Abstract
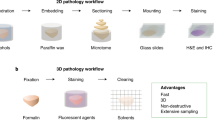
Histology images are the golden standard for medical diagnostic analysis. However, 2D images can lose some critical information, such as the spatial structure of blood vessels. Therefore, it is necessary to perform 3D reconstruction for the histology images. At the same time, due to the differences between institutions and hospitals, a general 3D reconstruction method is needed. In this work, we propose a 3D reconstruction pipeline that is compatible with Whole Slide Imaging (WSI) and can also be applied to other imaging modalities such as CT images, MRI images, and immunohistochemistry images. Through semantic segmentation, point cloud construction and registration, and 3D rendering, we can reconstruct serialized images into 3D models. By optimizing the pipeline workflow, we can significantly reduce the computation workload required for the 3D reconstruction of high-resolution images and thus save time. In clinical practice, our method helps pathologists triage and evaluate tumor tissues with real-time 3D visualization.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Histology images are golden standards for medical diagnosis and analysis, as they contain key information such as the cause and severity of the diseases. With the advancement of deep learning technology, computers are now capable of being applied in the analysis of medical images and in extracting key information. However, traditional 2D images can lose a lot of important information, such as the vascular structure in 3D space. Moreover, due to different task requirements and variations such as machine specifications among hospitals and institutions, there is a need to develop a general 3D reconstruction system. Current 3D reconstruction tasks, especially those involving high-resolution images, require extensive computational resources and are extremely time-consuming, with the registration and semantic segmentation tasks as the bottleneck of the real-time visualization for gigabyte WSIs [1]. In this work, we propose a computational-efficient method to reconstruct the pathology images for WSI 3D reconstruction using point clouds, a discrete set of data points in the 3D space. The process comprises semantic segmentation, point cloud sampling, point cloud registration, and 3D rendering. This process outperforms the existing reconstruction process as it combines the sampling and modeling processes by constructing point clouds. Subsequently, registration is performed, greatly reducing the computational and time costs required for the process.
2 Related Works
Many approaches have recently been proposed recently for 3D reconstruction. For example, [2] developed techniques to inspect the surface of organs by reconstruction from endoscope videos. A pipeline named CODA [1] perceives the spatial distribution of tumors such as the pancreas and liver. ITA3D reconstructs tissues through non-destructive 3D pathology images [3]. Comparative studies have published to reconstruct 3D organs in the disciplines of ultrasound [4, 5], radiology [6,7,8] and orthodontic [9, 10]. Notably, due to factors such as the quality of the loaded glass slides and manual operation during the preparation of pathological sections, the three-dimensional reconstruction must perform image registration, which makes the three-dimensional reconstruction method based on CT images, as in [11], unsuitable for direct application to WSI. Despite many AI-powered applications, accuracy and performance are still the dominant challenges for real-time diagnoses. In the setting of gigabyte pathology images, cellular-level segmentation and image registration are required to be produced in a short time to keep up with the high-throughput scanners and minimize the waiting time for the final confirmation by pathologists.
The 3D visualization pipeline of tumor tissue. For the visualization of tumor volume, the raw WSIs are firstly decoupled to image patches and processed by the binary segmentation network, with the gated axial transformer as the encoder and CNN as the decoder. Then the patches are rejoined to form segmentation maps representing tumor (positive) area. The color convention is applied to WSI binary image to visualize the entire tissue volume. (a) and (b) stand for one layer of point clouds generated from the binary images to represent the density of the tumor (red) and tissue (blue). (c) and (d) are the 3D visualization tissue volume generated from the representative point clouds. (Color figure online)
3 Method
WSI-Level Tissue Segmentation. The medical transformer, namely gated axial-attention transformer [12, 13] employs a position-sensitive axial-attention mechanism, with a shallow global branch and a deep local branch incorporated.
Inspired by this design, we trained a network with two branches of gated-axial transformer and a CNN-transformer hybrid architecture as the backbone to extract global and local information. The segmentation ground truths are derived from 2D WSI segmentation maps labeled manually by QuPath [14]. Then the 2D WSIs are cropped to image patches and curated to feed the segmentation network, as patch-based deep learning networks are currently the mainstream structures in the discipline of histology image analysis. The raw images and paired segmentation masks are cropped to \(128 \times 128\) pixel image patches for input. The network consists of two branches. The gated-axial transformer aims to learn global information by capturing feature correlations. The other branch of CNN-transformer hybrid architecture employs the transformer structure as the encoder and the CNN as the decoder, where the latter is deepened with multiple layers to allow a clear separation of tumor tissue (positive) and dense tissue (negative), as shown in Fig. 1. After the binary segmentation, the output patches are rejoined to form WSI for the later tumor visualization.
