Put another way, the genetic instrument selected should not have a direct effect on the endpoint above and beyond the effect through the intermediate trait.
In typical designs, genetic instruments are selected to have a strong reproducible effect on the trait of interest. Confounding is typically addressed by measuring additional associations with variables that may potentially confound. For example, if using a genetic instrument to test the causal relationship between raised triglycerides and coronary artery disease, we would want to know that the genetic variant is not also associated with raised blood pressure or increased BMI. In addition, instruments must also not be subject to genetic confounding, from unaccounted for differences in allele frequency due to population ancestry. Those instruments that have no obvious or reproducible association with confounders are subsequently advanced into further MR analysis (Fig. 24.3).
Variants identified by genetic studies of common variation are individually weak, which may raise a concern if the potency assumption is upheld. To address this concern, multiple genetic instruments are often combined together to perform a specific test. If multiple instruments are utilized, there are additional required assumptions (Johnson 2012):
-
1.
Uncorrelated. Each variant included in the genetic score should be mutually uncorrelated, i.e., should not be in strong linkage disequilibrium with one another (inherited on the same ancestral chromosomal segment).
-
2.
Equanimity. Individual variants included should not contribute excessively relative to other included SNPs.
-
3.
Additivity. For some tests, the dosage of alleles at each variant should influence the intermediate trait in an additive manner.
While this approach can address the potency assumption, using multiple variants still requires care to construct valid tests. The use of multiple variants increases the likelihood of pleiotropy confounding the results—that the accumulated genetic instrument will be related to additional traits. Furthermore, situations could arise where individual variants analyzed separately are more powerful or useful than collecting multiple instruments and evaluating collectively. Overall, the challenges and benefits of using multiple instruments have not been fully described, though research along these lines continues actively.
4 Highlighted Applications of Mendelian Randomization
With a broad overview of the approach and its underlying assumptions, we now turn to specific examples of MR studies from the literature, describing the question, approach, inferences made, and residual challenges. While here, our focus is on cardiovascular and T2D phenotypes, we point curious readers to additional examples of MR studies outside of these traits which exemplify the approach (Chen et al. 2008; Pfister et al. 2011).
4.1 Serum Plasma Cholesterol Levels and Heart Disease
Epidemiological studies have demonstrated reproducible association with serum levels of cholesterol and risk of CHD (Kannel et al. 1964). Specifically, elevation of LDL-C is associated with increased risk to CHD, while elevation of serum high-density lipoprotein cholesterol (HDL-C) is associated with protection against atherosclerosis. With the accumulated body of knowledge of genetic factors related to LDL-C and HDL-C (Teslovich et al. 2010), the stage was set to apply the framework of MR to both of these traits. For LDL-C, in one MR study of nine polymorphisms strongly associated with LDL-C levels (Ference et al. 2012), they reported a causal 54.5 % estimated risk reduction to CHD for each mmol/l lower LDL-C, consistent with another MR study (Voight et al. 2012). Collectively, these results are consistent with randomized controlled trials for LDL-C lowering therapies, which have been successful at reducing cardiovascular events (Baigent et al. 2005).
In contrast, evidence for a causal role for HDL-C is quite uncertain. Controlled trials for drug interventions (e.g., torcetrapib, dalcetrapib, niacin, and others) that raise plasma HDL-C over a range of mechanisms have not met with success in lowering rates of myocardial infarction (MI). To address the hypothesis that HDL-C is causally related to MI, one study applied a two-pronged MR approach (Voight et al. 2012). First, the authors took advantage of a low-frequency coding mutation in endothelial lipase, LIPG N396S, strongly associated with higher HDL-C (0.14 mmol/L), but not associated with other MI-related confounding factors. Association analysis in prospective cohorts, combined with data from case/control studies of MI, found no association (P = 0.85). Second, the authors collected a set of 14 genetic variants associated exclusively with HDL-C and performed a combined variant MR analysis. In contrast to LDL-C, the HDL-C instrument was not causally associated with risk to MI (P = 0.63). A conservative conclusion from this data is that some genetic mechanisms that elevate plasma HDL-C do not seem to confer protection against MI. These observations challenge the simple hypothesis that HDL-C elevation—by any means—will systematically lower risk to MI.
4.2 The Controversy of hsCRP
Subclinical inflammation is associated with many metabolic diseases and higher levels of adverse metabolic traits including raised cholesterol, fatty liver markers, and insulin resistance (Brunner et al. 2008). Furthermore, animal studies provide evidence that some inflammatory markers may causally influence obesity (Bachmaier et al. 1999). Studies in humans have identified a number of genetic variants associated with several markers for inflammation, including interleukin 6 signaling, C-reactive protein, soluble intercellular adhesion molecule-1, and P-selectin (Raman et al. 2013). One recent MR study has provided evidence that the interleukin 6 signaling pathway is causally linked to heart disease (IL6R MR Consortium et al. 2012). These observations strongly motivate translation work in humans, that is, do markers of inflammation, particularly hsCRP levels, cause metabolic disease or adverse metabolic conditions?