Point Clouds. Point clouds are applied for 3D modeling objects such as buildings [15] and human bodies [11, 16, 17]. This research generates the layered point clouds with down-sampled semantic segmentation results. The pixels of tumor (positive) masks are appended to the layered point cloud. The x and y coordinates of the points are generated from the segmented images, and the z coordinate is the interpolation of the stacked WSI. The computed point clouds are then reconstructed at the three dimensions for WSI registration. Compared with another commonly used 3D reconstruction tool, voxel-based 3D pixel representation using a 3D 0/1 matrix, the point cloud is more suitable for modeling high-resolution images with enormous data volumes thanks to its sparser data. In the current task, point cloud reconstruction also serves the function of extracting feature points. If registering the WSI, even when selecting only a few feature points and calculating simple translation and rotational coordinates, the entire WSI needs to be transformed accordingly, and the model needs to be re-sampled. By building the model first and then applying the transformation to it, only the coordinates of the points in the three-dimensional space need to be transformed, and a model that can be used for subsequent processing can be obtained directly (Fig. 2).
Axial Registration. Current registration methods employ Radon transform and cross-correlation, where WSIs are cropped and applied with rigid and elastic registration [1]. This computation workload is often massive and also redundant for unimportant regions. Moreover, elastic segmentation may cause image distortion and inaccuracy in segmentation. By contrast, we optimize the overall framework by bringing forward the segmentation and the point cloud generation before the registration.
Specifically, we incorporate the ICP (Iterative Closest Point) strategy to register for the layered point clouds generated from the segmentation output. As each point cloud for registration uses exclusively one layer, we apply point-to-point strategy [18] without employing the normal vectors. A brief review of the point-to-point strategy is formulated as follows:
\(P_{fix}\) and \(P_{mov}\) are the fixed and moving point clouds. R and T are the rotation matrix and translation vector.
\(P_{i,fix}\) and \(P_{i,mov}\) \((1 \le i \le N)\) are the paired-points in the point cloud; \(C_{fix}\) and \(C_{mov}\) are the center of the two point clouds; and \(V_{i,fix}\) and \(V_{i,mov}\) are the vectors from point to the center.
N is the number of points in \(P_{mov}\), and \(\mathscr {L}\) is the loss of the registration. Expand the equation and eliminate terms with zero means, \(V_{i,fix}\) and \(V_{i,mov}\) particularly and we obtain the following formula to calculate the final values of R and T in order to minimize the loss value.
where \(R^*\) and \(T^*\) are the computed rotation matrix and translation vector with minimized loss. The minimum value is achieved through SVD or nonlinear optimization.
An example of registration processing. The fixed WSI (pink point clouds) and the corresponding moving WSI (green) are computed in the current iteration, and the registration is iteratively performed from bottom (grey) to top (silver). Our selective algorithm pinpoints the essential points (blue) for matrix computation for the ICP translation speedup. (Color figure online)
Innovatively, to speed up the processing, we select the representative layered point cloud, determined by the spatial density and 2D coordinate, to apply the transformation to the entire layer. In each iteration from bottom to top, we select horizontal and vertical band-shaped areas in the moving point cloud, as shown in Fig. 3. For a consistent spatial presentation of the tumor tissue, interpolation is required upon the different resolutions of x, y, z. In this case study, the z value of the points are multiplied by a factor of 4 to map with the x, y resolution. The point cloud is interpolated based on the nearest layered point cloud. The layered point clouds are then re-registered iteratively in the same manner.
4 Implementation
We employ Open3D library [19] to generate point clouds to visualize spatial tissue distribution. The model presents point arrays with x, y, z coordinates, and the functions models produce color point clouds and 3D meshes. The 3D visualization allows the demonstration of comprehensive information interpreted by deep learning structures, including the spatial distribution of tumors and tissues.
5 Quantitative Results
Segmentation. The loss and training time of the segmentation network are demonstrated in Fig. 4. WSIs are cropped to \(128 \times 128\) image patches to feed the network, then rejoined to generate the layered point clouds, as shown in the segmentation image in Fig. 3.