Mendelian Randomization studies have now provided a very strong case that increased levels of subclinical (non-acute phase) C-reactive protein do not causally increase the risk of heart disease (Casas et al. 2006, CCGC et al. 2011). Known to be a marker of heart disease and other traits, considerable research efforts have been made to understand its role as both a biomarker and causal factor. Fortunately, the presence of common genetic variants that alter circulating CRP levels, in the gene that encodes the C-reactive protein, provided an exquisite, early example of the power of the MR approach (Timpson et al. 2005). Additional MR studies have examined the role of hsCRP in a range of metabolic diseases. These variants are not associated with any metabolic traits, and the instrumental variable analyses using them provide no evidence of a causal relationship between higher hsCRP and metabolic disease and traits (Brunner et al. 2008).
4.3 Serum Urate , Heart Disease , and Type 2 Diabetes
An end product of purine metabolism is uric acid, which circulates in blood serum as the anion urate. Patients with gout have shown elevated levels of serum urate, and consistent with a causal role in gout, randomized clinical trials with urate lowering therapies have demonstrated these drugs as effective therapeutics (Tayar et al. 2012). Epidemiologic studies have also correlated serum urate levels with a number of cardiometabolic traits, including blood pressure, lipids, as well as endpoint disease (Hozawa et al. 2006; Holme et al. 2009; Kim et al. 2010). A number of genetic variants that result in population level variation in serum urate levels have been identified (Köttgen et al. 2013). Predictably, these variants are also risk factors for gout. With such genetic factors in hand, several studies have looked for association at markers related to serum urate levels with heart disease (Stark et al. 2009; Yang et al. 2010). Altogether, these studies indicate no causal relationship. A particularly strong variant (rs12398742) at the SLC2A9 locus is associated with a profound change in serum urate levels (0.37 mg/dl). However, this variant is not associated with T2D (odds ratio (OR) = 0.99, P = 0.52). This contrasts to the effect expected from epidemiology (expected OR = 1.06, given a 0.37 mg/dl change in serum urate), which have demonstrated a positive correlation between urate levels and T2D (Kodama et al. 2009). These data strongly de-emphasize the need for a randomized clinical trial for urate versus CHD and T2D endpoints. But additional work to evaluate other endpoints—including kidney disease, stroke, or heart failure, with MR analyses that utilize multiple genetic variants for urate levels—is certainly possible and source of active investigation.
4.4 Adiponectin and Insulin Resistance
MR studies have recently provided much needed insight into the relationship between insulin resistance and adiponectin. Adiponectin is a large, multimeric, circulating protein produced and secreted exclusively from adipose tissue (Stefan and Stumvoll 2002). Adiponectin is strongly and inversely correlated with insulin resistance (Stefan and Stumvoll 2002). Studies in mice, including the adipose tissue-specific knockout of the adiponectin receptors, has led many to suggest adiponectin is an important mediator of insulin resistance (Okada-Iwabu et al. 2013). In human subjects, however, the evidence is less clear cut. For example, individuals with severe mutations that cause a primary disorder of insulin resistance have reduced adiponectin levels, providing evidence that altered adiponectin levels are a consequence, not cause of insulin resistance (Semple et al. 2008). Several studies have identified common genetic variants within the gene that encodes adiponectin (ADIPOQ) as associated with circulating levels. A combination of four variants at this locus explain approximately 4 % of the population variance in adiponectin levels, more than sufficient to meet the potency assumption required for MR studies (Yaghootkar et al. 2013). Furthermore, because these variants are situated in and around the gene that encodes the protein of interest, the chances that they have pleiotropic effects on other phenotypes—which are not a consequence of altered adiponectin levels—are greatly reduced compared to variants elsewhere in the genome. Consequently, these genetic variants provide a great tool to evaluate the causal consequences of altered adiponectin levels on important clinical endpoints and are excellent proof of principle examples of MR. A recent MR study (Yaghootkar et al. 2013) of more than 29,000 individuals with both adiponectin and fasting insulin measurements found no causal evidence between lower adiponectin and higher insulin resistance (P = 0.60), or with T2D (P = 0.77). While studies of proportionally fewer samples have suggestive evidence to the contrary (Gao et al. 2013), the overall current weight of evidence appears to be that in humans, altered adiponectin levels are more likely to be a consequence rather than cause of insulin resistance in the general population.
4.5 Circulating Sex Hormone-Binding Globulin and Type 2 Diabetes
Common genetic variants in the gene that encodes an important carrier protein—sex hormone-binding globulin (SHBG)—provide another example of an excellent instrument for measuring the causal effects of the SHBG protein. SHBG binds to and transports testosterone and, to a lesser extent, estrogen, around the circulation to its target tissues (Petra 1991). Produced by the liver, it is inversely correlated with insulin resistance (Ding et al. 2006), and as with adiponectin, higher levels are associated with a healthy metabolic profile and with lower risk of T2D. In contrast to adiponectin, most people had assumed that lower SHBG levels were merely a consequence of insulin resistance and T2D, not a cause. However, two independent studies have recently shown that common variants in the SHBG gene are associated with T2D (Ding et al. 2009; Perry et al. 2010). MR effect estimates (OR per SHBG raising allele: 0.94, 95 % confidence interval (CI): 0.91–0.97) are consistent to the extent expected given the association between the gene variants and circulating SHBG and the phenotypic association between SHBG and T2D as observed from epidemiologic studies (expected OR 0.92, 95 % CI: 0.88–0.96) (Perry et al. 2010). While further studies are needed to establish the mechanism behind this association, MR studies have changed conventional thinking about the role of SHBG in diabetes.
4.6 FTO : A Good Instrument for Evaluating Adiposity to Related Traits and Endpoints?
In 2007, several groups described the first reproducible association between a common genetic variant and normal variation in BMI. An allele in an intron of the “fatso” gene, FTO, was associated with a 0.5 kg/m2 increase in BMI. Carriers of two copies of the BMI-increasing allele (approximately 16 % of Europeans) were 1 kg/m2 larger than individuals who carried zero copies of this allele. This finding potentially offered a superb opportunity for Mendelian Randomization studies. Epidemiologic data has shown that variation in BMI is correlated with multiple metabolic traits and disease endpoints, including susceptibility to T2D, CHD, some cancers, hypertension, raised circulating inflammatory markers, lower adiponectin, and SHBG. The difficulty in dissecting cause and effect in obesity is further illustrated by the fact that more than half of all human genes are differentially expressed in the adipose tissue of larger individuals compared to smaller individuals, presumably, mostly as a consequence rather than cause of obesity (Emilsson et al. 2008). Furthermore, many of these measures are correlated with each other independently of BMI. Multiple, phenotypic correlations between many (and potentially confounding) traits make understanding the causal factors that link obesity to increased risk of disease difficult, at best. A genetic variant that effectively randomizes people to higher or lower BMI could be extremely useful for disentangling some of these relationships.
To this end, several studies have performed MR experiments using FTO as an instrument for altered BMI (Fall et al. 2013; Holmes et al. 2014). These studies have suggested that higher BMI is causally related to susceptibility to T2D, heart failure, and several circulating biomarkers of public health importance including lower vitamin D levels (Vimaleswaran et al. 2013). However, some scientists have raised concerns that we must be cautious when using FTO as an instrument for BMI. First of all BMI is used as a surrogate of adiposity, but more importantly, little is known about how FTO alters BMI—we are still uncertain as to whether or not the genetic variant targets FTO itself or a nearby gene, and even if FTO is the target, we do not know exactly how it alters BMI. These are legitimate concerns, but several lines of evidence suggest that the variant in FTO can help improve our knowledge of causal relationships between adiposity and other traits. First, DEXA scans in children show that it is specifically associated with an alteration in fat mass, not increased skeletal or muscle mass (Frayling et al. 2007). Second, most studies show that the associations between the FTO variant and related traits are entirely consistent with that expected given the association between the FTO variant and BMI and the association between BMI and the “outcome” related trait. For example, FTO variants are associated with raised triglycerides and insulin levels (a marker of insulin resistance) to a greater extent than blood pressure and cholesterol levels—in keeping with the stronger phenotypic associations between BMI and triglycerides and insulin compared to BMI and blood pressure and cholesterol (Freathy et al. 2008).
4.7 Triglycerides and Fasting Insulin , Glucose , or Type 2 Diabetes
Elevated circulating triglyceride levels are strongly correlated with a poor metabolic state, including insulin resistance, higher levels of fasting glucose, and overt type 2 diabetes. Observational data suggest that elevated circulating triglycerides could be causal for this state, for example, through accumulation in adipose or liver resulting in lipotoxicity or impairment of hepatic insulin signaling resulting in insulin resistance (Trauner et al. 2010). Because genome-wide studies had accumulated associations with plasma triglycerides along with lipid levels, a Mendelian Randomization study was once again possible. De Silva et al. (2011) identified ten genetic variants strongly associated with triglyceride levels (P < 2 × 10−72) and, when aggregated in a score, resulted in a 0.59 SD change in triglyceride levels (upper vs. lower quintile). But in contrast to observational epidemiology (the Go-DARTS study), where a 1-SD increase in log10-triglyceride level was associated with elevated fasting glucose levels, HOMA-B, fasting insulin, HOMA-IR, and type 2 diabetes, the genetic risk score was not associated with any of these outcomes in diabetic or in control populations. Furthermore, the score was also not associated with frank type 2 diabetes (De Silva et al. 2011). While additional clarifying work is required, particularly to consider confounding in the context of this analysis, this raises significant questions about the expected efficacy of triglyceride control in prediabetic populations as a strategy to control or prevent type 2 diabetes onset.
5 Closing Summary
The simple idea underlying Mendelian Randomization is that a genetic variant, strongly related to a biomarker of interest, by the transitive property facilitates an appropriate framework to test the hypothesis that the given biomarker is causal for the endpoint of interest. Instrumental variable analysis—a procedure utilized in great detail in the field of econometrics—provides the precise framework by which this test can be formally implemented. Today, the wide abundance of human genetics data across a great range of phenotypes provides a unique opportunity to test a range of hypotheses that, until today, could not have been evaluated rigorously by the clinical research community, owing to the prohibitive and time-consuming costs of randomized controlled trials. For example, a potential application of great relevance to the T2D and CHD field, as yet untested, is whether hyperglycemia is pathogenic for T2D, given the controversy of conflicting RCTs in T1D and T2D.