Registration Speedup. Two metrics of speedup and accuracy evaluate the registration performance, and the latter is measured by Root Mean Square Error (RMSE) of the point pairs. For the axial registration example demonstrated in Fig. 5, the representative points are sampled in x value from 2,250 to 2,750, or y value from 6,750 to 7,250 at the bottom layer, about 1/3 of the total points employed for registration. Overall, the axial registration is with smaller RMSE on average, as shown in Fig. 5.
This pipeline attempts a significant decrease in registration computation, with 1.54 s per layer required, which is about 10.94% the time required for the regular ICP registration [18], and is a tremendous advantage compared with WSI-level registration [1] taking about 40 min per image. Overall, processing the WSI stack registration workflow takes only several minutes on average, whereas the state-of-the-art approach requires a couple of hours [1], as shown in Fig. 5. Consequently, the registration processing will not be the bottleneck of the 3D tissue reconstruction.
6 Conclusion and Future Work
In this task, we have optimized and integrated existing 3D reconstruction pipelines for WSI (Whole Slide Imaging) and CT (Computed Tomography), resulting in a more efficient pipeline for 3D reconstruction of high-resolution images. By utilizing point cloud merging and assisted registration processes, this pipeline significantly reduces redundant computations, decreases data volume in comparison to voxel methods, and minimizes time consumption during the registration process. While this pipeline is specifically designed for the unique requirements of Whole Slide Imaging (WSI), it also has the potential to adapt to CT and MRI images through semantic segmentation and point cloud sampling, 3D rendering, and omitting the registration. The 3D reconstruction section in [11, 20, 21] also utilized a similar method of acquiring layered images, stacking and aligning them to generate a 3D model. Although there were some differences in the specific implementation, it also demonstrated that our method theoretically could be applied to the 3D reconstruction of other medical images, such as immunohistochemistry images. Therefore, as long as there are appropriate training models and data available, this pipeline can be adaptable to 3D reconstruction tasks for different types of images and tissues.
References
Kiemen, A.L., Braxton, A.M., Grahn, M.P., et al.: CODA: quantitative 3D reconstruction of large tissues at cellular resolution. Nat. Meth. 19, 1490–1499 (2022). https://doi.org/10.1038/s41592-022-01650-9
Ma, R., Wang, R., Pizer, S., Rosenman, J., McGill, S.K., Frahm, J.-M.: Real-time 3D reconstruction of colonoscopic surfaces for determining missing regions. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11768, pp. 573–582. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32254-0_64
Xie, W., et al.: Prostate cancer risk stratification via nondestructive 3D pathology with deep learning-assisted gland analysis. Cancer Res. 82(2), 334–345 (2022). https://doi.org/10.1158/0008-5472.CAN-21-2843
Chen, C., et al.: Region proposal network with graph prior and IoU-balance loss for landmark detection in 3D ultrasound. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), pp. 1–5 (2020). https://doi.org/10.1109/ISBI45749.2020.9098368
Wiskin, J., et al.: Full wave 3D inverse scattering transmission ultrasound tomography: breast and whole body imaging. In: 2019 IEEE International Ultrasonics Symposium (IUS), pp. 951–958 (2019). https://doi.org/10.1109/ULTSYM.2019.8925778
Kamencay, P., Zachariasova, M., Hudec, R., Benco, M., Radil, R.: 3D image reconstruction from 2D CT slices. In: 2014 3DTV-Conference: The True Vision - Capture, Transmission and Display of 3D Video (3DTV-CON), pp. 1–4 (2014). https://doi.org/10.1109/3DTV.2014.6874742
Kermi, A., Djennelbaroud, H.C., Khadir, M.T.: A deep learning-based 3D CNN for automated Covid-19 lung lesions segmentation from 3D chest CT scans. In: 2022 5th International Symposium on Informatics and its Applications (ISIA), pp. 1–5 (2022). https://doi.org/10.1109/ISIA55826.2022.9993505