Excitingly, genetic information across human subjects—hundreds of thousands of individuals—is accruing at a rapid pace. This growth is following in step with separate advances in sequencing technologies, which is enabling the characterization of less-common genetic variation and association studies with human disease. This next-generation of data sets will facilitate the application of a great deal of useful and important MR studies to understand—and develop targeted therapies for—disease endpoints. While the initial model and framework for MR is a solid advance for the field, a great deal of methodological work still remains to improve the approach to widen applicability, to take advantage of these upcoming data sets. That said, MR studies have been, and will continue to be, an essential tool in the arsenal of translational studies in human disease.
References
1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491(7422):56–65
Arrowsmith J, Miller P (2013) Trial watch: phase II and phase III attrition rates 2011–2012. Nat Rev Drug Discov 12(8):569
Bachmaier K, Neu N, Yeung RS, Mak TW, Liu P, Penninger JM (1999) Generation of humanized mice susceptible to peptide-induced inflammatory heart disease. Circulation 99(14):1885–1891
Baigent C, Keech A, Kearney PM, Blackwell L, Buck G, Pollicino C, Kirby A, Sourjina T, Peto R, Collins R, Simes R, Cholesterol Treatment Trialists’ (CTT) Collaborators (2005) Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet 366(9493):1267–1278
Brunner EJ, Kivimäki M, Witte DR, Lawlor DA, Davey Smith G, Cooper JA, Miller M, Lowe GD, Rumley A, Casas JP, Shah T, Humphries SE, Hingorani AD, Marmot MG, Timpson NJ, Kumari M (2008) Inflammation, insulin resistance, and diabetes—Mendelian randomization using CRP haplotypes points upstream. PLoS Med 5(8), e155. doi:10.1371/journal.pmed.0050155
Burgess S, Thompson SG (2013) Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol 42(4):1134–1144
Casas JP, Shah T, Cooper J, Hawe E, McMahon AD, Gaffney D, Packard CJ, O’Reilly DS, Juhan-Vague I, Yudkin JS, Tremoli E, Margaglione M, Di Minno G, Hamsten A, Kooistra T, Stephens JW, Hurel SJ, Livingstone S, Colhoun HM, Miller GJ, Bautista LE, Meade T, Sattar N, Humphries SE, Hingorani AD (2006) Insight into the nature of the CRP-coronary event association using Mendelian randomization. Int J Epidemiol 35(4):922–931
Chen L, Davey Smith G, Harbord RM, Lewis SJ (2008) Alcohol intake and blood pressure: a systematic review implementing a Mendelian randomization approach. PLoS Med 5(3), e52
Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH (2006) Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 354(12):1264–1272
C-Reactive Protein Coronary Heart Disease Genetics Collaboration (CCGC), Wensley F, Gao P, Burgess S, Kaptoge S, Di Angelantonio E, Shah T, Engert JC, Clarke R, Davey-Smith G, Nordestgaard BG, Saleheen D, Samani NJ, Sandhu M, Anand S, Pepys MB, Smeeth L, Whittaker J, Casas JP, Thompson SG, Hingorani AD, Danesh J (2011) Association between C reactive protein and coronary heart disease: Mendelian randomisation analysis based on individual participant data. BMJ 15(342):d548
Danesh J, Collins R, Appleby P, Peto R (1998) Association of fibrinogen, C-reactive protein, albumin, or leukocyte count with coronary heart disease: meta-analyses of prospective studies. JAMA 279(18):1477–1482
De Silva NM, Freathy RM, Palmer TM, Donnelly LA, Luan J, Gaunt T, Langenberg C, Weedon MN, Shields B, Knight BA, Ward KJ, Sandhu MS, Harbord RM, McCarthy MI, Smith GD, Ebrahim S, Hattersley AT, Wareham N, Lawlor DA, Morris AD, Palmer CN, Frayling TM (2011) Mendelian randomization studies do not support a role for raised circulating triglyceride levels influencing type 2 diabetes, glucose levels, or insulin resistance. Diabetes 60(3):1008–1018
Ding EL, Song Y, Malik VS, Liu S (2006) Sex differences of endogenous sex hormones and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA 295(11):1288–1299
Ding EL, Song Y, Manson JE, Hunter DJ, Lee CC, Rifai N, Buring JE, Gaziano JM, Liu S (2009) Sex hormone-binding globulin and risk of type 2 diabetes in women and men. N Engl J Med 361(12):1152–1163
Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K (2008) Genetics of gene expression and its effect on disease. Nature 452(7186):423–428
Fall T, Hägg S, Mägi R, Ploner A, Fischer K, Horikoshi M, Sarin AP, Thorleifsson G, Ladenvall C, Kals M, Kuningas M, Draisma HH, Ried JS, van Zuydam NR, Huikari V, Mangino M, Sonestedt E, Benyamin B, Nelson CP, Rivera NV, Kristiansson K, Shen HY, Havulinna AS, Dehghan A, Donnelly LA, Kaakinen M, Nuotio ML, Robertson N, de Bruijn RF, Ikram MA, Amin N, Balmforth AJ, Braund PS, Doney AS, Döring A, Elliott P, Esko T, Franco OH, Gretarsdottir S, Hartikainen AL, Heikkilä K, Herzig KH, Holm H, Hottenga JJ, Hyppönen E, Illig T, Isaacs A, Isomaa B, Karssen LC, Kettunen J, Koenig W, Kuulasmaa K, Laatikainen T, Laitinen J, Lindgren C, Lyssenko V, Läärä E, Rayner NW, Männistö S, Pouta A, Rivadeneira F, Ruokonen A, Savolainen MJ, Sijbrands EJ, Small KS, Smit JH, Steinthorsdottir V, Syvänen AC, Taanila A, Tobin MD, Uitterlinden AG, Willems SM, Willemsen G, Witteman J, Perola M, Evans A, Ferrières J, Virtamo J, Kee F, Tregouet DA, Arveiler D, Amouyel P, Ferrario MM, Brambilla P, Hall AS, Heath AC, Madden PA, Martin NG, Montgomery GW, Whitfield JB, Jula A, Knekt P, Oostra B, van Duijn CM, Penninx BW, Davey Smith G, Kaprio J, Samani NJ, Gieger C, Peters A, Wichmann HE, Boomsma DI, de Geus EJ, Tuomi T, Power C, Hammond CJ, Spector TD, Lind L, Orho-Melander M, Palmer CN, Morris AD, Groop L, Järvelin MR, Salomaa V, Vartiainen E, Hofman A, Ripatti S, Metspalu A, Thorsteinsdottir U, Stefansson K, Pedersen NL, McCarthy MI, Ingelsson E, Prokopenko I (2013) European network for genetic and genomic epidemiology (ENGAGE) consortium. The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS Med 10(6), e1001474
Ference BA, Yoo W, Alesh I, Mahajan N, Mirowska KK, Mewada A, Kahn J, Afonso L, Williams KA Sr, Flack JM (2012) Effect of long-term exposure to lower low-density lipoprotein cholesterol beginning early in life on the risk of coronary heart disease: a Mendelian randomization analysis. J Am Coll Cardiol 60(25):2631–2639
Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI (2007) A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316(5826):889–894
Freathy RM, Timpson NJ, Lawlor DA, Pouta A, Ben-Shlomo Y, Ruokonen A, Ebrahim S, Shields B, Zeggini E, Weedon MN, Lindgren CM, Lango H, Melzer D, Ferrucci L, Paolisso G, Neville MJ, Karpe F, Palmer CN, Morris AD, Elliott P, Jarvelin MR, Smith GD, McCarthy MI, Hattersley AT, Frayling TM (2008) Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes 57(5):1419–1426
Gao H, Kim YM, Chen P, Igase M, Kawamoto R, Kim MK, Kohara K, Lee J, Miki T, Ong RT, Onuma H, Osawa H, Sim X, Teo YY, Tabara Y, Tai ES, van Dam RM (2013) Genetic variation in CDH13 is associated with lower plasma adiponectin levels but greater adiponectin sensitivity in East Asian populations. Diabetes 62(12):4277–4283
Gray R, Manola J, Saxman S, Wright J, Dutcher J, Atkins M, Carducci M, See W, Sweeney C, Liu G, Stein M, Dreicer R, Wilding G, DiPaola RS (2006) Phase II clinical trial design: methods in translational research from the Genitourinary Committee at the Eastern Cooperative Oncology Group. Clin Cancer Res 12(7 Pt 1):1966–1969
Hernán MA, Robins JM (2006) Instruments for causal inference. An epidemiologist’s dream? Epidemiology 17(4):360–372
Holme I, Aastveit AH, Hammar N, Jungner I, Walldius G (2009) Uric acid and risk of myocardial infarction, stroke and congestive heart failure in 417,734 men and women in the apolipoprotein MOrtality RISk study (AMORIS). J Intern Med 266(6):558–570
Holmes MV, Lange LA, Palmer T, Lanktree MB, North KE, Almoguera B, Buxbaum S, Chandrupatla HR, Elbers CC, Guo Y, Hoogeveen RC, Li J, Li YR, Swerdlow DI, Cushman M, Price TS, Curtis SP, Fornage M, Hakonarson H, Patel SR, Redline S, Siscovick DS, Tsai MY, Wilson JG, van der Schouw YT, Fitzgerald GA, Hingorani AD, Casas JP, de Bakker PI, Rich SS, Schadt EE, Asselbergs FW, Reiner AP, Keating BJ (2014) Causal effects of body mass index on cardiometabolic traits and events: a Mendelian randomization analysis. Am J Hum Genet 94(2):198–208, pii: S0002-9297(13)00584-3