Ueda, D., et al.: Deep learning for MR angiography: automated detection of cerebral aneurysms. Radiology 290(1), 187–194 (2019). pMID: 30351253. https://doi.org/10.1148/radiol.2018180901
Tang, H., Hsung, T.C., Lam, W.Y., Cheng, L.Y.Y., Pow, E.H.: On 2D–3D image feature detections for image-to-geometry registration in virtual dental model. In: 2020 IEEE International Conference on Visual Communications and Image Processing (VCIP), pp. 140–143 (2020). https://doi.org/10.1109/VCIP49819.2020.9301774
Zhang, L.z., Shen, K.: A volumetric measurement algorithm of defects in 3D CT image based on spatial intuitionistic fuzzy c-means. In: 2021 IEEE Far East NDT New Technology & Application Forum (FENDT), pp. 78–82 (2021). https://doi.org/10.1109/FENDT54151.2021.9749668
Leonardi, V., Vidal, V., Mari, J.L., Daniel, M.: 3D reconstruction from CT-scan volume dataset application to kidney modeling. In: Proceedings of the 27th Spring Conference on Computer Graphics, SCCG 2011, pp. 111–120. Association for Computing Machinery, New York (2011). https://doi.org/10.1145/2461217.2461239
Valanarasu, J.M.J., Oza, P., Hacihaliloglu, I., Patel, V.M.: Medical transformer: gated axial-attention for medical image segmentation. In: de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C. (eds.) MICCAI 2021. LNCS, vol. 12901, pp. 36–46. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87193-2_4
Wang, H., Zhu, Y., Green, B., Adam, H., Yuille, A., Chen, L.-C.: Axial-DeepLab: stand-alone axial-attention for panoptic segmentation. In: Vedaldi, A., Bischof, H., Brox, T., Frahm, J.-M. (eds.) ECCV 2020. LNCS, vol. 12349, pp. 108–126. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-58548-8_7
Bankhead, P., et al.: QuPath: open source software for digital pathology image analysis. Sci. Rep. 7(1), 16878 (2017). https://doi.org/10.1038/s41598-017-17204-5
Chauhan, I., Rawat, A., Chauhan, M., Garg, R.: Fusion of low-cost UAV point cloud with TLS point cloud for complete 3D visualisation of a building. In: 2021 IEEE International India Geoscience and Remote Sensing Symposium (InGARSS), pp. 234–237 (2021). https://doi.org/10.1109/InGARSS51564.2021.9792104
Chen, M., Miao, Y., Gong, Y., Mao, X.: Convolutional neural network powered identification of the location and orientation of human body via human form point cloud. In: 2021 15th European Conference on Antennas and Propagation (EuCAP), pp. 1–5 (2021). https://doi.org/10.23919/EuCAP51087.2021.9410980
Wen, Z., Yan, Y., Cui, H.: Study on segmentation of 3D human body based on point cloud data. In: 2012 Second International Conference on Intelligent System Design and Engineering Application, pp. 657–660 (2012). https://doi.org/10.1109/ISdea.2012.676
Arun, K.S., Huang, T.S., Blostein, S.D.: Least-squares fitting of two 3-D point sets. IEEE Trans. Pattern Anal. Mach. Intell. (PAMI) 9(5), 698–700 (1987). https://doi.org/10.1109/TPAMI.1987.4767965
Zhou, Q.Y., Park, J., Koltun, V.: Open3D: a modern library for 3D data processing. arXiv:1801.09847 (2018)
Alsaid, B., et al.: Coexistence of adrenergic and cholinergic nerves in the inferior hypogastric plexus: anatomical and immunohistochemical study with 3D reconstruction in human male fetus. J. Anat. 214(5), 645–654 (2009). https://doi.org/10.1111/j.1469-7580.2009.01071.x. https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1469-7580.2009.01071.x
Karam, I., Droupy, S., Abd-Alsamad, I., Uhl, J.F., Benoît, G., Delmas, V.: Innervation of the female human urethral sphincter: 3D reconstruction of immunohistochemical studies in the fetus. Eur. Urol. 47(5), 627–634 (2005). https://doi.org/10.1016/j.eururo.2005.01.001. https://www.sciencedirect.com/science/article/pii/S0302283805000060
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Wu, Q., Shen, Y., Ke, J. (2023). A General Computationally-Efficient 3D Reconstruction Pipeline for Multiple Images with Point Clouds. In: Celebi, M.E., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2023 Workshops . MICCAI 2023. Lecture Notes in Computer Science, vol 14393. Springer, Cham. https://doi.org/10.1007/978-3-031-47401-9_19
Download citation
DOI: https://doi.org/10.1007/978-3-031-47401-9_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-47400-2
Online ISBN: 978-3-031-47401-9
eBook Packages: Computer ScienceComputer Science (R0)