Hozawa A, Folsom AR, Ibrahim H, Nieto FJ, Rosamond WD, Shahar E (2006) Serum uric acid and risk of ischemic stroke: the ARIC study. Atherosclerosis 187(2):401–407
Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium, Hingorani AD, Casas JP (2012) The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet 379(9822):1214–1224
Johnson T (2012) Efficient calculation for Multi-SNP genetic risk scores. Poster presented at the American society of human genetics annual meeting, San Francisco, 6–10 November 2012
Kannel WB, Dawber TR, Friedman GD, Glennon WE, McNamara PM (1964) Risk factors in coronary heart disease. An evaluation of several serum lipids as predictors of coronary heart disease; the Framingham Study. Ann Intern Med 61:888–899
Katan MB (1986) Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet 1(8479):507–508
Kim SY, Guevara JP, Kim KM, Choi HK, Heitjan DF, Albert DA (2010) Hyperuricemia and coronary heart disease: a systematic review and meta-analysis. Arthritis Care Res (Hoboken) 62(2):170–180
Kodama S, Saito K, Yachi Y, Asumi M, Sugawara A, Totsuka K, Saito A, Sone H (2009) Association between serum uric acid and development of type 2 diabetes. Diabetes Care 32(9):1737–1742
Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, Pistis G, Ruggiero D, O’Seaghdha CM, Haller T, Yang Q, Tanaka T, Johnson AD, Kutalik Z, Smith AV, Shi J, Struchalin M, Middelberg RP, Brown MJ, Gaffo AL, Pirastu N, Li G, Hayward C, Zemunik T, Huffman J, Yengo L, Zhao JH, Demirkan A, Feitosa MF, Liu X, Malerba G, Lopez LM, van der Harst P, Li X, Kleber ME, Hicks AA, Nolte IM, Johansson A, Murgia F, Wild SH, Bakker SJ, Peden JF, Dehghan A, Steri M, Tenesa A, Lagou V, Salo P, Mangino M, Rose LM, Lehtimäki T, Woodward OM, Okada Y, Tin A, Müller C, Oldmeadow C, Putku M, Czamara D, Kraft P, Frogheri L, Thun GA, Grotevendt A, Gislason GK, Harris TB, Launer LJ, McArdle P, Shuldiner AR, Boerwinkle E, Coresh J, Schmidt H, Schallert M, Martin NG, Montgomery GW, Kubo M, Nakamura Y, Tanaka T, Munroe PB, Samani NJ, Jacobs DR Jr, Liu K, D’Adamo P, Ulivi S, Rotter JI, Psaty BM, Vollenweider P, Waeber G, Campbell S, Devuyst O, Navarro P, Kolcic I, Hastie N, Balkau B, Froguel P, Esko T, Salumets A, Khaw KT, Langenberg C, Wareham NJ, Isaacs A, Kraja A, Zhang Q, Wild PS, Scott RJ, Holliday EG, Org E, Viigimaa M, Bandinelli S, Metter JE, Lupo A, Trabetti E, Sorice R, Döring A, Lattka E, Strauch K, Theis F, Waldenberger M, Wichmann HE, Davies G, Gow AJ, Bruinenberg M, LifeLines Cohort Study, Stolk RP, Kooner JS, Zhang W, Winkelmann BR, Boehm BO, Lucae S, Penninx BW, Smit JH, Curhan G, Mudgal P, Plenge RM, Portas L, Persico I, Kirin M, Wilson JF, Mateo Leach I, van Gilst WH, Goel A, Ongen H, Hofman A, Rivadeneira F, Uitterlinden AG, Imboden M, von Eckardstein A, Cucca F, Nagaraja R, Piras MG, Nauck M, Schurmann C, Budde K, Ernst F, Farrington SM, Theodoratou E, Prokopenko I, Stumvoll M, Jula A, Perola M, Salomaa V, Shin SY, Spector TD, Sala C, Ridker PM, Kähönen M, Viikari J, Hengstenberg C, Nelson CP, CARDIoGRAM Consortium, DIAGRAM Consortium, ICBP Consortium, MAGIC Consortium, Meschia JF, Nalls MA, Sharma P, Singleton AB, Kamatani N, Zeller T, Burnier M, Attia J, Laan M, Klopp N, Hillege HL, Kloiber S, Choi H, Pirastu M, Tore S, Probst-Hensch NM, Völzke H, Gudnason V, Parsa A, Schmidt R, Whitfield JB, Fornage M, Gasparini P, Siscovick DS, Polašek O, Campbell H, Rudan I, Bouatia-Naji N, Metspalu A, Loos RJ, van Duijn CM, Borecki IB, Ferrucci L, Gambaro G, Deary IJ, Wolffenbuttel BH, Chambers JC, März W, Pramstaller PP, Snieder H, Gyllensten U, Wright AF, Navis G, Watkins H, Witteman JC, Sanna S, Schipf S, Dunlop MG, Tönjes A, Ripatti S, Soranzo N, Toniolo D, Chasman DI, Raitakari O, Kao WH, Ciullo M, Fox CS, Caulfield M, Bochud M, Gieger C (2013) Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 45(2):145–154
Okada-Iwabu M, Yamauchi T, Iwabu M, Honma T, Hamagami K, Matsuda K, Yamaguchi M, Tanabe H, Kimura-Someya T, Shirouzu M, Ogata H, Tokuyama K, Ueki K, Nagano T, Tanaka A, Yokoyama S, Kadowaki T (2013) A small-molecule AdipoR agonist for type 2 diabetes and short life in obesity. Nature 503(7477):493–499
Perry JR, Weedon MN, Langenberg C, Jackson AU, Lyssenko V, Sparsø T, Thorleifsson G, Grallert H, Ferrucci L, Maggio M, Paolisso G, Walker M, Palmer CN, Payne F, Young E, Herder C, Narisu N, Morken MA, Bonnycastle LL, Owen KR, Shields B, Knight B, Bennett A, Groves CJ, Ruokonen A, Jarvelin MR, Pearson E, Pascoe L, Ferrannini E, Bornstein SR, Stringham HM, Scott LJ, Kuusisto J, Nilsson P, Neptin M, Gjesing AP, Pisinger C, Lauritzen T, Sandbaek A, Sampson M, MAGIC, Zeggini E, Lindgren CM, Steinthorsdottir V, Thorsteinsdottir U, Hansen T, Schwarz P, Illig T, Laakso M, Stefansson K, Morris AD, Groop L, Pedersen O, Boehnke M, Barroso I, Wareham NJ, Hattersley AT, McCarthy MI, Frayling TM (2010) Genetic evidence that raised sex hormone binding globulin (SHBG) levels reduce the risk of type 2 diabetes. Hum Mol Genet 19(3):535–544
Petra PH (1991) The plasma sex steroid binding protein (SBP or SHBG). A critical review of recent developments on the structure, molecular biology and function. J Steroid Biochem Mol Biol 40(4–6):735–753
Pfister R, Sharp S, Luben R, Welsh P, Barroso I, Salomaa V, Meirhaeghe A, Khaw KT, Sattar N, Langenberg C, Wareham NJ (2011) Mendelian randomization study of B-type natriuretic peptide and type 2 diabetes: evidence of causal association from population studies. PLoS Med 8(10), e1001112
Raman K, Chong M, Akhtar-Danesh GG, D’Mello M, Hasso R, Ross S, Xu F, Paré G (2013) Genetic markers of inflammation and their role in cardiovascular disease. Can J Cardiol 29(1):67–74
Semple RK, Cochran EK, Soos MA, Burling KA, Savage DB, Gorden P, O’Rahilly S (2008) Plasma adiponectin as a marker of insulin receptor dysfunction: clinical utility in severe insulin resistance. Diabetes Care 31(5):977–979
Sheehan NA, Didelez V, Burton PR, Tobin MD (2008) Mendelian randomisation and causal inference in observational epidemiology. PLoS Med 5(8), e177
Smith GD, Ebrahim S (2003) ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32(1):1–22
Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, D’Agostino RB Sr, Fox CS, Larson MG, Murabito JM, O’Donnell CJ, Vasan RS, Wolf PA, Levy D (2007) The third generation cohort of the national heart, lung, and blood institute’s Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol 165(11):1328–1335
Stark K, Reinhard W, Grassl M, Erdmann J, Schunkert H, Illig T, Hengstenberg C (2009) Common polymorphisms influencing serum uric acid levels contribute to susceptibility to gout, but not to coronary artery disease. PLoS One 4(11), e7729
Stefan N, Stumvoll M (2002) Adiponectin—its role in metabolism and beyond. Horm Metab Res 34(9):469–474
Tatsioni A, Bonitis NG, Ioannidis JPA (2007) Persistence of contradicted claims in the literature. JAMA 298:2517–2526
Tayar JH, Lopez-Olivo MA, Suarez-Almazor ME (2012) Febuxostat for treating chronic gout. Cochrane Database Syst Rev 11, CD008653
Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466(7307):707–713
Thanassoulis G, O’Donnell CJ (2009) Mendelian randomization: nature’s randomized trial in the post-genome era. JAMA 301(22):2386–2388
Timpson NJ, Lawlor DA, Harbord RM, Gaunt TR, Day IN, Palmer LJ, Hattersley AT, Ebrahim S, Lowe GD, Rumley A, Davey Smith G (2005) C-reactive protein and its role in metabolic syndrome: Mendelian randomisation study. Lancet 366(9501):1954–1959
Trauner M, Arrese M, Wagner M (2010) Fatty liver and lipotoxicity. Biochim Biophys Acta 1801:299–310
Vimaleswaran KS, Berry DJ, Lu C, Tikkanen E, Pilz S, Hiraki LT, Cooper JD, Dastani Z, Li R, Houston DK, Wood AR, Michaëlsson K, Vandenput L, Zgaga L, Yerges-Armstrong LM, McCarthy MI, Dupuis J, Kaakinen M, Kleber ME, Jameson K, Arden N, Raitakari O, Viikari J, Lohman KK, Ferrucci L, Melhus H, Ingelsson E, Byberg L, Lind L, Lorentzon M, Salomaa V, Campbell H, Dunlop M, Mitchell BD, Herzig KH, Pouta A, Hartikainen AL, Genetic Investigation of Anthropometric Traits-GIANT Consortium, Streeten EA, Theodoratou E, Jula A, Wareham NJ, Ohlsson C, Frayling TM, Kritchevsky SB, Spector TD, Richards JB, Lehtimäki T, Ouwehand WH, Kraft P, Cooper C, März W, Power C, Loos RJ, Wang TJ, Järvelin MR, Whittaker JC, Hingorani AD, Hyppönen E (2013) Causal relationship between obesity and vitamin D status: bi-directional Mendelian randomization analysis of multiple cohorts. PLoS Med 10(2):e1001383
Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Hólm H, Ding EL, Johnson T, Schunkert H, Samani NJ, Clarke R, Hopewell JC, Thompson JF, Li M, Thorleifsson G, Newton-Cheh C, Musunuru K, Pirruccello JP, Saleheen D, Chen L, Stewart A, Schillert A, Thorsteinsdottir U, Thorgeirsson G, Anand S, Engert JC, Morgan T, Spertus J, Stoll M, Berger K, Martinelli N, Girelli D, McKeown PP, Patterson CC, Epstein SE, Devaney J, Burnett MS, Mooser V, Ripatti S, Surakka I, Nieminen MS, Sinisalo J, Lokki ML, Perola M, Havulinna A, de Faire U, Gigante B, Ingelsson E, Zeller T, Wild P, de Bakker PI, Klungel OH, der Zee AH M-v, Peters BJ, de Boer A, Grobbee DE, Kamphuisen PW, Deneer VH, Elbers CC, Onland-Moret NC, Hofker MH, Wijmenga C, Verschuren WM, Boer JM, van der Schouw YT, Rasheed A, Frossard P, Demissie S, Willer C, Do R, Ordovas JM, Abecasis GR, Boehnke M, Mohlke KL, Daly MJ, Guiducci C, Burtt NP, Surti A, Gonzalez E, Purcell S, Gabriel S, Marrugat J, Peden J, Erdmann J, Diemert P, Willenborg C, König IR, Fischer M, Hengstenberg C, Ziegler A, Buysschaert I, Lambrechts D, Van de Werf F, Fox KA, El Mokhtari NE, Rubin D, Schrezenmeir J, Schreiber S, Schäfer A, Danesh J, Blankenberg S, Roberts R, McPherson R, Watkins H, Hall AS, Overvad K, Rimm E, Boerwinkle E, Tybjaerg-Hansen A, Cupples LA, Reilly MP, Melander O, Mannucci PM, Ardissino D, Siscovick D, Elosua R, Stefansson K, O’Donnell CJ, Salomaa V, Rader DJ, Peltonen L, Schwartz SM, Altshuler D, Kathiresan S (2012) Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet 572(9841):572–580
Whincup PH, Gilg JA, Papacosta O, Seymour C, Miller GJ, Alberti KG, Cook DG (2002) Early evidence of ethnic differences in cardiovascular risk: cross sectional comparison of British South Asian and white children. BMJ 324(7338):635
Yaghootkar H, Lamina C, Scott RA, Dastani Z, Hivert MF, Warren LL, Stancáková A, Buxbaum SG, Lyytikäinen LP, Henneman P, Wu Y, Cheung CY, Pankow JS, Jackson AU, Gustafsson S, Zhao JH, Ballantyne CM, Xie W, Bergman RN, Boehnke M, el Bouazzaoui F, Collins FS, Dunn SH, Dupuis J, Forouhi NG, Gillson C, Hattersley AT, Hong J, Kähönen M, Kuusisto J, Kedenko L, Kronenberg F, Doria A, Assimes TL, Ferrannini E, Hansen T, Hao K, Häring H, Knowles JW, Lindgren CM, Nolan JJ, Paananen J, Pedersen O, Quertermous T, Smith U, GENESIS Consortium, RISC Consortium, Lehtimäki T, Liu CT, Loos RJ, McCarthy MI, Morris AD, Vasan RS, Spector TD, Teslovich TM, Tuomilehto J, van Dijk KW, Viikari JS, Zhu N, Langenberg C, Ingelsson E, Semple RK, Sinaiko AR, Palmer CN, Walker M, Lam KS, Paulweber B, Mohlke KL, van Duijn C, Raitakari OT, Bidulescu A, Wareham NJ, Laakso M, Waterworth DM, Lawlor DA, Meigs JB, Richards JB, Frayling TM (2013) Mendelian randomization studies do not support a causal role for reduced circulating adiponectin levels in insulin resistance and type 2 diabetes. Diabetes 62(10):3589–3598
Yang Q, Köttgen A, Dehghan A, Smith AV, Glazer NL, Chen MH, Chasman DI, Aspelund T, Eiriksdottir G, Harris TB, Launer L, Nalls M, Hernandez D, Arking DE, Boerwinkle E, Grove ML, Li M, Linda Kao WH, Chonchol M, Haritunians T, Li G, Lumley T, Psaty BM, Shlipak M, Hwang SJ, Larson MG, O’Donnell CJ, Upadhyay A, van Duijn CM, Hofman A, Rivadeneira F, Stricker B, Uitterlinden AG, Paré G, Parker AN, Ridker PM, Siscovick DS, Gudnason V, Witteman JC, Fox CS, Coresh J (2010) Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ Cardiovasc Genet 3(6):523–530
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Frayling, T.M., Voight, B.F. (2016). Causal Inference in Medicine via Mendelian Randomization. In: Florez, J. (eds) The Genetics of Type 2 Diabetes and Related Traits. Springer, Cham. https://doi.org/10.1007/978-3-319-01574-3_24
Download citation
DOI: https://doi.org/10.1007/978-3-319-01574-3_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-01573-6
Online ISBN: 978-3-319-01574-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)